SYNcro#
#
Authors: Chris Rorden & Steffen Bollmann
Date: 8 Jan 2026
#
License:
Note: If this notebook uses neuroimaging tools from Neurocontainers, those tools retain their original licenses. Please see Neurodesk citation guidelines for details.
Citation:#
Tools included in this workflow#
SynthStrip:
SynthStrip: Skull-Stripping for Any Brain Image; Andrew Hoopes, Jocelyn S. Mora, Adrian V. Dalca, Bruce Fischl*, Malte Hoffmann* (*equal contribution); NeuroImage 260, 2022, 119474; https://doi.org/10.1016/j.neuroimage.2022.119474
Boosting skull-stripping performance for pediatric brain images; William Kelley, Nathan Ngo, Adrian V. Dalca, Bruce Fischl, Lilla Zöllei*, Malte Hoffmann* (*equal contribution); IEEE International Symposium on Biomedical Imaging (ISBI), 2024, forthcoming; https://arxiv.org/abs/2402.16634
SynthStrip tool: https://w3id.org/synthstrip
Dataset#
Opensource Data from OpenNeuro:
Makayla Gibson, Roger Newman-Norlund, Leonardo Bonilha, Julius Fridriksson, Gregory Hickok, Argye E. Hillis, Dirk-Bart den Ouden, and Chris Rorden (2023). Aphasia Recovery Cohort (ARC) Dataset. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds004884.v1.0.0
ANTs Brain Templates:
Avants, Brian; Tustison, Nick (2018). ANTs/ANTsR Brain Templates. figshare. Dataset. https://doi.org/10.6084/m9.figshare.915436.v2
Load Software#
import module
await module.load('syncro/0.1.1')
await module.list()
['syncro/0.1.1']
Download Data#
%%bash
datalad install https://github.com/OpenNeuroDatasets/ds004884.git
cd ds004884 && datalad get "sub-M2304/ses-262/anat"
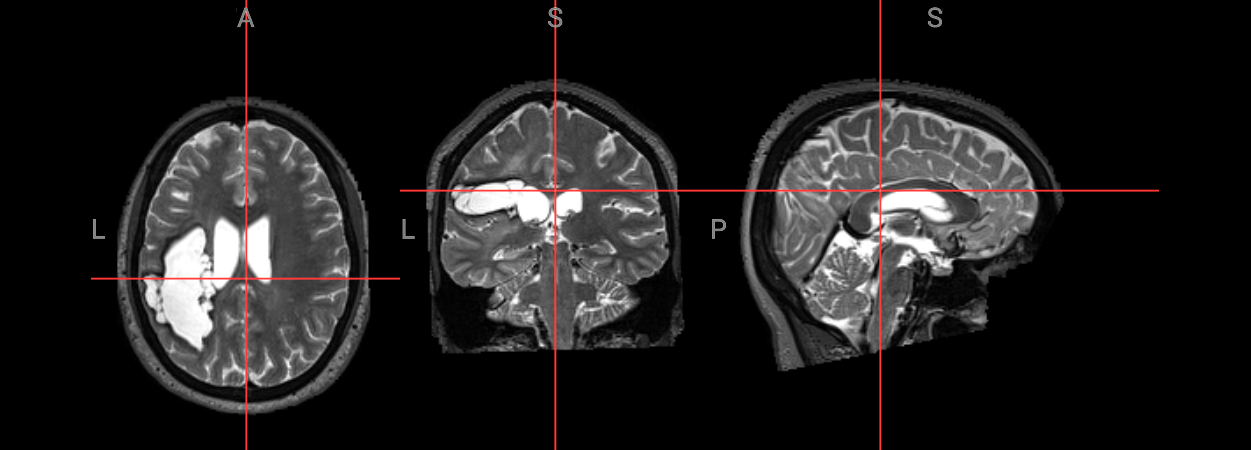
input_image = 'ds004884/sub-M2304/ses-262/anat/sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz'
from ipyniivue import NiiVue
nv_subject_space = NiiVue()
nv_subject_space.load_volumes([{"path": input_image}])
nv_subject_space
from IPython.display import Image
Image(filename='../../static/examples/structural_imaging/SYNcro/subject_space.png')
!SYNcro.py -h
usage: SYNcro.py [-h] [--force-gpu {true,false,auto}] [-b] [-c] [-d DIRECTORY]
[--log {silent,verbose,debug}] [-v]
N [N ...]
Normalize NIfTI images with lesion maps.
positional arguments:
N NIfTI images: first is anatomical (required), second
(optional) lesion map, third (optional) pathological
options:
-h, --help show this help message and exit
--force-gpu {true,false,auto}
Force GPU usage (true), disable GPU (false), or auto-
detect (auto, default)
-b, --bet images are already brain extracted (default: False)
-c, --ct images are CT scans (default: False)
-d DIRECTORY, --directory DIRECTORY
output directory (default: same as input)
--log {silent,verbose,debug}
Set log level: silent (default), verbose, or debug
-v, --version show program's version number and exit
!SYNcro.py ds004884/sub-M2304/ses-262/anat/sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz
INFO - Running: /usr/local/bin/py_synthsr --i /home/jovyan/workspace/books/examples/structural_imaging/SYNcro_a24fiuhq/sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz --o /home/jovyan/workspace/books/examples/structural_imaging/SYNcro_a24fiuhq/t1sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz --cpu --threads 31
using CPU, hiding all CUDA_VISIBLE_DEVICES
Using general model from January 2023 (version 2)
/freesurfer/models/synthsr_v20_230130.h5
using 31 threads
predicting 1/1
Prediction without flipping
1/1 ━━━━━━━━━━━━━━━━━━━━ 0s 9s/step
1/1 ━━━━━━━━━━━━━━━━━━━━ 9s 9s/step
Prediction with flipping
1/1 ━━━━━━━━━━━━━━━━━━━━ 8s 8s/step
prediction saved in: /home/jovyan/workspace/books/examples/structural_imaging/SYNcro_a24fiuhq/t1sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz
If you use this tool in a publication, please cite:
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical
MRI exams with scans of different orientation, resolution and contrast
JE Iglesias, B Billot, Y Balbastre, A Tabari, J Conklin, RG Gonzalez, DC Alexander,
P Golland, BL Edlow, B Fischl, for the ADNI
NeuroImage, 118206 (2021)
SynthSR: a public AI tool to turn heterogeneous clinical brain scans into
high-resolution T1-weighted images for 3D morphometry
JE Iglesias, B Billot, Y Balbastre, C Magdamo, S Arnold, S Das, B Edlow, D Alexander,
P Golland, B Fischl
Science Advances, 9(5), eadd3607 (2023)
If you use the low-field (Hyperfine) version, please cite also:
Quantitative Brain Morphometry of Portable Low-Field-Strength MRI Using
Super-Resolution Machine Learning
JE Iglesias, R Schleicher, S Laguna, B Billot, P Schaefer, B McKaig, JN Goldstein,
KN Sheth, MS Rosen, WT Kimberly
Radiology, 220522 (2022)
Configuring model on the CPU
Running SynthStrip model version 1
Input image read from: /home/jovyan/workspace/books/examples/structural_imaging/SYNcro_a24fiuhq/t1sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz
Processing frame (of 1): 1
done
Set background to: 0
Masked image saved to: /home/jovyan/workspace/books/examples/structural_imaging/SYNcro_a24fiuhq/bt1sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz
If you use SynthStrip in your analysis, please cite:
----------------------------------------------------
SynthStrip: Skull-Stripping for Any Brain Image
A Hoopes, JS Mora, AV Dalca, B Fischl, M Hoffmann
NeuroImage 206 (2022), 119474
https://doi.org/10.1016/j.neuroimage.2022.119474
Website: https://synthstrip.io
INFO - sub-M2304_ses-262_acq-spc3p2_run-5_T2w.nii.gz: min=0.0, max=571.0, interp=linear
INFO - SYNcro time: 83873 ms
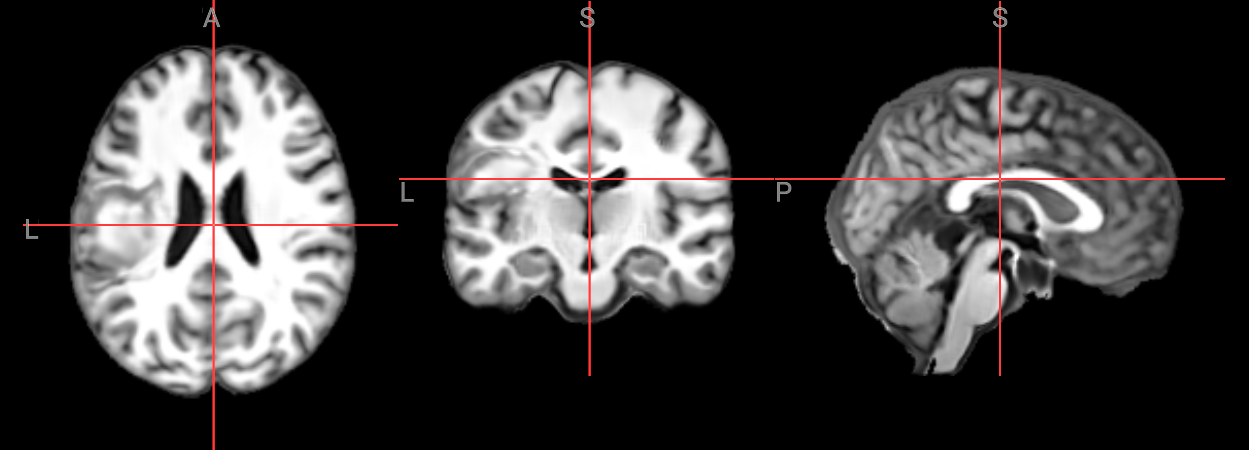
nv_mni_space = NiiVue()
nv_mni_space.load_volumes([{"url": "https://huggingface.co/datasets/neurodeskorg/neurodeskedu/resolve/main/data/examples/structural_imaging/SYNcro/wbt1sub-M2304_ses-262_acq-spc3p2_run-5_T2w_e75b1be55e40.nii.gz"}])
nv_mni_space
Image(filename='../../static/examples/structural_imaging/SYNcro/mni_wbt1sub-M2304.png')
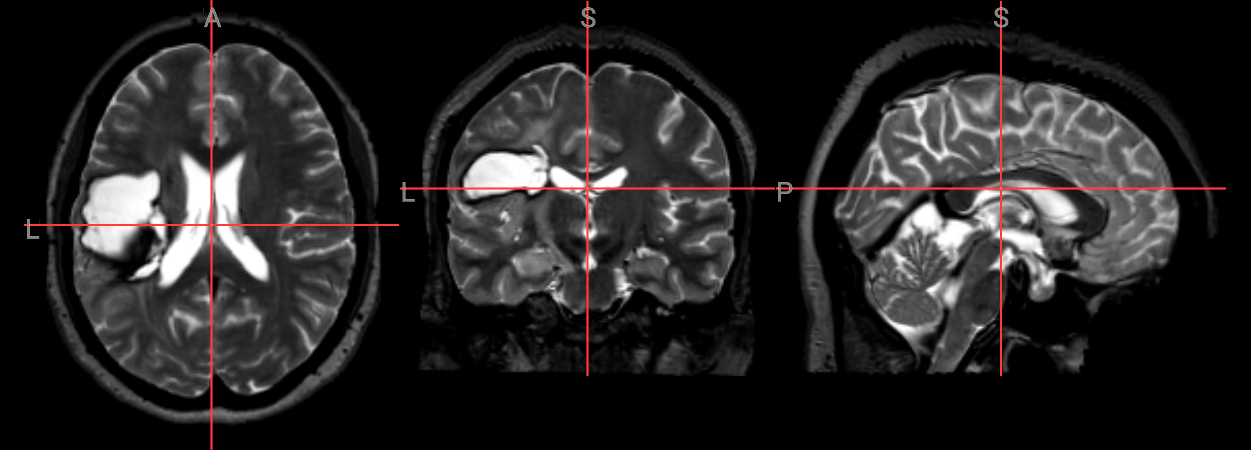
nv_mni_space = NiiVue()
nv_mni_space.load_volumes([{"url": "https://huggingface.co/datasets/neurodeskorg/neurodeskedu/resolve/main/data/examples/structural_imaging/SYNcro/wsub-M2304_ses-262_acq-spc3p2_run-5_T2w_be6e40f42d9c.nii.gz"}])
nv_mni_space
Image(filename='../../static/examples/structural_imaging/SYNcro/mni_wsub-M2304.png')
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2026-02-26T02:44:26.216685+00:00
Python implementation: CPython
Python version : 3.13.11
IPython version : 9.9.0
Compiler : GCC 14.3.0
OS : Linux
Release : 5.15.0-170-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
IPython : 9.9.0
ipyniivue: 2.4.4
