MRtrix3Tissue#
#
Authors: Thuy Dao, Monika Doerig
Date: 18 Nov 2024
#
License:
Note: If this notebook uses neuroimaging tools from Neurocontainers, those tools retain their original licenses. Please see Neurodesk citation guidelines for details.
Citation and Resources:#
Tools included in this workflow#
MRtrix3Tissue:
MRtrix3Tissue, a fork of MRtrix3 (Tournier et al. 2019)
Tournier, J.-D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., Christiaens, D., Jeurissen, B., Yeh, C.-H., & Connelly, A. (2019). MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 202, 116137. https://doi.org/10.1016/j.neuroimage.2019.116137
Dataset#
Dataset from OpenNeuro: BTC_postop
Hannelore Aerts and Daniele Marinazzo (2022). BTC_postop. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds002080.v4.0.0
Load software tools and import python libraries#
%%capture
!pip install numpy nibabel
import subprocess
import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
import tempfile
import os
import module
await module.load('mrtrix3tissue/5.2.8')
await module.list()
['mrtrix3tissue/5.2.8']
Data preparation#
# download data
!datalad install https://github.com/OpenNeuroDatasets/ds002080.git
!cd ds002080 && datalad get sub-CON02
Cloning: 0%| | 0.00/2.00 [00:00<?, ? candidates/s]
Enumerating: 0.00 Objects [00:00, ? Objects/s]
Counting: 0%| | 0.00/20.6k [00:00<?, ? Objects/s]
Compressing: 0%| | 0.00/12.7k [00:00<?, ? Objects/s]
Receiving: 0%| | 0.00/22.6k [00:00<?, ? Objects/s]
Receiving: 60%|████████████▌ | 13.5k/22.6k [00:00<00:00, 133k Objects/s]
Resolving: 0%| | 0.00/5.65k [00:00<?, ? Deltas/s]
[INFO ] scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
install(ok): /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/ds002080 (dataset)
Total: 0%| | 0.00/97.7M [00:00<?, ? Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 0%| | 0.00/8.06M [00:00<?, ? Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 0%| | 33.3k/8.06M [00:00<00:26, 307k Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 1%| | 68.1k/8.06M [00:00<00:25, 316k Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 2%| | 173k/8.06M [00:00<00:12, 614k Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 4%|▏ | 347k/8.06M [00:00<00:07, 1.01M Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 9%|▎ | 712k/8.06M [00:00<00:03, 1.86M Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 18%|▌ | 1.44M/8.06M [00:00<00:01, 3.54M Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 36%|█ | 2.91M/8.06M [00:00<00:00, 6.83M Bytes/s]
Get sub-CON0 .. p_T1w.nii.gz: 65%|█▉ | 5.23M/8.06M [00:00<00:00, 11.5M Bytes/s]
Total: 8%|██▏ | 8.06M/97.7M [00:04<00:51, 1.75M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 0%| | 0.00/48.5M [00:00<?, ? Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 5%|▏ | 2.32M/48.5M [00:00<00:04, 10.8M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 9%|▎ | 4.50M/48.5M [00:00<00:02, 14.8M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 14%|▍ | 6.88M/48.5M [00:00<00:02, 17.5M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 19%|▌ | 9.20M/48.5M [00:00<00:02, 18.9M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 23%|▋ | 11.2M/48.5M [00:00<00:01, 18.9M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 28%|▊ | 13.7M/48.5M [00:00<00:01, 20.3M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 33%|▉ | 16.0M/48.5M [00:00<00:01, 20.6M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 38%|█▏ | 18.3M/48.5M [00:00<00:01, 20.7M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 43%|█▎ | 20.7M/48.5M [00:01<00:01, 21.1M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 48%|█▍ | 23.0M/48.5M [00:01<00:01, 21.4M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 52%|█▌ | 25.4M/48.5M [00:01<00:01, 21.6M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 57%|█▋ | 27.7M/48.5M [00:01<00:00, 21.6M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 62%|█▊ | 30.1M/48.5M [00:01<00:00, 21.6M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 67%|██ | 32.5M/48.5M [00:01<00:00, 21.7M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 72%|██▏| 34.7M/48.5M [00:01<00:00, 21.3M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 76%|██▎| 37.0M/48.5M [00:01<00:00, 21.4M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 81%|██▍| 39.2M/48.5M [00:01<00:00, 21.5M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 86%|██▌| 41.5M/48.5M [00:02<00:00, 22.1M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 91%|██▋| 43.9M/48.5M [00:02<00:00, 22.5M Bytes/s]
Get sub-CON0 .. P_dwi.nii.gz: 98%|██▉| 47.7M/48.5M [00:02<00:00, 20.8M Bytes/s]
Total: 58%|███████████████ | 56.5M/97.7M [00:07<00:05, 7.54M Bytes/s]
Get sub-CON0 .. A_dwi.nii.gz: 0%| | 0.00/1.18M [00:00<?, ? Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 0%| | 0.00/40.0M [00:00<?, ? Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 4%|▏ | 1.73M/40.0M [00:00<00:04, 7.95M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 10%|▎ | 3.87M/40.0M [00:00<00:02, 12.9M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 15%|▍ | 5.97M/40.0M [00:00<00:02, 15.3M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 20%|▌ | 8.10M/40.0M [00:00<00:01, 16.9M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 26%|▊ | 10.2M/40.0M [00:00<00:01, 17.9M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 31%|▉ | 12.4M/40.0M [00:00<00:01, 18.8M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 36%|█ | 14.6M/40.0M [00:00<00:01, 19.1M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 42%|█▎ | 16.7M/40.0M [00:00<00:01, 19.6M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 47%|█▍ | 18.8M/40.0M [00:01<00:01, 19.9M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 53%|█▌ | 21.1M/40.0M [00:01<00:00, 20.1M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 58%|█▊ | 23.4M/40.0M [00:01<00:00, 20.3M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 64%|█▉ | 25.6M/40.0M [00:01<00:00, 20.5M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 70%|██ | 27.8M/40.0M [00:01<00:00, 20.6M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 75%|██▎| 30.1M/40.0M [00:01<00:00, 20.8M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 81%|██▍| 32.2M/40.0M [00:01<00:00, 21.0M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 91%|██▋| 36.4M/40.0M [00:01<00:00, 20.9M Bytes/s]
Get sub-CON0 .. _bold.nii.gz: 96%|██▉| 38.5M/40.0M [00:02<00:00, 20.9M Bytes/s]
get(ok): sub-CON02/ses-postop/anat/sub-CON02_ses-postop_T1w.nii.gz (file)
get(ok): sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.nii.gz (file)
get(ok): sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-PA_dwi.nii.gz (file)
get(ok): sub-CON02/ses-postop/func/sub-CON02_ses-postop_task-rest_bold.nii.gz (file)
get(ok): sub-CON02 (directory)
action summary:
get (ok: 5)
Convert to .mif file#
Once data is downloaded, it is recommended to import and store it as .mif file(s), the so-called “MRtrix Image Format”.
mrconvert is the tool to convert between all sorts of image formats (or extract parts of images or change properties) and enables you to read from e.g. DICOM folders or NIfTI (.nii) images and store an image as a .mif file.
If your diffusion MRI data comes as a NIfTI (.nii) image instead, the gradient orientations and b-values will typically be stored in 2 separate files (a “bvecs” and “bvals” file).
%%bash
mrconvert ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.nii.gz AP.mif -fslgrad ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.bvec ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.bval
mrconvert ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-PA_dwi.nii.gz PA.mif
mrconvert PA.mif -fslgrad ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-PA_dwi.bvec ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-PA_dwi.bval - | mrmath - mean mean_b0_PA.mif -axis 3
mrconvert: [100%] uncompressing image "ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.nii.gz"?7h?7l
mrconvert: [100%] copying from "ds002080/s...es-postop_acq-AP_dwi.nii.gz" to "AP.mif"?7h?7l?7l?7l?7l?7l
mrconvert: [100%] uncompressing image "ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-PA_dwi.nii.gz"?7h?7l
mrconvert: [100%] copying from "ds002080/s...es-postop_acq-PA_dwi.nii.gz" to "PA.mif"?7h?7l
mrconvert: [100%] copying from "PA.mif" to "/tmp/mrtrix-tmp-wDqyZO.mif"?7h?7l
mrmath: [100%] preloading data for "/tmp/mrtrix-tmp-wDqyZO.mif"?7h
mrmath: [100%] computing mean along axis 3...?7h?7l
Preprocessing#
The following are common preprocessing steps done with MRtrix.
Denoising and Gibbs ringing removal (“unringing”)#
The first preprocessing step is denoise the data by using dwidenoise command. This requires an input and an output argument, and you also have the option to output the noise map with the -noise option.
The next step is to run mrdegibbs, which removes Gibbs’ ringing artifacts from the data.
%%bash
dwidenoise AP.mif dwi_denoised.mif -noise noise.mif
mrdegibbs dwi_denoised.mif dwi_denoised_unringed.mif
dwidenoise: [100%] preloading data for "AP.mif"?7h?7l?7l?7l?7l?7l?7l
dwidenoise: [ 22%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [ 37%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [ 51%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [ 63%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [ 76%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [ 89%] running MP-PCA denoising... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
dwidenoise: [100%] running MP-PCA denoising?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
mrdegibbs: [ 56%] performing Gibbs ringing removal... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lIOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
mrdegibbs: [100%] performing Gibbs ringing removal?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
Next, we extract the b-values from the primary phase-encoded image using dwiextract.
If you have also scanned a pair of reverse phase-encoded b=0 images, you can correct for susceptibility-induced EPI distortions by concatenating the reverse phase-encoded b=0 images using mrcat to create a new image b0_pair.mif
!dwiextract dwi_denoised.mif - -bzero | mrmath - mean mean_b0_AP.mif -axis 3
!mrcat mean_b0_AP.mif mean_b0_PA.mif -axis 3 b0_pair.mif
dwiextract: [100%] extracting volumes?7h?7l?7l
mrmath: [100%] preloading data for "/tmp/mrtrix-tmp-Ail9FL.mif"?7h?7l
mrmath: [100%] computing mean along axis 3...?7h?7l
mrcat: [100%] concatenating "mean_b0_AP.mif"?7h?7l
mrcat: [100%] concatenating "mean_b0_PA.mif"?7h?7l
Motion and distortion correction#
Motion and distortion correction are performed by the dwifslpreproc command, most of the heavy lifting is done automatically by FSL’s topup and eddy tools.
-pe_dir option is used to tell dwifslpreproc the phase-encoding direction of the scan.
-se_epi is an additional image series consisting of spin-echo EPI images used by topup for estimating the inhomogeneity field
-eddy_options is an additional command-line options to the eddy
!dwifslpreproc dwi_denoised_unringed.mif dwi_denoised_unringed_preproc.mif -pe_dir AP -rpe_pair -se_epi b0_pair.mif -eddy_options " --slm=linear --data_is_shelled"
dwifslpreproc:
dwifslpreproc: Note that this script makes use of commands / algorithms that have relevant articles for citation; INCLUDING FROM EXTERNAL SOFTWARE PACKAGES. Please consult the help page (-help option) for more information.
dwifslpreproc:
dwifslpreproc: Generated scratch directory: /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/
Command: mrconvert /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi_denoised_unringed.mif /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/dwi.mif -json_export /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/dwi.json
Command: mrconvert /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/b0_pair.mif /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/se_epi.mif
dwifslpreproc: Changing to scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/)
dwifslpreproc: Total readout time not provided at command-line; assuming sane default of 0.1
Command: mrinfo dwi.mif -export_grad_mrtrix grad.b
Command: mrconvert se_epi.mif topup_in.nii -import_pe_table se_epi_manual_pe_scheme.txt -strides -1,+2,+3,+4 -export_pe_table topup_datain.txt
Command: topup --imain=topup_in.nii --datain=topup_datain.txt --out=field --fout=field_map.nii.gz --config=/opt/fsl-6.0.3/etc/flirtsch/b02b0.cnf --verbose
Command: mrconvert dwi.mif -import_pe_table dwi_manual_pe_scheme.txt - | mrinfo - -export_pe_eddy applytopup_config.txt applytopup_indices.txt
Command: dwiextract dwi.mif -import_pe_table dwi_manual_pe_scheme.txt -pe 0.0,-1.0,0.0,0.1 - | mrconvert - dwi_pe_1.nii -json_export dwi_pe_1.json
Command: applytopup --imain=dwi_pe_1.nii --datain=applytopup_config.txt --inindex=1 --topup=field --out=dwi_pe_1_applytopup.nii --method=jac
Command: mrconvert dwi_pe_1_applytopup.nii.gz dwi_pe_1_applytopup.mif -json_import dwi_pe_1.json
Command: dwi2mask dwi_pe_1_applytopup.mif - | maskfilter - dilate - | mrconvert - eddy_mask.nii -datatype float32 -strides -1,+2,+3
Command: mrconvert dwi.mif -import_pe_table dwi_manual_pe_scheme.txt eddy_in.nii -strides -1,+2,+3,+4 -export_grad_fsl bvecs bvals -export_pe_eddy eddy_config.txt eddy_indices.txt
Command: eddy_cuda9.1 --imain=eddy_in.nii --mask=eddy_mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --slm=linear --data_is_shelled --out=dwi_post_eddy --verbose
dwifslpreproc: CUDA version of 'eddy' was not successful; attempting OpenMP version
Command: eddy_openmp --imain=eddy_in.nii --mask=eddy_mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --slm=linear --data_is_shelled --out=dwi_post_eddy --verbose
Command: mrconvert dwi_post_eddy.nii.gz result.mif -strides -1,2,3,4 -fslgrad dwi_post_eddy.eddy_rotated_bvecs bvals
Command: mrconvert result.mif /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi_denoised_unringed_preproc.mif
dwifslpreproc: Changing back to original directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging)
dwifslpreproc: Deleting scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwifslpreproc-tmp-OMRCOI/)
Bias field correction#
You can correct for bias fields at this point in the pipeline with the dwibiascorrect ants command
!dwibiascorrect ants dwi_denoised_unringed_preproc.mif dwi_denoised_unringed_preproc_unbiased.mif
dwibiascorrect:
dwibiascorrect: Note that this script makes use of commands / algorithms that have relevant articles for citation; INCLUDING FROM EXTERNAL SOFTWARE PACKAGES. Please consult the help page (-help option) for more information.
dwibiascorrect:
dwibiascorrect: Generated scratch directory: /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwibiascorrect-tmp-Y7JHHU/
Command: mrconvert /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi_denoised_unringed_preproc.mif /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwibiascorrect-tmp-Y7JHHU/in.mif
dwibiascorrect: Changing to scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwibiascorrect-tmp-Y7JHHU/)
Command: dwi2mask in.mif mask.mif
Command: dwiextract in.mif - -bzero | mrmath - mean mean_bzero.mif -axis 3
Command: mrconvert mean_bzero.mif mean_bzero.nii -strides +1,+2,+3
Command: mrconvert mask.mif mask.nii -strides +1,+2,+3
Command: N4BiasFieldCorrection -d 3 -i mean_bzero.nii -w mask.nii -o [corrected.nii,init_bias.nii] -s 4 -b [100,3] -c [1000,0.0]
Command: mrcalc mean_bzero.mif mask.mif -mult - | mrmath - sum - -axis 0 | mrmath - sum - -axis 1 | mrmath - sum - -axis 2 | mrdump -
Command: mrcalc corrected.nii mask.mif -mult - | mrmath - sum - -axis 0 | mrmath - sum - -axis 1 | mrmath - sum - -axis 2 | mrdump -
Command: mrcalc init_bias.nii 0.531319575981 -mult bias.mif
Command: mrcalc in.mif bias.mif -div result.mif
Command: mrconvert result.mif /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi_denoised_unringed_preproc_unbiased.mif
dwibiascorrect: Changing back to original directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging)
dwibiascorrect: Deleting scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwibiascorrect-tmp-Y7JHHU/)
Brain mask estimation#
A brain mask can be estimated automatically from the diffusion MRI data using dwi2mask as follows:
!dwi2mask dwi_denoised_unringed_preproc_unbiased.mif dwi_mask.mif
dwi2mask: [100%] preloading data for "dwi_denoised_unringed_preproc_unbiased.mif"?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
dwi2mask: [100%] finding min/max of "mean b=0 image"?7h?7l
dwi2mask: [done] optimising threshold?7h
dwi2mask: [100%] thresholding?7h?7l
dwi2mask: [100%] finding min/max of "mean b=700 image"?7h?7l
dwi2mask: [done] optimising threshold?7h
dwi2mask: [100%] thresholding?7h?7l
dwi2mask: [100%] finding min/max of "mean b=1200 image"?7h?7l
dwi2mask: [done] optimising threshold?7h
dwi2mask: [100%] thresholding?7h?7l
dwi2mask: [100%] finding min/max of "mean b=2800 image"?7h?7l
dwi2mask: [done] optimising threshold?7h
dwi2mask: [100%] thresholding?7h?7l
dwi2mask: [done] computing dwi brain mask?7h?7l?7l
dwi2mask: [done] applying mask cleaning filter?7h
Decomposition#
3-tissue response function estimation#
An method to obtain 3-tissue response functions representing single-fibre white matter (WM), grey matter (GM) and CSF from the data itself, is available as the so-called dwi2response dhollander command from MRtrix3Tissue package.
!dwi2response dhollander dwi_denoised_unringed_preproc_unbiased.mif response_wm.txt response_gm.txt response_csf.txt
dwi2response:
dwi2response: Note that this script makes use of commands / algorithms that have relevant articles for citation. Please consult the help page (-help option) for more information.
dwi2response:
dwi2response: Generated scratch directory: /home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi2response-tmp-5QHWDZ/
dwi2response: Importing DWI data (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi_denoised_unringed_preproc_unbiased.mif)...
dwi2response: Changing to scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi2response-tmp-5QHWDZ/)
dwi2response: Computing brain mask (dwi2mask)...
dwi2response: -------
dwi2response: 4 unique b-value(s) detected: 0,700,1200,2800 with 6,16,30,50 volumes
dwi2response: -------
dwi2response: Preparation:
dwi2response: * Eroding brain mask by 3 pass(es)...
dwi2response: [ mask: 96467 -> 69356 ]
dwi2response: * Computing signal decay metric (SDM):
dwi2response: * b=0...
dwi2response: * b=700...
dwi2response: * b=1200...
dwi2response: * b=2800...
dwi2response: * Removing erroneous voxels from mask and correcting SDM...
dwi2response: [ mask: 69356 -> 69356 ]
dwi2response: -------
dwi2response: Crude segmentation:
dwi2response: * Crude WM versus GM-CSF separation (at FA=0.2)...
dwi2response: [ 69356 -> 37105 (WM) & 32251 (GM-CSF) ]
dwi2response: * Crude GM versus CSF separation...
dwi2response: [ 32251 -> 21468 (GM) & 10783 (CSF) ]
dwi2response: -------
dwi2response: Refined segmentation:
dwi2response: * Refining WM...
dwi2response: [ WM: 37105 -> 33591 ]
dwi2response: * Refining GM...
dwi2response: [ GM: 21468 -> 11977 ]
dwi2response: * Refining CSF...
dwi2response: [ CSF: 10783 -> 5380 ]
dwi2response: -------
dwi2response: Final voxel selection and response function estimation:
dwi2response: * CSF:
dwi2response: * Selecting final voxels (10.0% of refined CSF)...
dwi2response: [ CSF: 5380 -> 538 ]
dwi2response: * Estimating response function...
dwi2response: * GM:
dwi2response: * Selecting final voxels (2.0% of refined GM)...
dwi2response: [ GM: 11977 -> 240 ]
dwi2response: * Estimating response function...
dwi2response: * single-fibre WM:
dwi2response: * Selecting final voxels (0.5% of refined WM)...
dwi2response: [ WM: 33591 -> 168 (single-fibre) ]
dwi2response: * Estimating response function...
dwi2response: -------
dwi2response: Generating outputs...
dwi2response: -------
dwi2response: Changing back to original directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging)
dwi2response: Deleting scratch directory (/home/jovyan/Git_repositories/neurodeskedu/books/diffusion_imaging/dwi2response-tmp-5QHWDZ/)
Upsampling#
Upsampling your preprocessed diffusion MRI data before the 3-tissue constrained spherical deconvolution (3-tissue CSD) step to get a higher quality result. Upsampling is useful if you’re after a higher quality visualisation, which reveals finer details using mrgrid.
!mrgrid dwi_denoised_unringed_preproc_unbiased.mif regrid dwi_denoised_unringed_preproc_unbiased_upsampled.mif -voxel 1.5
mrgrid: [ 45%] reslicing "dwi_denoised_unringed_preproc_unbiased.mif"... ?7h?7l?7l?7lbiased.mif"... ?7h?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l?7l?7l11%] reslicing "dwi_denoised_unringed_preproc_unbiased.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l23%] reslicing "dwi_denoised_unringed_preproc_unbiased.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l
mrgrid: [100%] reslicing "dwi_denoised_unringed_preproc_unbiased.mif"?7h?7l?7l?7l?7lbiased.mif"... ?7h?7lbiased.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l?7l?7l?7l?7l?7lbiased.mif"... ?7h?7l?7l?7lbiased.mif"... ?7h?7l
IOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
After upsampling the image, you need to regrid the brain mask to the exact same voxel grid as the image via the -template option to mrgrid. If the regridding is effectively upsampling, you can avoid jagged edges around masks by performing linear interpolation -interp linear. Then a median filter the result using the maskfilter command. To perform all of this at once (without storing an intermediate image), you an directly “pipe” the output of mrgrid into the input of maskfilter, by replacing the output in the one command and the input in the other command by a dash (“-”), and typing a “pipe” (“|”) between both commands.
!mrgrid dwi_mask.mif regrid - -template dwi_denoised_unringed_preproc_unbiased_upsampled.mif -interp linear -datatype bit | maskfilter - median dwi_mask_upsampled.mif
mrgrid: [100%] reslicing "dwi_mask.mif"?7h?7lng "dwi_mask.mif"... ?7h?7l
maskfilter: [100%] applying median filter to image -?7h?7l?7l?7l
3-tissue CSD modelling#
Your data is now ready for 3-tissue CSD modelling with the previously obtained 3-tissue response functions, which will result in modelling the diffusion MRI data using WM-like (FOD), GM-like and CSF-like compartments. There are 2 different methods (or algorithms) available to perform 3-tissue CSD. The choice between both depends on what (part of your) data you intend to perform 3-tissue CSD modelling for.
If you want to perform 3-tissue CSD modelling for multi-shell data, this can be achieved using the multi-shell multi-tissue CSD (MSMT-CSD) method or algorithm, as follows:
!dwi2fod msmt_csd dwi_denoised_unringed_preproc_unbiased_upsampled.mif response_wm.txt wmfod.mif response_gm.txt gm.mif response_csf.txt csf.mif -mask dwi_mask_upsampled.mif
dwi2fod: [100%] preloading data for "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"?7h?7l?7l.mif"... ?7h?7l?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7ldwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7l?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7l?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7lfor "dwi_denoised_unringed_preproc_unbiased_upsampled.mif"... ?7h?7l?7l
dwi2fod: [ 6%] performing multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l
dwi2fod: [ 19%] performing multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
dwi2fod: [ 24%] performing multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
dwi2fod: [100%] performing multi-shell, multi-tissue CSD?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7l?7l?7li-tissue CSD... ?7h?7li-tissue CSD... ?7h?7l?7li-tissue CSD... ?7h?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7lforming multi-shell, multi-tissue CSD... ?7h?7l?7l?7l?7l
IOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
IOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
IOPub data rate exceeded.
The Jupyter server will temporarily stop sending output
to the client in order to avoid crashing it.
To change this limit, set the config variable
`--ServerApp.iopub_data_rate_limit`.
Current values:
ServerApp.iopub_data_rate_limit=1000000.0 (bytes/sec)
ServerApp.rate_limit_window=3.0 (secs)
3-tissue bias field and intensity normalisation#
Bias field correction and global intensity normalisation can be performed using the mtnormalise command, as follows:
!mtnormalise wmfod.mif wmfod_norm.mif gm.mif gm_norm.mif csf.mif csf_norm.mif -mask dwi_mask_upsampled.mif
mtnormalise: [done] loading input images?7h?7l?7l
mtnormalise: [100%] performing log-domain intensity normalisation?7h?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l?7l
mtnormalise: [done] writing output images?7h?7l
mtnormalise: [100%] performing log-domain intensity normalisation
mtnormalise: [done] loading input images
Temperature computation#
Get CSF mask#
You first need to get 2 CSF masks from 2 criteria:
csf_mask1.miffrom CSF spherical harmonic coefficients applying the first criteria (aka the csf.mif file)csf_mask2.miffrom second criteria (note: 0.141047 might be half of max response function)
Then find the overlap of both masks resulting in the final mask csf_maskcombined.mif.
%%bash
mrconvert wmfod_norm.mif -coord 3 0 -axes 0,1,2 - | mrcalc csf_norm.mif gm_norm.mif - -add 5 -mult -gt csf_mask1.mif -datatype bit
mrthreshold csf_norm.mif csf_mask2.mif -abs 0.141047
mrcalc csf_mask1.mif csf_mask2.mif -mult csf_maskcombined.mif -datatype bit
mrconvert: [100%] copying from "wmfod_norm.mif" to "/tmp/mrtrix-tmp-7Z4XgQ.mif"?7h?7l
mrcalc: [100%] computing: (csf_norm.mif > ((gm_norm.mif + /tmp/mrtrix-tmp-7Z4XgQ.mif) * 5))?7h?7l
mrthreshold: [100%] Determining and applying per-volume thresholds?7h
mrcalc: [100%] computing: (csf_mask1.mif * csf_mask2.mif)?7h?7l
Calculate diffusion coefficient in CSF#
First, we filter the processed image to exclude b-values over 1000 into a new image reduced.mif.
Next, estimate the Apparent Diffusion Coefficient (ADC) using dwi2adc.
Then we extract the coefficient from the second array in dwi_adc.mif to get D values.
Finally, we filter the D values within CSF using generated mask.
# Get the index of b-values < 1000 from table and write to txt file for next step
bvals = np.loadtxt("ds002080/sub-CON02/ses-postop/dwi/sub-CON02_ses-postop_acq-AP_dwi.bval")
bvals_filtered = np.where(bvals < 1000)
with open("bval_less_1000.txt", "w") as txt_file:
txt_file.write(",".join((str(x) for x in bvals_filtered[0])))
%%bash
mrconvert dwi_denoised_unringed_preproc_unbiased_upsampled.mif reduced.mif -coord 3 $(cat bval_less_1000.txt)
dwi2adc reduced.mif dwi_adc.mif
mrconvert dwi_adc.mif extracted_adc.mif -coord 3 1
mrcalc extracted_adc.mif csf_maskcombined.mif -mult masked_adc.mif
mrconvert: [100%] copying from "dwi_denois...proc_unbiased_upsampled.mif" to "reduced.mif"?7h?7l?7l?7l?7l?7l
dwi2adc: [100%] computing ADC values?7h?7l?7l?7l
mrconvert: [100%] copying from "dwi_adc.mif" to "extracted_adc.mif"?7h?7l
mrcalc: [100%] computing: (extracted_adc.mif * csf_maskcombined.mif)?7h?7l
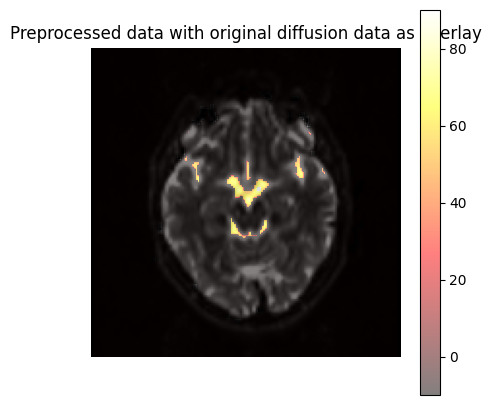
Temperature estimation#
We calculate the temperature of cerebrospinal fluid (CSF) using the following equation from this study:
where ( D ) is the diffusion constant.
!mrcalc 2256.74 4.39221 masked_adc.mif -divide -log -divide 273.15 -subtract masked_temperature.mif
mrcalc: [100%] computing: ((2256.74 / log ((4.39221 / masked_adc.mif))) - 273.15)?7h?7l?7l
Visualization#
# File paths for both images
mif_file_path1 = 'dwi_denoised_unringed_preproc_unbiased_upsampled.mif'
mif_file_path2 = 'masked_temperature.mif'
# Convert both .mif files to temporary .nii.gz files with -force flag
with tempfile.NamedTemporaryFile(suffix=".nii.gz") as temp_file1, \
tempfile.NamedTemporaryFile(suffix=".nii.gz") as temp_file2:
# Convert main image
result1 = subprocess.run(["mrconvert", mif_file_path1, temp_file1.name, "-force"], capture_output=True, text=True)
if result1.returncode != 0:
print(f"Error in mrconvert for preprocessed image: {result1.stderr}")
exit()
# Convert overlay image
result2 = subprocess.run(["mrconvert", mif_file_path2, temp_file2.name, "-force"], capture_output=True, text=True)
if result2.returncode != 0:
print(f"Error in mrconvert for original image: {result2.stderr}")
exit()
# Load the converted images
nii_image1 = nib.load(temp_file1.name)
nii_image2 = nib.load(temp_file2.name)
data1 = nii_image1.get_fdata()
data2 = nii_image2.get_fdata()
# Select middle slice
slice_index = data1.shape[2] // 3
fig, axes = plt.subplots(1, 1, figsize=(5, 5))
# Plot original diffusion data as overlay
axes.imshow(np.rot90(data1[:, :, slice_index, 0]), cmap="Greys_r", vmin=0, vmax=1000)
overlay = axes.imshow(np.rot90(data2[:, :, slice_index, 0]), cmap="hot", alpha=0.5, vmin=-10, vmax=90)
axes.set_title("Preprocessed data with original diffusion data as overlay")
axes.axis("off")
fig.colorbar(overlay, ax=axes, location='right')
plt.show()
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-11-06T02:30:05.908666+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
matplotlib: 3.8.4
nibabel : 5.3.2
numpy : 2.2.4
