QSMxT#
Author: Ashley Stewart
Date: 25 Aug 2025
Citation and Resources:#
Tools included in this workflow#
QSMxT: Stewart AW, Robinson SD, O’Brien K, et al. QSMxT: Robust masking and artifact reduction for quantitative susceptibility mapping. Magnetic Resonance in Medicine. 2022;87(3):1289-1300. doi:10.1002/mrm.29048
QSMxT: Stewart AW, Bollman S, et al. QSMxT/QSMxT. GitHub; 2022. https://github.com/QSMxT/QSM
Dataset#
Bollmann, S., & Stewart, A. (2023, August 7). QSM DICOM testdata for QSMxT pipeline. Retrieved from osf.io/ru43c
QSMxT Interactive Notebook#
This interactive notebook estimates Quantitative Susceptibility Maps (QSMs) for two gradient-echo (GRE) MRI acquisitions using QSMxT provided by the Neurodesk project.
What is QSM?#
QSM is a form of quantitative MRI (qMRI) that estimates the magnetic susceptibility distribution across an imaged object. Magnetic susceptibility is the degree to which a material becomes magnetised by an external magnetic field. Major contributors to susceptibility include iron, calcium, and myelin, with the susceptibility of water typically approximating a zero-reference, though it is slightly diamagnetic. Read more about QSM here.
What is QSMxT?#
QSMxT is a suite of tools for building and running automated pipelines for QSM that:
is available open-source without any licensing required;
is distributed as a software container making it straightforward to access and install (Neurodesk!)
scales its processing to execute across many acquisitions through jobs parallelisation (using multiple processors or HPCs) provided by Nipype;
automates steps that usually require manual intervention and scripting, including:
DICOM to BIDS conversion;
QSM reconstruction using a range of algorithms;
segmentation using FastSurfer;
group space generation using ANTs;
export of susceptibility statistics by subject and region of interest (ROI) to CSV.

How do I access QSMxT?#
There are a few ways you can access QSMxT:
This notebook: You can access QSMxT in this notebook right now!
If you are running this on a Neurodesk Play instance, you can upload your own data into the sidebar via drag-and-drop.
Neurodesktop: QSMxT is in the applications menu of Neurodesktop.
On Neurodesk Play, upload your own data into the desktop via drag-and-drop.
On a local install of Neurodesk, bring any necessary files into the shared
~/neurodesktop-storagedirectory
Local install: QSMxT can also be installed via the Docker container
HPC install: QSMxT can also be installed via the Singularity container for use on HPCs
Download example DICOMs#
Here, we download some example DICOMs from our OSF repository for QSMxT.
These data include GRE and T1-weighted acquisitions for one subject (duplicated to act as two subjects).
!osf -p ru43c clone . > /dev/null 2>&1
!tar xf osfstorage/dicoms-unsorted.tar
!rm -rf osfstorage/
!tree dicoms-unsorted | head
!echo -e "...\nThere are `ls dicoms-unsorted | wc -l` unsorted DICOMs in ./dicoms-unsorted/"
dicoms-unsorted
├── MR.1.1.dcm
├── MR.1.10.dcm
├── MR.1.100.dcm
├── MR.1.101.dcm
├── MR.1.102.dcm
├── MR.1.103.dcm
├── MR.1.104.dcm
├── MR.1.105.dcm
├── MR.1.106.dcm
...
There are 1216 unsorted DICOMs in ./dicoms-unsorted/
Load QSMxT#
To load QSMxT inside a notebook, we can use the available module system:
import module
await module.load('qsmxt/8.1.1')
!qsmxt --version
[INFO]: QSMxT v8.1.1
Data standardisation#
QSMxT requires input data to conform to the Brain Imaging Data Structure (BIDS).
Luckily, QSMxT also provides scripts that can convert unorganised NIfTI or DICOM images to BIDS. If you are using NIfTI images and do not have DICOMs, see nifti-convert.
Convert DICOM to BIDS#
The DICOM to BIDS conversion must identify which series should be used for QSM reconstruction (e.g. MEGRE), and which series should be used for segmentation (T1-weighted). Because this information is not stored in the DICOM header in a standardised way, it can only be estimated based on other fields, or the user must identify series themselves.
If QSMxT is run interactively, the user will be prompted to identify the relevant series. However, because we are running QSMxT in a notebook, we disable the interactivity using --auto_yes and allow QSMxT to guess the intended purpose of each acquisition. In this case, it makes the right guess:
!dicom-convert dicoms-unsorted bids --auto_yes
/opt/miniconda-4.12.0/lib/python3.8/site-packages/dicompare/io.py:30: UserWarning: The DICOM readers are highly experimental, unstable, and only work for Siemens time-series at the moment
Please use with caution. We would be grateful for your help in improving them
from nibabel.nicom.csareader import get_csa_header
[INFO]: Running QSMxT 8.1.1
[INFO]: Command: /opt/miniconda-4.12.0/bin/dicom-convert dicoms-unsorted bids --auto_yes
[INFO]: Python interpreter: /opt/miniconda-4.12.0/bin/python3.8
Loading DICOM files: 100%|█████████████████| 1216/1216 [00:11<00:00, 102.57it/s]
[INFO]: Loaded DICOM session in 12.07 seconds
/opt/miniconda-4.12.0/lib/python3.8/site-packages/dicompare/acquisition.py:144: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
session_df["BaseAcquisition"] = "acq-" + session_df[acquisition_fields].apply(
/opt/miniconda-4.12.0/lib/python3.8/site-packages/dicompare/acquisition.py:195: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
session_df.loc[param_group.index, "AcquisitionSignature"] = signature
/opt/miniconda-4.12.0/lib/python3.8/site-packages/dicompare/acquisition.py:218: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
session_df["RunNumber"] = 1
/opt/miniconda-4.12.0/lib/python3.8/site-packages/dicompare/acquisition.py:301: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
session_df["Acquisition"] = session_df["AcquisitionSignature"].fillna("Unknown").astype(str)
[INFO]: Auto-assigning initial labels
[INFO]: Auto-assigned selections:
Acquisition ... Description
0 acq-mp2ragehighres0p5isoslab ...
1 acq-qsmp21mmisote20 ...
2 acq-qsmp21mmisote20 ...
[3 rows x 7 columns]
[INFO]: Merging selections into dataframe...
[INFO]: Processing base group: ('dev_siemens^SB', 1, 20170705, 'acq-mp2ragehighres0p5isoslab', 'mp2rage_highRes_0p5iso_slab_UNI-DEN', 1, '1.2.276.0.7230010.3.1.3.0.4480.1677042698.772469', 'NA')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: T1w with 288 rows
[INFO]: Found multiple DICOM paths for Type 'T1w', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 288 rows
[INFO]: Converting group with 288 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 288 DICOM file(s)
Convert 288 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output (378x420x288x1)
Conversion required 1.654690 seconds (1.637657 for core code).
[INFO]: Processing base group: ('dev_siemens^SB', 1, 20170705, 'acq-qsmp21mmisote20', 'QSM_p2_1mmIso_TE20', 1, '1.3.12.2.1107.5.2.43.167001.2017070513471948579426649.0.0.0', 'HC1-7')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: Mag with 160 rows
[INFO]: Found multiple DICOM paths for Type 'Mag', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 160 rows
[INFO]: Converting group with 160 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 160 DICOM file(s)
Convert 160 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output (224x224x160x1)
Conversion required 0.246731 seconds (0.245992 for core code).
[INFO]: Processing base group: ('dev_siemens^SB', 1, 20170705, 'acq-qsmp21mmisote20', 'QSM_p2_1mmIso_TE20', 1, '1.3.12.2.1107.5.2.43.167001.2017070513471948580526650.0.0.0', 'HC1-7')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: Phase with 160 rows
[INFO]: Found multiple DICOM paths for Type 'Phase', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 160 rows
[INFO]: Converting group with 160 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 160 DICOM file(s)
Convert 160 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output_ph (224x224x160x1)
Conversion required 0.298057 seconds (0.297147 for core code).
[INFO]: Processing base group: ('dev_siemens^SB', 2, 20170705, 'acq-mp2ragehighres0p5isoslab', 'mp2rage_highRes_0p5iso_slab_UNI-DEN', 1, 2, 'NA')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: T1w with 288 rows
[INFO]: Found multiple DICOM paths for Type 'T1w', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 288 rows
[INFO]: Converting group with 288 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 288 DICOM file(s)
Convert 288 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output (378x420x288x1)
Conversion required 1.788802 seconds (1.779442 for core code).
[INFO]: Processing base group: ('dev_siemens^SB', 2, 20170705, 'acq-qsmp21mmisote20', 'QSM_p2_1mmIso_TE20', 1, 1, 'HC1-7')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: Mag with 160 rows
[INFO]: Found multiple DICOM paths for Type 'Mag', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 160 rows
[INFO]: Converting group with 160 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 160 DICOM file(s)
Convert 160 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output (224x224x160x1)
Conversion required 0.323626 seconds (0.323518 for core code).
[INFO]: Processing base group: ('dev_siemens^SB', 2, 20170705, 'acq-qsmp21mmisote20', 'QSM_p2_1mmIso_TE20', 1, 3, 'HC1-7')
[INFO]: Found 1 unique Types in this group
[INFO]: Processing Type group: Phase with 160 rows
[INFO]: Found multiple DICOM paths for Type 'Phase', grouping by EchoNumber
[INFO]: Processing Echo group: 1.0 with 160 rows
[INFO]: Converting group with 160 rows
[INFO]: Running command: '"dcm2niix" -o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert" -f "temp_output" -z n -m o "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert"'
Chris Rorden's dcm2niiX version v1.0.20240202 (JP2:OpenJPEG) (JP-LS:CharLS) GCC11.4.0 x86-64 (64-bit Linux)
Found 160 DICOM file(s)
Convert 160 DICOM as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/temp_convert/temp_output_ph (224x224x160x1)
Conversion required 0.296623 seconds (0.296486 for core code).
[INFO]: Scanning for multi-coil data to merge...
[INFO]: Scanning for multi-coil runs to combine...
[INFO]: Checking for complex or polar data requiring fixing...
Loading NIfTIs: 100%|███████████████████████████| 6/6 [00:00<00:00, 1135.23it/s]
[INFO]: Finished
!tree bids
bids
├── log_2025-10-30_01-40-18013068.txt
├── references.txt
├── sub-1
│ └── ses-20170705
│ └── anat
│ ├── sub-1_ses-20170705_acq-acqmp2ragehighres0p5isoslab_part-mag_T1w.json
│ ├── sub-1_ses-20170705_acq-acqmp2ragehighres0p5isoslab_part-mag_T1w.nii
│ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_part-mag_T2starw.json
│ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_part-mag_T2starw.nii
│ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_part-phase_T2starw.json
│ └── sub-1_ses-20170705_acq-acqqsmp21mmisote20_part-phase_T2starw.nii
└── sub-2
└── ses-20170705
└── anat
├── sub-2_ses-20170705_acq-acqmp2ragehighres0p5isoslab_part-mag_T1w.json
├── sub-2_ses-20170705_acq-acqmp2ragehighres0p5isoslab_part-mag_T1w.nii
├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_part-mag_T2starw.json
├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_part-mag_T2starw.nii
├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_part-phase_T2starw.json
└── sub-2_ses-20170705_acq-acqqsmp21mmisote20_part-phase_T2starw.nii
6 directories, 14 files
Inspect input data#
Here we define a function we will use to visualise NIfTI images so we can view some of the input data:
%%capture
!pip install seaborn numpy nibabel pandas
from glob import glob
from matplotlib import pyplot as plt
import numpy as np
import nibabel as nib
def show_nii(nii_path, title=None, figsize=(4,5), cmap='gray', **imshow_args):
# load data\n",
data_1 = nib.load(nii_path).get_fdata()
# get middle slices\n",
slc_data1 = np.rot90(data_1[np.shape(data_1)[0]//2,:,:])
slc_data2 = np.rot90(data_1[:,np.shape(data_1)[1]//2,:])
slc_data3 = np.rot90(data_1[:,:,np.shape(data_1)[2]//2])
# show slices\n",
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=figsize)
if title: plt.suptitle(title)
axes[0].imshow(slc_data1, cmap=cmap, **imshow_args)
axes[1].imshow(slc_data2, cmap=cmap, **imshow_args)
axes[2].imshow(slc_data3, cmap=cmap, **imshow_args)
axes[0].axis('off')
axes[1].axis('off')
axes[2].axis('off')
fig.tight_layout()
fig.subplots_adjust(top=1.55)
plt.show()
show_nii(glob("bids/sub-*/ses-*/anat/*mag*T2starw*nii*")[0], title="Magnitude", vmax=500)
show_nii(glob("bids/sub-*/ses-*/anat/*phase*T2starw*nii*")[0], title="Phase")
show_nii(glob("bids/sub-*/ses-*/anat/*T1w*nii*")[0], title="T1-weighted")
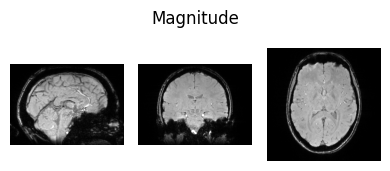

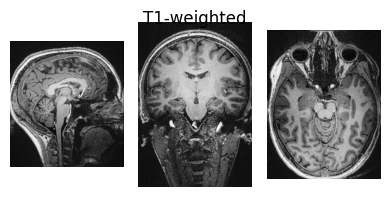
Interactive Display using Niivue#
Uncomment the next cell if you want to inspect the T1-weighted image interactively
# from ipyniivue import NiiVue
# nv_T1 = NiiVue()
# nv_T1.load_volumes([{"path": glob("bids/sub-*/ses-*/anat/*T1w*nii*")[0]}])
# nv_T1
Run QSMxT#
We are now ready to run QSMxT! We will generate susceptibility maps and segmentations, and export analysis CSVs to file.
The usual way of running QSMxT is to use qsmxt bids_dir. This will launch an interactive command-line interface (CLI) to setup your desired pipelines. However, since we are running this in a notebook, we need to use command-line arguments to by-pass the interface and execute a pipeline.
But first, let’s consider our pipeline settings. For QSM reconstruction, QSMxT provides a range of sensible defaults fit for different purposes. We can list the premade QSM pipelines using --list_premades. For the full pipeline details used for each premade pipeline, see qsm_pipelines.json.
!qsmxt --list_premades
=== Premade pipelines ===
default: Default QSMxT settings (GRE; assumes human brain)
gre: Applies suggested settings for 3D-GRE images
epi: Applies suggested settings for 3D-EPI images (assumes human brain)
bet: Applies a traditional BET-masking approach (artefact reduction unavailable; assumes human brain)
fast: Applies a set of fast algorithms
body: Applies suggested settings for non-brain applications
nextqsm: Applies suggested settings for running the NeXtQSM algorithm (assumes human brain)
[INFO]: Finished
For this demonstration, we will go with the fast pipeline. To export segmentations and analysis results, we will use --do_segmentation and --do_analysis. The --auto_yes option avoid the interactive CLI interface that cannot be used in a notebook:
!qsmxt bids \
--premade fast \
--do_qsm \
--do_segmentation \
--do_analysis \
--auto_yes
[INFO]: QSMxT v8.1.1
[INFO]: Python interpreter: /opt/miniconda-4.12.0/bin/python3.8
[INFO]: Command: qsmxt /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids --premade 'fast' --do_qsm --do_segmentation --do_analysis --auto_yes
[WARNING]: Pipeline is NOT guidelines compliant (see https://doi.org/10.1002/mrm.30006):; Phase-quality-based masking recommended
[INFO]: Available memory: 195.188 GB
[INFO]: Creating QSMxT workflow for sub-1.ses-20170705.acq-acqmp2ragehighres0p5isoslab.T1w...
[INFO]: Creating QSMxT workflow for sub-1.ses-20170705.acq-acqqsmp21mmisote20.T2starw...
[INFO]: Creating QSMxT workflow for sub-2.ses-20170705.acq-acqmp2ragehighres0p5isoslab.T1w...
[INFO]: Creating QSMxT workflow for sub-2.ses-20170705.acq-acqqsmp21mmisote20.T2starw...
[INFO]: Running using MultiProc plugin with n_procs=32
251030-02:01:26,143 nipype.workflow INFO:
Workflow qsmxt-workflow settings: ['check', 'execution', 'logging', 'monitoring']
251030-02:01:26,393 nipype.workflow INFO:
Running in parallel.
251030-02:01:26,407 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 24 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
251030-02:01:27,160 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-me" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_read-json-me".
251030-02:01:27,163 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_read-nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_read-nii".
251030-02:01:27,171 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-se" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_read-json-se".
251030-02:01:27,191 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/fastsurfer_segment-t1".
251030-02:01:27,197 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/ants_register-t1-to-qsm".
251030-02:01:27,208 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_as-canonical" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_as-canonical".
251030-02:01:27,219 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_read-nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_read-nii".
251030-02:01:27,254 nipype.workflow INFO:
[Node] Executing "nibabel_read-nii" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,266 nipype.workflow INFO:
[Node] Executing "func_read-json-se" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,274 nipype.workflow INFO:
[Node] Executing "ants_register-t1-to-qsm" <nipype.interfaces.ants.registration.RegistrationSynQuick>
251030-02:01:27,274 nipype.workflow INFO:
[Node] Executing "fastsurfer_segment-t1" <qsmxt.interfaces.nipype_interface_fastsurfer.FastSurferInterface>
251030-02:01:27,226 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/fastsurfer_segment-t1".
251030-02:01:27,169 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_as-canonical" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_as-canonical".
251030-02:01:27,209 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-me" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_read-json-me".
251030-02:01:27,301 nipype.workflow INFO:
[Node] Executing "nibabel_read-nii" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,301 nipype.workflow INFO:
[Node] Finished "nibabel_read-nii", elapsed time 0.028565s.
251030-02:01:27,313 nipype.workflow INFO:
[Node] Finished "func_read-json-se", elapsed time 0.015641s.
251030-02:01:27,319 nipype.workflow INFO:
[Node] Executing "fastsurfer_segment-t1" <qsmxt.interfaces.nipype_interface_fastsurfer.FastSurferInterface>
251030-02:01:27,285 nipype.workflow INFO:
[Node] Executing "nibabel_as-canonical" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,191 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-me" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_read-json-me".
251030-02:01:27,336 nipype.workflow INFO:
[Node] Executing "nibabel_as-canonical" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,214 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-se" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_read-json-se".
251030-02:01:27,350 nipype.workflow INFO:
[Node] Executing "func_read-json-me" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,358 nipype.workflow INFO:
[Node] Finished "nibabel_read-nii", elapsed time 0.031016s.
251030-02:01:27,192 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-se" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_read-json-se".
251030-02:01:27,392 nipype.workflow INFO:
[Node] Finished "func_read-json-me", elapsed time 0.025958s.
251030-02:01:27,403 nipype.workflow INFO:
[Node] Executing "func_read-json-se" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,416 nipype.workflow INFO:
[Node] Finished "func_read-json-se", elapsed time 0.009522s.
251030-02:01:27,285 nipype.workflow INFO:
[Node] Executing "func_read-json-me" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,390 nipype.workflow INFO:
[Node] Executing "func_read-json-se" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,392 nipype.workflow INFO:
[Node] Executing "func_read-json-me" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:27,457 nipype.workflow INFO:
[Node] Finished "func_read-json-me", elapsed time 0.008337s.
251030-02:01:27,458 nipype.workflow INFO:
[Node] Finished "func_read-json-me", elapsed time 0.007277s.
251030-02:01:27,457 nipype.workflow INFO:
[Node] Finished "func_read-json-se", elapsed time 0.007988s.
251030-02:01:27,945 nipype.workflow INFO:
[Node] Finished "nibabel_as-canonical", elapsed time 0.594592s.
251030-02:01:27,946 nipype.workflow INFO:
[Node] Finished "nibabel_as-canonical", elapsed time 0.608775s.
251030-02:01:28,408 nipype.workflow INFO:
[Job 0] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-me).
251030-02:01:28,412 nipype.workflow INFO:
[Job 1] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-se).
251030-02:01:28,413 nipype.workflow INFO:
[Job 2] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_read-nii).
251030-02:01:28,415 nipype.workflow INFO:
[Job 3] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_as-canonical).
251030-02:01:28,416 nipype.workflow INFO:
[Job 6] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-me).
251030-02:01:28,417 nipype.workflow INFO:
[Job 7] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-se).
251030-02:01:28,419 nipype.workflow INFO:
[Job 8] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_read-nii).
251030-02:01:28,420 nipype.workflow INFO:
[Job 9] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_as-canonical).
251030-02:01:28,422 nipype.workflow INFO:
[Job 12] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-me).
251030-02:01:28,423 nipype.workflow INFO:
[Job 13] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_read-json-se).
251030-02:01:28,426 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 14 jobs ready. Free memory (GB): 187.48/219.48, Free processors: 10/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:28,647 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_as-canonical" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_as-canonical".
251030-02:01:28,655 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-se" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_read-json-se".
251030-02:01:28,656 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-me" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_read-json-me".
251030-02:01:28,657 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/ants_register-t1-to-qsm".
251030-02:01:28,694 nipype.workflow INFO:
[Node] Executing "nibabel_as-canonical" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:28,701 nipype.workflow INFO:
[Node] Executing "func_read-json-se" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:28,655 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_read-nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_read-nii".
251030-02:01:28,721 nipype.workflow INFO:
[Node] Executing "ants_register-t1-to-qsm" <nipype.interfaces.ants.registration.RegistrationSynQuick>
251030-02:01:28,729 nipype.workflow INFO:
[Node] Finished "func_read-json-se", elapsed time 0.017923s.
251030-02:01:28,701 nipype.workflow INFO:
[Node] Executing "func_read-json-me" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:28,752 nipype.workflow INFO:
[Node] Executing "nibabel_read-nii" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:28,780 nipype.workflow INFO:
[Node] Finished "nibabel_as-canonical", elapsed time 0.066453s.
251030-02:01:28,794 nipype.workflow INFO:
[Node] Finished "func_read-json-me", elapsed time 0.014706s.
251030-02:01:28,822 nipype.workflow INFO:
[Node] Finished "nibabel_read-nii", elapsed time 0.044388s.
251030-02:01:30,408 nipype.workflow INFO:
[Job 14] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_read-nii).
251030-02:01:30,410 nipype.workflow INFO:
[Job 15] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_as-canonical).
251030-02:01:30,411 nipype.workflow INFO:
[Job 18] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-me).
251030-02:01:30,413 nipype.workflow INFO:
[Job 19] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_read-json-se).
251030-02:01:30,415 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 10 jobs ready. Free memory (GB): 179.48/219.48, Free processors: 4/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:30,499 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_as-canonical" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_as-canonical".
251030-02:01:30,500 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_getfirst-canonical-magnitude" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_getfirst-canonical-magnitude".
251030-02:01:30,513 nipype.workflow INFO:
[Node] Executing "nibabel_as-canonical" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:30,515 nipype.workflow INFO:
[Node] Executing "func_getfirst-canonical-magnitude" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:30,517 nipype.workflow INFO:
[Node] Finished "func_getfirst-canonical-magnitude", elapsed time 0.000813s.
251030-02:01:30,524 nipype.workflow INFO:
[Node] Finished "nibabel_as-canonical", elapsed time 0.008307s.
251030-02:01:30,499 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_read-nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_read-nii".
251030-02:01:30,500 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_scale-phase".
251030-02:01:30,556 nipype.workflow INFO:
[Node] Executing "nibabel_read-nii" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:30,558 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_scale-phase" <qsmxt.interfaces.nipype_interface_processphase.ScalePhaseInterface>
251030-02:01:30,563 nipype.workflow INFO:
[Node] Finished "nibabel_read-nii", elapsed time 0.005143s.
251030-02:01:32,424 nipype.workflow INFO:
[Job 20] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_read-nii).
251030-02:01:32,431 nipype.workflow INFO:
[Job 21] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_as-canonical).
251030-02:01:32,433 nipype.workflow INFO:
[Job 24] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_getfirst-canonical-magnitude).
251030-02:01:32,435 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 8 jobs ready. Free memory (GB): 177.48/219.48, Free processors: 3/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:32,584 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_getfirst-canonical-magnitude" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_getfirst-canonical-magnitude".
251030-02:01:32,593 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_scale-phase".
251030-02:01:32,592 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_getfirst-canonical-magnitude" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/func_getfirst-canonical-magnitude".
251030-02:01:32,667 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_scale-phase" <qsmxt.interfaces.nipype_interface_processphase.ScalePhaseInterface>
251030-02:01:32,668 nipype.workflow INFO:
[Node] Executing "func_getfirst-canonical-magnitude" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:32,677 nipype.workflow INFO:
[Node] Finished "func_getfirst-canonical-magnitude", elapsed time 0.001213s.
251030-02:01:32,686 nipype.workflow INFO:
[Node] Executing "func_getfirst-canonical-magnitude" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:32,701 nipype.workflow INFO:
[Node] Finished "func_getfirst-canonical-magnitude", elapsed time 0.006979s.
251030-02:01:33,267 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_scale-phase", elapsed time 2.706454s.
251030-02:01:34,420 nipype.workflow INFO:
[Job 26] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase).
251030-02:01:34,428 nipype.workflow INFO:
[Job 27] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_getfirst-canonical-magnitude).
251030-02:01:34,429 nipype.workflow INFO:
[Job 29] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.func_getfirst-canonical-magnitude).
251030-02:01:34,436 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 6 jobs ready. Free memory (GB): 177.48/219.48, Free processors: 3/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:34,564 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_scale-phase", elapsed time 1.889243s.
251030-02:01:34,582 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_axial-resampling" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_axial-resampling".
251030-02:01:34,589 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_axial-resampling" <qsmxt.interfaces.nipype_interface_axialsampling.AxialSamplingInterface>
251030-02:01:34,596 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_axial-resampling", elapsed time 0.004848s.
251030-02:01:34,582 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_getfirst-canonical-magnitude" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/func_getfirst-canonical-magnitude".
251030-02:01:34,639 nipype.workflow INFO:
[Node] Executing "func_getfirst-canonical-magnitude" <nipype.interfaces.utility.wrappers.Function>
251030-02:01:34,641 nipype.workflow INFO:
[Node] Finished "func_getfirst-canonical-magnitude", elapsed time 0.00079s.
251030-02:01:36,424 nipype.workflow INFO:
[Job 31] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_scale-phase).
251030-02:01:36,425 nipype.workflow INFO:
[Job 32] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.func_getfirst-canonical-magnitude).
251030-02:01:36,427 nipype.workflow INFO:
[Job 36] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_axial-resampling).
251030-02:01:36,430 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 7 jobs ready. Free memory (GB): 179.48/219.48, Free processors: 4/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:36,515 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_axial-resampling" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_axial-resampling".
251030-02:01:36,517 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/mrt_romeo".
251030-02:01:36,523 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_axial-resampling" <qsmxt.interfaces.nipype_interface_axialsampling.AxialSamplingInterface>
251030-02:01:36,525 nipype.workflow INFO:
[Node] Executing "mrt_romeo" <qsmxt.interfaces.nipype_interface_romeo.RomeoB0Interface>
251030-02:01:36,530 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_axial-resampling", elapsed time 0.004856s.
251030-02:01:38,423 nipype.workflow INFO:
[Job 41] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_axial-resampling).
251030-02:01:38,425 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 7 jobs ready. Free memory (GB): 177.15/219.48, Free processors: 3/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:01:38,557 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/mrt_romeo".
251030-02:01:38,643 nipype.workflow INFO:
[Node] Executing "mrt_romeo" <qsmxt.interfaces.nipype_interface_romeo.RomeoB0Interface>
251030-02:01:40,425 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 6 jobs ready. Free memory (GB): 174.81/219.48, Free processors: 2/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:04:27,72 nipype.workflow INFO:
[Node] Finished "ants_register-t1-to-qsm", elapsed time 178.333576s.
251030-02:04:28,679 nipype.workflow INFO:
[Job 11] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm).
251030-02:04:28,682 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 6 jobs ready. Free memory (GB): 182.81/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:04:28,775 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/fastsurfer_segment-t1".
251030-02:04:28,789 nipype.workflow INFO:
[Node] Executing "fastsurfer_segment-t1" <qsmxt.interfaces.nipype_interface_fastsurfer.FastSurferInterface>
251030-02:04:30,683 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 5 jobs ready. Free memory (GB): 170.81/219.48, Free processors: 0/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:09:33,926 nipype.workflow INFO:
[Node] Finished "ants_register-t1-to-qsm", elapsed time 486.632802s.
251030-02:09:35,496 nipype.workflow INFO:
[Job 5] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm).
251030-02:09:35,509 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 5 jobs ready. Free memory (GB): 178.81/219.48, Free processors: 6/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:09:35,719 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/ants_register-t1-to-qsm".
251030-02:09:35,802 nipype.workflow INFO:
[Node] Executing "ants_register-t1-to-qsm" <nipype.interfaces.ants.registration.RegistrationSynQuick>
251030-02:09:37,499 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 4 jobs ready. Free memory (GB): 170.81/219.48, Free processors: 0/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:12:37,199 nipype.workflow INFO:
[Node] Finished "mrt_romeo", elapsed time 658.543122s.
251030-02:12:37,206 nipype.workflow INFO:
[Node] Finished "mrt_romeo", elapsed time 660.680345s.
251030-02:12:37,838 nipype.workflow INFO:
[Job 46] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo).
251030-02:12:37,850 nipype.workflow INFO:
[Job 50] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.mrt_romeo).
251030-02:12:37,851 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 6 jobs ready. Free memory (GB): 175.48/219.48, Free processors: 2/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:12:38,84 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.nibabel-numpy_normalize-phase" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/nibabel-numpy_normalize-phase".
251030-02:12:38,246 nipype.workflow INFO:
[Node] Executing "nibabel-numpy_normalize-phase" <qsmxt.interfaces.nipype_interface_processphase.PhaseToNormalizedInterface>
251030-02:12:39,208 nipype.workflow INFO:
[Node] Finished "nibabel-numpy_normalize-phase", elapsed time 0.953909s.
251030-02:12:39,840 nipype.workflow INFO:
[Job 53] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.nibabel-numpy_normalize-phase).
251030-02:12:39,842 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 5 jobs ready. Free memory (GB): 175.48/219.48, Free processors: 2/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:12:39,935 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.nibabel-numpy_normalize-phase" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/nibabel-numpy_normalize-phase".
251030-02:12:39,963 nipype.workflow INFO:
[Node] Executing "nibabel-numpy_normalize-phase" <qsmxt.interfaces.nipype_interface_processphase.PhaseToNormalizedInterface>
251030-02:12:40,144 nipype.workflow INFO:
[Node] Finished "nibabel-numpy_normalize-phase", elapsed time 0.179565s.
251030-02:12:41,845 nipype.workflow INFO:
[Job 55] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.nibabel-numpy_normalize-phase).
251030-02:12:41,847 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 4 jobs ready. Free memory (GB): 175.48/219.48, Free processors: 2/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:16:47,604 nipype.workflow INFO:
[Node] Finished "ants_register-t1-to-qsm", elapsed time 431.799179s.
251030-02:16:50,358 nipype.workflow INFO:
[Job 17] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_register-t1-to-qsm).
251030-02:16:50,364 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 4 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:16:50,595 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/fastsurfer_segment-t1".
251030-02:16:50,651 nipype.workflow INFO:
[Node] Executing "fastsurfer_segment-t1" <qsmxt.interfaces.nipype_interface_fastsurfer.FastSurferInterface>
251030-02:16:52,362 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 3 jobs ready. Free memory (GB): 171.48/219.48, Free processors: 0/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:16:57,826 nipype.workflow INFO:
[Node] Finished "fastsurfer_segment-t1", elapsed time 930.488019s.
251030-02:16:57,849 nipype.workflow INFO:
[Node] Finished "fastsurfer_segment-t1", elapsed time 930.553816s.
251030-02:16:58,383 nipype.workflow INFO:
[Job 4] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1).
251030-02:16:58,391 nipype.workflow INFO:
[Job 10] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1).
251030-02:16:58,402 nipype.workflow INFO:
[MultiProc] Running 2 tasks, and 5 jobs ready. Free memory (GB): 195.48/219.48, Free processors: 16/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:16:58,589 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/ants_register-t1-to-qsm".
251030-02:16:58,610 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.numpy_numpy_nibabel_mgz2nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/numpy_numpy_nibabel_mgz2nii".
251030-02:16:58,630 nipype.workflow INFO:
[Node] Executing "ants_register-t1-to-qsm" <nipype.interfaces.ants.registration.RegistrationSynQuick>
251030-02:16:58,638 nipype.workflow INFO:
[Node] Executing "numpy_numpy_nibabel_mgz2nii" <qsmxt.interfaces.nipype_interface_mgz2nii.Mgz2NiiInterface>
251030-02:16:58,602 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/mask_workflow/fsl-bet".
251030-02:16:58,615 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.numpy_numpy_nibabel_mgz2nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/numpy_numpy_nibabel_mgz2nii".
251030-02:16:58,689 nipype.workflow INFO:
[Node] Executing "fsl-bet" <qsmxt.interfaces.nipype_interface_bet2.Bet2Interface>
251030-02:16:58,690 nipype.workflow INFO:
[Node] Executing "numpy_numpy_nibabel_mgz2nii" <qsmxt.interfaces.nipype_interface_mgz2nii.Mgz2NiiInterface>
251030-02:16:59,257 nipype.workflow INFO:
[Node] Finished "numpy_numpy_nibabel_mgz2nii", elapsed time 0.617801s.
251030-02:16:59,259 nipype.workflow INFO:
[Node] Finished "numpy_numpy_nibabel_mgz2nii", elapsed time 0.56043s.
251030-02:17:00,379 nipype.workflow INFO:
[Job 25] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.numpy_numpy_nibabel_mgz2nii).
251030-02:17:00,381 nipype.workflow INFO:
[Job 28] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.numpy_numpy_nibabel_mgz2nii).
251030-02:17:00,385 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 5 jobs ready. Free memory (GB): 185.48/219.48, Free processors: 2/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:00,493 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/ants_transform-segmentation-to-qsm".
251030-02:17:00,500 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_numpy_nilearn_t1w-seg-resampled".
251030-02:17:00,521 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_t1w-seg-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:17:00,526 nipype.workflow INFO:
[Node] Executing "ants_transform-segmentation-to-qsm" <nipype.interfaces.ants.resampling.ApplyTransforms>
251030-02:17:02,390 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 3 jobs ready. Free memory (GB): 181.48/219.48, Free processors: 0/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:04,171 nipype.workflow INFO:
[Node] Finished "fsl-bet", elapsed time 5.480773s.
251030-02:17:04,384 nipype.workflow INFO:
[Job 45] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet).
251030-02:17:04,386 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 4 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:04,475 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_t1w-seg-resampled".
251030-02:17:04,505 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_transform-segmentation-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/ants_transform-segmentation-to-qsm".
251030-02:17:04,522 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_t1w-seg-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:17:04,546 nipype.workflow INFO:
[Node] Executing "ants_transform-segmentation-to-qsm" <nipype.interfaces.ants.resampling.ApplyTransforms>
251030-02:17:04,488 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.scipy_numpy_nibabel_bet_erode" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/mask_workflow/scipy_numpy_nibabel_bet_erode".
251030-02:17:04,566 nipype.workflow INFO:
[Node] Executing "scipy_numpy_nibabel_bet_erode" <qsmxt.interfaces.nipype_interface_erode.ErosionInterface>
251030-02:17:05,557 nipype.workflow INFO:
[Node] Finished "scipy_numpy_nibabel_bet_erode", elapsed time 0.989792s.
251030-02:17:06,401 nipype.workflow INFO:
[Job 52] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.scipy_numpy_nibabel_bet_erode).
251030-02:17:06,422 nipype.workflow INFO:
[MultiProc] Running 7 tasks, and 2 jobs ready. Free memory (GB): 179.48/219.48, Free processors: 6/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_transform-segmentation-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:06,598 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/qsmjl_vsharp".
251030-02:17:06,672 nipype.workflow INFO:
[Node] Executing "qsmjl_vsharp" <qsmxt.interfaces.nipype_interface_qsmjl.VsharpInterface>
251030-02:17:06,848 nipype.workflow INFO:
[Node] Finished "ants_transform-segmentation-to-qsm", elapsed time 2.300818s.
251030-02:17:08,301 nipype.workflow INFO:
[Node] Finished "ants_transform-segmentation-to-qsm", elapsed time 7.760466s.
251030-02:17:08,396 nipype.workflow INFO:
[Job 35] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm).
251030-02:17:08,398 nipype.workflow INFO:
[Job 38] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_transform-segmentation-to-qsm).
251030-02:17:08,400 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 2 jobs ready. Free memory (GB): 181.48/219.48, Free processors: 6/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:08,506 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists2" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/combine_lists2".
251030-02:17:08,539 nipype.workflow INFO:
[Node] Executing "combine_lists2" <nipype.interfaces.utility.wrappers.Function>
251030-02:17:08,555 nipype.workflow INFO:
[Node] Finished "combine_lists2", elapsed time 0.000945s.
251030-02:17:08,610 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_t1w-seg-resampled", elapsed time 8.080448s.
251030-02:17:10,405 nipype.workflow INFO:
[Job 34] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled).
251030-02:17:10,411 nipype.workflow INFO:
[Job 47] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists2).
251030-02:17:10,413 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 2 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 7/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:10,674 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.copyfile" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/copyfile".
251030-02:17:10,723 nipype.workflow INFO:
[Node] Executing "copyfile" <qsmxt.interfaces.nipype_interface_copyfile.DynamicCopyFiles>
251030-02:17:11,575 nipype.workflow INFO:
[Node] Finished "copyfile", elapsed time 0.848245s.
251030-02:17:12,400 nipype.workflow INFO:
[Job 44] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.copyfile).
251030-02:17:12,403 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 1 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 7/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:12,632 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_t1w-seg-resampled", elapsed time 8.108064s.
251030-02:17:14,407 nipype.workflow INFO:
[Job 37] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled).
251030-02:17:14,409 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 185.48/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:14,637 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/mask_workflow/fsl-bet".
251030-02:17:14,806 nipype.workflow INFO:
[Node] Executing "fsl-bet" <qsmxt.interfaces.nipype_interface_bet2.Bet2Interface>
251030-02:17:16,433 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 0/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:26,117 nipype.workflow INFO:
[Node] Finished "fsl-bet", elapsed time 11.288238s.
251030-02:17:26,443 nipype.workflow INFO:
[Job 49] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.fsl-bet).
251030-02:17:26,452 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 185.48/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:26,758 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.scipy_numpy_nibabel_bet_erode" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/mask_workflow/scipy_numpy_nibabel_bet_erode".
251030-02:17:26,908 nipype.workflow INFO:
[Node] Executing "scipy_numpy_nibabel_bet_erode" <qsmxt.interfaces.nipype_interface_erode.ErosionInterface>
251030-02:17:28,446 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 7/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.scipy_numpy_nibabel_bet_erode
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:29,151 nipype.workflow INFO:
[Node] Finished "scipy_numpy_nibabel_bet_erode", elapsed time 2.2210739999999998s.
251030-02:17:30,447 nipype.workflow INFO:
[Job 54] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.mask_workflow.scipy_numpy_nibabel_bet_erode).
251030-02:17:30,450 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 185.48/219.48, Free processors: 8/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:30,544 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/qsmjl_vsharp".
251030-02:17:30,553 nipype.workflow INFO:
[Node] Executing "qsmjl_vsharp" <qsmxt.interfaces.nipype_interface_qsmjl.VsharpInterface>
251030-02:17:32,451 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 183.48/219.48, Free processors: 6/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1
251030-02:17:41,342 nipype.workflow INFO:
[Node] Finished "fastsurfer_segment-t1", elapsed time 792.549331s.
251030-02:17:42,460 nipype.workflow INFO:
[Job 16] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.fastsurfer_segment-t1).
251030-02:17:42,468 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 195.48/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:17:42,575 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.numpy_numpy_nibabel_mgz2nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/numpy_numpy_nibabel_mgz2nii".
251030-02:17:42,596 nipype.workflow INFO:
[Node] Executing "numpy_numpy_nibabel_mgz2nii" <qsmxt.interfaces.nipype_interface_mgz2nii.Mgz2NiiInterface>
251030-02:17:42,924 nipype.workflow INFO:
[Node] Finished "numpy_numpy_nibabel_mgz2nii", elapsed time 0.325727s.
251030-02:17:44,461 nipype.workflow INFO:
[Job 30] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.numpy_numpy_nibabel_mgz2nii).
251030-02:17:44,463 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 2 jobs ready. Free memory (GB): 195.48/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:17:44,575 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/ants_transform-segmentation-to-qsm".
251030-02:17:44,576 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/nibabel_numpy_nilearn_t1w-seg-resampled".
251030-02:17:44,598 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_t1w-seg-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:17:44,600 nipype.workflow INFO:
[Node] Executing "ants_transform-segmentation-to-qsm" <nipype.interfaces.ants.resampling.ApplyTransforms>
251030-02:17:46,464 nipype.workflow INFO:
[MultiProc] Running 6 tasks, and 0 jobs ready. Free memory (GB): 191.48/219.48, Free processors: 12/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:17:47,662 nipype.workflow INFO:
[Node] Finished "ants_transform-segmentation-to-qsm", elapsed time 3.060115s.
251030-02:17:48,463 nipype.workflow INFO:
[Job 40] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.ants_transform-segmentation-to-qsm).
251030-02:17:48,477 nipype.workflow INFO:
[MultiProc] Running 5 tasks, and 0 jobs ready. Free memory (GB): 193.48/219.48, Free processors: 13/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:17:51,517 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_t1w-seg-resampled", elapsed time 6.916225s.
251030-02:17:52,467 nipype.workflow INFO:
[Job 39] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.nibabel_numpy_nilearn_t1w-seg-resampled).
251030-02:17:52,469 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 1 jobs ready. Free memory (GB): 195.48/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:17:52,553 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.copyfile" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqmp2ragehighres0p5isoslab_T1w/copyfile".
251030-02:17:52,613 nipype.workflow INFO:
[Node] Executing "copyfile" <qsmxt.interfaces.nipype_interface_copyfile.DynamicCopyFiles>
251030-02:17:53,530 nipype.workflow INFO:
[Node] Finished "copyfile", elapsed time 0.907036s.
251030-02:17:54,469 nipype.workflow INFO:
[Job 48] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqmp2ragehighres0p5isoslab_T1w.copyfile).
251030-02:17:54,471 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 0 jobs ready. Free memory (GB): 195.48/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:18:57,672 nipype.workflow INFO:
[Node] Finished "qsmjl_vsharp", elapsed time 110.992297s.
251030-02:18:58,580 nipype.workflow INFO:
[Job 56] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp).
251030-02:18:58,583 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 1 jobs ready. Free memory (GB): 197.48/219.48, Free processors: 16/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:18:58,695 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/qsmjl_rts".
251030-02:18:58,742 nipype.workflow INFO:
[Node] Executing "qsmjl_rts" <qsmxt.interfaces.nipype_interface_qsmjl.RtsQsmInterface>
251030-02:19:00,582 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 0 jobs ready. Free memory (GB): 195.08/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:07,509 nipype.workflow INFO:
[Node] Finished "qsmjl_vsharp", elapsed time 96.954908s.
251030-02:19:08,589 nipype.workflow INFO:
[Job 57] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_vsharp).
251030-02:19:08,591 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 1 jobs ready. Free memory (GB): 197.08/219.48, Free processors: 16/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:08,694 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/qsm_workflow/qsmjl_rts".
251030-02:19:08,730 nipype.workflow INFO:
[Node] Executing "qsmjl_rts" <qsmxt.interfaces.nipype_interface_qsmjl.RtsQsmInterface>
251030-02:19:10,593 nipype.workflow INFO:
[MultiProc] Running 4 tasks, and 0 jobs ready. Free memory (GB): 194.67/219.48, Free processors: 14/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:14,22 nipype.workflow INFO:
[Node] Finished "ants_register-t1-to-qsm", elapsed time 135.383104s.
251030-02:19:14,597 nipype.workflow INFO:
[Job 23] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_register-t1-to-qsm).
251030-02:19:14,600 nipype.workflow INFO:
[MultiProc] Running 3 tasks, and 0 jobs ready. Free memory (GB): 202.67/219.48, Free processors: 20/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:47,482 nipype.workflow INFO:
[Node] Finished "qsmjl_rts", elapsed time 48.738805s.
251030-02:19:48,106 nipype.workflow INFO:
[Node] Finished "qsmjl_rts", elapsed time 39.368049s.
251030-02:19:48,638 nipype.workflow INFO:
[Job 58] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts).
251030-02:19:48,641 nipype.workflow INFO:
[Job 59] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.qsm_workflow.qsmjl_rts).
251030-02:19:48,645 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 2 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:48,734 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-average" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_qsm-average".
251030-02:19:48,771 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_qsm-average" <qsmxt.interfaces.nipype_interface_nonzeroaverage.NonzeroAverageInterface>
251030-02:19:48,772 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_qsm-average", elapsed time 0.000347s.
251030-02:19:48,734 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-average" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_qsm-average".
251030-02:19:48,799 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_qsm-average" <qsmxt.interfaces.nipype_interface_nonzeroaverage.NonzeroAverageInterface>
251030-02:19:48,801 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_qsm-average", elapsed time 0.00041s.
251030-02:19:50,637 nipype.workflow INFO:
[Job 60] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-average).
251030-02:19:50,639 nipype.workflow INFO:
[Job 61] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-average).
251030-02:19:50,640 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 2 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:50,732 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_qsm-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_qsm-resampled".
251030-02:19:50,741 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_qsm-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_qsm-resampled".
251030-02:19:50,777 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_qsm-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:19:50,780 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_qsm-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:19:50,782 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_qsm-resampled", elapsed time 0.003792s.
251030-02:19:50,785 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_qsm-resampled", elapsed time 0.00363s.
251030-02:19:52,639 nipype.workflow INFO:
[Job 62] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_qsm-resampled).
251030-02:19:52,641 nipype.workflow INFO:
[Job 63] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_qsm-resampled).
251030-02:19:52,642 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 4 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:52,716 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-referenced" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_qsm-referenced".
251030-02:19:52,738 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/combine_lists1".
251030-02:19:52,745 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists1" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/combine_lists1".
251030-02:19:52,762 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_qsm-referenced" <qsmxt.interfaces.nipype_interface_qsm_referencing.ReferenceQSMInterface>
251030-02:19:52,778 nipype.workflow INFO:
[Node] Executing "combine_lists1" <nipype.interfaces.utility.wrappers.Function>
251030-02:19:52,780 nipype.workflow INFO:
[Node] Executing "combine_lists1" <nipype.interfaces.utility.wrappers.Function>
251030-02:19:52,788 nipype.workflow INFO:
[Node] Finished "combine_lists1", elapsed time 0.007968s.
251030-02:19:52,789 nipype.workflow INFO:
[Node] Finished "combine_lists1", elapsed time 0.000789s.
251030-02:19:52,738 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-referenced" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_qsm-referenced".
251030-02:19:52,801 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_qsm-referenced" <qsmxt.interfaces.nipype_interface_qsm_referencing.ReferenceQSMInterface>
251030-02:19:53,925 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_qsm-referenced", elapsed time 1.161757s.
251030-02:19:53,934 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_qsm-referenced", elapsed time 1.13148s.
251030-02:19:54,643 nipype.workflow INFO:
[Job 64] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-referenced).
251030-02:19:54,645 nipype.workflow INFO:
[Job 65] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists1).
251030-02:19:54,647 nipype.workflow INFO:
[Job 66] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_qsm-referenced).
251030-02:19:54,648 nipype.workflow INFO:
[Job 67] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists1).
251030-02:19:54,649 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 3 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:54,769 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copy_qsm_json_sidecar" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/copy_qsm_json_sidecar".
251030-02:19:54,773 nipype.workflow INFO:
[Node] Executing "copy_qsm_json_sidecar" <qsmxt.interfaces.nipype_interface_copy_json_sidecar.CopyJsonSidecarInterface>
251030-02:19:54,775 nipype.workflow INFO:
[Node] Finished "copy_qsm_json_sidecar", elapsed time 0.000943s.
251030-02:19:54,785 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copy_qsm_json_sidecar" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/copy_qsm_json_sidecar".
251030-02:19:54,790 nipype.workflow INFO:
[Node] Executing "copy_qsm_json_sidecar" <qsmxt.interfaces.nipype_interface_copy_json_sidecar.CopyJsonSidecarInterface>
251030-02:19:54,802 nipype.workflow INFO:
[Node] Finished "copy_qsm_json_sidecar", elapsed time 0.010119s.
251030-02:19:54,769 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.create_permutations" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/create_permutations".
251030-02:19:54,823 nipype.workflow INFO:
[Node] Executing "create_permutations" <nipype.interfaces.utility.wrappers.Function>
251030-02:19:54,831 nipype.workflow INFO:
[Node] Finished "create_permutations", elapsed time 0.007179s.
251030-02:19:56,643 nipype.workflow INFO:
[Job 68] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copy_qsm_json_sidecar).
251030-02:19:56,645 nipype.workflow INFO:
[Job 69] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.create_permutations).
251030-02:19:56,646 nipype.workflow INFO:
[Job 70] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copy_qsm_json_sidecar).
251030-02:19:56,647 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 1 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:19:56,739 nipype.workflow INFO:
[Node] Setting-up "_nibabel_numpy_analyse-qsm0" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_analyse-qsm/mapflow/_nibabel_numpy_analyse-qsm0".
251030-02:19:56,741 nipype.workflow INFO:
[Node] Executing "_nibabel_numpy_analyse-qsm0" <qsmxt.interfaces.nipype_interface_analyse.AnalyseInterface>
251030-02:19:58,646 nipype.workflow INFO:
[MultiProc] Running 2 tasks, and 0 jobs ready. Free memory (GB): 205.48/219.48, Free processors: 23/32.
Currently running:
* qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_analyse-qsm
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:21:27,616 nipype.workflow INFO:
[Node] Finished "_nibabel_numpy_analyse-qsm0", elapsed time 90.873416s.
251030-02:21:28,770 nipype.workflow INFO:
[Job 72] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_analyse-qsm).
251030-02:21:28,773 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 1 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:21:28,882 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copyfile" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-1/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/copyfile".
251030-02:21:28,906 nipype.workflow INFO:
[Node] Executing "copyfile" <qsmxt.interfaces.nipype_interface_copyfile.DynamicCopyFiles>
251030-02:21:29,472 nipype.workflow INFO:
[Node] Finished "copyfile", elapsed time 0.564569s.
251030-02:21:30,771 nipype.workflow INFO:
[Job 74] Completed (qsmxt-workflow.sub-1.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copyfile).
251030-02:21:30,773 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 207.48/219.48, Free processors: 24/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1
251030-02:23:26,750 nipype.workflow INFO:
[Node] Finished "fastsurfer_segment-t1", elapsed time 396.090615s.
251030-02:23:26,905 nipype.workflow INFO:
[Job 22] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.fastsurfer_segment-t1).
251030-02:23:26,907 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
251030-02:23:26,986 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.numpy_numpy_nibabel_mgz2nii" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/numpy_numpy_nibabel_mgz2nii".
251030-02:23:26,997 nipype.workflow INFO:
[Node] Executing "numpy_numpy_nibabel_mgz2nii" <qsmxt.interfaces.nipype_interface_mgz2nii.Mgz2NiiInterface>
251030-02:23:27,393 nipype.workflow INFO:
[Node] Finished "numpy_numpy_nibabel_mgz2nii", elapsed time 0.394689s.
251030-02:23:28,907 nipype.workflow INFO:
[Job 33] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.numpy_numpy_nibabel_mgz2nii).
251030-02:23:28,909 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 2 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
251030-02:23:28,988 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_nilearn_t1w-seg-resampled".
251030-02:23:29,6 nipype.workflow INFO:
[Node] Executing "nibabel_numpy_nilearn_t1w-seg-resampled" <qsmxt.interfaces.nipype_interface_resample_like.ResampleLikeInterface>
251030-02:23:28,998 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_transform-segmentation-to-qsm" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/ants_transform-segmentation-to-qsm".
251030-02:23:29,24 nipype.workflow INFO:
[Node] Executing "ants_transform-segmentation-to-qsm" <nipype.interfaces.ants.resampling.ApplyTransforms>
251030-02:23:30,449 nipype.workflow INFO:
[Node] Finished "ants_transform-segmentation-to-qsm", elapsed time 1.414784s.
251030-02:23:30,909 nipype.workflow INFO:
[Job 43] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.ants_transform-segmentation-to-qsm).
251030-02:23:30,911 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 1 jobs ready. Free memory (GB): 217.48/219.48, Free processors: 31/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
251030-02:23:30,987 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists2" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/combine_lists2".
251030-02:23:30,992 nipype.workflow INFO:
[Node] Executing "combine_lists2" <nipype.interfaces.utility.wrappers.Function>
251030-02:23:31,0 nipype.workflow INFO:
[Node] Finished "combine_lists2", elapsed time 0.000689s.
251030-02:23:32,911 nipype.workflow INFO:
[Job 51] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.combine_lists2).
251030-02:23:32,913 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 1 jobs ready. Free memory (GB): 217.48/219.48, Free processors: 31/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled
251030-02:23:32,983 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.create_permutations" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/create_permutations".
251030-02:23:32,987 nipype.workflow INFO:
[Node] Executing "create_permutations" <nipype.interfaces.utility.wrappers.Function>
251030-02:23:32,989 nipype.workflow INFO:
[Node] Finished "create_permutations", elapsed time 0.000541s.
251030-02:23:34,366 nipype.workflow INFO:
[Node] Finished "nibabel_numpy_nilearn_t1w-seg-resampled", elapsed time 5.354482s.
251030-02:23:34,914 nipype.workflow INFO:
[Job 42] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_nilearn_t1w-seg-resampled).
251030-02:23:34,915 nipype.workflow INFO:
[Job 71] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.create_permutations).
251030-02:23:34,916 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
251030-02:23:34,990 nipype.workflow INFO:
[Node] Setting-up "_nibabel_numpy_analyse-qsm0" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/nibabel_numpy_analyse-qsm/mapflow/_nibabel_numpy_analyse-qsm0".
251030-02:23:34,992 nipype.workflow INFO:
[Node] Executing "_nibabel_numpy_analyse-qsm0" <qsmxt.interfaces.nipype_interface_analyse.AnalyseInterface>
251030-02:23:36,916 nipype.workflow INFO:
[MultiProc] Running 1 tasks, and 0 jobs ready. Free memory (GB): 217.48/219.48, Free processors: 31/32.
Currently running:
* qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_analyse-qsm
251030-02:24:46,223 nipype.workflow INFO:
[Node] Finished "_nibabel_numpy_analyse-qsm0", elapsed time 71.229384s.
251030-02:24:46,989 nipype.workflow INFO:
[Job 73] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.nibabel_numpy_analyse-qsm).
251030-02:24:46,991 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 1 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
251030-02:24:47,75 nipype.workflow INFO:
[Node] Setting-up "qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copyfile" in "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/bids/derivatives/workflow/qsmxt-workflow/sub-2/ses-20170705/qsm_acq-acqqsmp21mmisote20_T2starw/copyfile".
251030-02:24:47,91 nipype.workflow INFO:
[Node] Executing "copyfile" <qsmxt.interfaces.nipype_interface_copyfile.DynamicCopyFiles>
251030-02:24:47,878 nipype.workflow INFO:
[Node] Finished "copyfile", elapsed time 0.785195s.
251030-02:24:48,991 nipype.workflow INFO:
[Job 75] Completed (qsmxt-workflow.sub-2.ses-20170705.qsm_acq-acqqsmp21mmisote20_T2starw.copyfile).
251030-02:24:48,993 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 0 jobs ready. Free memory (GB): 219.48/219.48, Free processors: 32/32.
[INFO]: Warnings occurred!
[INFO]: Finished
View results#
Let’s have a look at the generated qsm folder:
!tree bids/derivatives/qsmxt-*-* --dirsfirst
bids/derivatives/qsmxt-2025-10-30-020038
├── sub-1
│ └── ses-20170705
│ ├── anat
│ │ ├── sub-1_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_space-orig_dseg.nii
│ │ ├── sub-1_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_space-qsm_dseg.nii
│ │ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_Chimap.json
│ │ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_Chimap.nii
│ │ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_space-orig_dseg.nii
│ │ └── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_space-qsm_dseg.nii
│ └── extra_data
│ ├── sub-1_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_desc-t1w-to-qsm_transform.mat
│ ├── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_desc-t1w-to-qsm_transform.mat
│ └── sub-1_ses-20170705_acq-acqqsmp21mmisote20_T2starw_qsm-analysis.csv
├── sub-2
│ └── ses-20170705
│ ├── anat
│ │ ├── sub-2_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_space-orig_dseg.nii
│ │ ├── sub-2_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_space-qsm_dseg.nii
│ │ ├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_Chimap.json
│ │ ├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_Chimap.nii
│ │ ├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_space-orig_dseg.nii
│ │ └── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_space-qsm_dseg.nii
│ └── extra_data
│ ├── sub-2_ses-20170705_acq-acqmp2ragehighres0p5isoslab_T1w_desc-t1w-to-qsm_transform.mat
│ ├── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_desc-t1w-to-qsm_transform.mat
│ └── sub-2_ses-20170705_acq-acqqsmp21mmisote20_T2starw_qsm-analysis.csv
├── command.txt
├── pypeline.log
├── qsmxt.log
├── references.txt
└── settings.json
8 directories, 23 files
The references.txt file contains a list of all the algorithms used and relevant citations:
!cat bids/derivatives/qsmxt-*-*/references.txt
== References ==
- QSMxT: Stewart AW, Robinson SD, O'Brien K, et al. QSMxT: Robust masking and artifact reduction for quantitative susceptibility mapping. Magnetic Resonance in Medicine. 2022;87(3):1289-1300. doi:10.1002/mrm.29048
- QSMxT: Stewart AW, Bollman S, et al. QSMxT/QSMxT. GitHub; 2022. https://github.com/QSMxT/QSMxT
- Python package - Nipype: Gorgolewski K, Burns C, Madison C, et al. Nipype: A Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python. Frontiers in Neuroinformatics. 2011;5. Accessed April 20, 2022. doi:10.3389/fninf.2011.00013
- Brain extraction: Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143-155. doi:10.1002/hbm.10062
- Brain extraction: Liangfu Chen. liangfu/bet2 - Standalone Brain Extraction Tool. GitHub; 2015. https://github.com/liangfu/bet2
- Unwrapping algorithm - ROMEO: Dymerska B, Eckstein K, Bachrata B, et al. Phase unwrapping with a rapid opensource minimum spanning tree algorithm (ROMEO). Magnetic Resonance in Medicine. 2021;85(4):2294-2308. doi:10.1002/mrm.28563
- Background field removal - V-SHARP: Wu B, Li W, Guidon A et al. Whole brain susceptibility mapping using compressed sensing. Magnetic resonance in medicine. 2012 Jan;67(1):137-47. doi:10.1002/mrm.23000
- QSM algorithm - RTS: Kames C, Wiggermann V, Rauscher A. Rapid two-step dipole inversion for susceptibility mapping with sparsity priors. Neuroimage. 2018 Feb 15;167:276-83. doi:10.1016/j.neuroimage.2017.11.018
- Julia package - QSM.jl: kamesy. GitHub; 2022. https://github.com/kamesy/QSM.jl
- Julia package - MriResearchTools: Eckstein K. korbinian90/MriResearchTools.jl. GitHub; 2022. https://github.com/korbinian90/MriResearchTools.jl
- Python package - nibabel: Brett M, Markiewicz CJ, Hanke M, et al. nipy/nibabel. GitHub; 2019. https://github.com/nipy/nibabel
- Python package - scipy: Virtanen P, Gommers R, Oliphant TE, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17(3):261-272. doi:10.1038/s41592-019-0686-2
- Python package - numpy: Harris CR, Millman KJ, van der Walt SJ, et al. Array programming with NumPy. Nature. 2020;585(7825):357-362. doi:10.1038/s41586-020-2649-2
- FastSurfer: Henschel L, Conjeti S, Estrada S, Diers K, Fischl B, Reuter M. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. NeuroImage. 2020;219:117012. doi:10.1016/j.neuroimage.2020.117012
- ANTs: Avants BB, Tustison NJ, Johnson HJ. Advanced Normalization Tools. GitHub; 2022. https://github.com/ANTsX/ANTs
Let’s view one of the QSM results:
QSM results#
show_nii(glob("bids/derivatives/qsmxt-*/sub-*/ses-*/anat/*Chimap.nii*")[0], title="Chimap", figsize=(10,8), cmap='gray', vmin=-0.1, vmax=0.1)
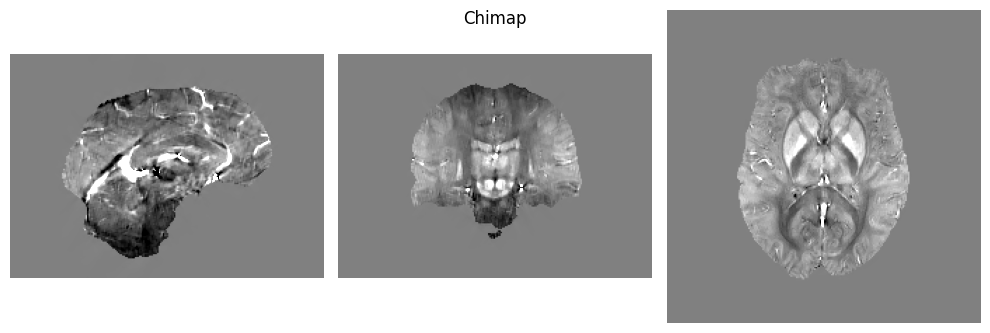
Segmentations#
Segmentations are generated in both the QSM space and the T1-weighted space. Transformations are also made available.
show_nii(glob("bids/derivatives/qsmxt-*/sub-*/ses-*/anat/*qsm_dseg.nii*")[0], title="Segmentation", figsize=(10,8), cmap='Set3', vmin=0, vmax=96)
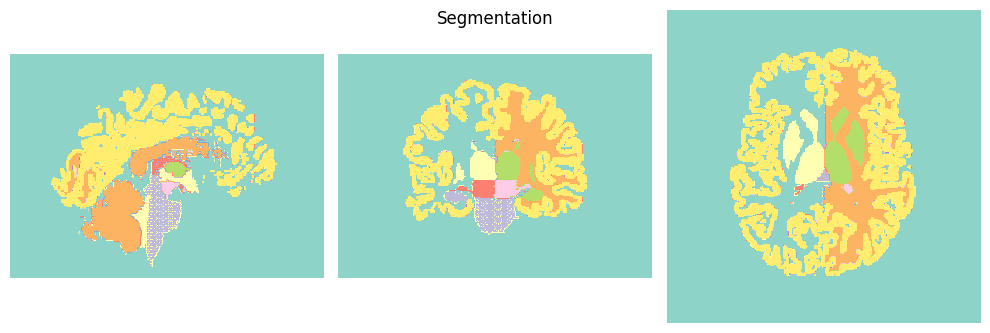
Analysis CSVs#
CSV files have been exported containing susceptibility values in regions of interest for each subject.
Here we will load the CSVs, inspect the data and generate figures:
# import modules
import pandas as pd
import seaborn as sns
The raw CSV files use names from FreeSurfer as exported by FastSurfer. The full list of regions is available here.
pd.read_csv(glob("bids/derivatives/qsmxt-*-*/sub*/ses*/extra*/*.csv")[0])
| roi | num_voxels | min | max | median | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | 3rd-Ventricle | 587 | -0.234321 | 0.054945 | -0.018367 | -0.021842 | 0.033371 |
| 1 | 4th-Ventricle | 1544 | -0.126117 | 0.052944 | -0.044766 | -0.045198 | 0.030768 |
| 2 | Brain-Stem | 18687 | -0.750579 | 0.184913 | -0.045976 | -0.042085 | 0.029045 |
| 3 | CSF | 834 | -1.393693 | 0.945752 | 0.030098 | 0.033957 | 0.140176 |
| 4 | Left-Accumbens-area | 524 | -0.141920 | 0.128334 | 0.025588 | 0.022306 | 0.031363 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 91 | ctx-rh-superiorfrontal | 29626 | -0.133274 | 0.251870 | -0.003875 | -0.009282 | 0.024289 |
| 92 | ctx-rh-superiorparietal | 12072 | -0.084225 | 0.306628 | -0.015670 | -0.014582 | 0.022277 |
| 93 | ctx-rh-superiortemporal | 16251 | -0.090982 | 0.248737 | 0.006754 | 0.012995 | 0.024299 |
| 94 | ctx-rh-supramarginal | 11992 | -0.056940 | 0.239688 | 0.019331 | 0.019976 | 0.022774 |
| 95 | ctx-rh-transversetemporal | 1255 | -0.004825 | 0.129559 | 0.034543 | 0.035700 | 0.013951 |
96 rows × 7 columns
We will select a subset of these ROIs and give them more readable names:
# define regions of interest
# see https://github.com/QSMxT/QSMxT/blob/main/qsmxt/aseg_labels.csv for a full list
rois = {
"Thalamus" : [9, 10, 48, 49],
"Pallidum" : [12, 13, 52, 53],
"Caudate" : [11, 50],
"Putamen" : [12, 51],
"Brain stem" : [16],
"CSF" : [24, 122, 257, 701],
"White matter" : [2, 7, 41, 46, 177]
}
roi_names = { value: key for key in rois for value in rois[key] }
roi_ids = [value for roi in rois.values() for value in roi]
# load a reconstruction
qsm = nib.load(glob("bids/derivatives/qsmxt-*-*/sub*/ses*/anat*/*Chimap.nii*")[0]).get_fdata().flatten()
seg = nib.load(glob("bids/derivatives/qsmxt-*-*/sub*/ses*/anat*/*space-qsm*dseg.nii*")[0]).get_fdata().flatten()
# retain only the rois
qsm = qsm[np.isin(seg, roi_ids)]
seg = seg[np.isin(seg, roi_ids)]
# convert to a dataframe for plotting purposes
seg = pd.Series(seg).map(roi_names)
data = pd.DataFrame({ 'qsm' : qsm, 'seg' : seg })
# summarise data by region including the average and standard deviation
data.groupby('seg')['qsm'].agg(['mean', 'std']).sort_values('mean').round(decimals=3)
| mean | std | |
|---|---|---|
| seg | ||
| Brain stem | -0.042 | 0.029 |
| White matter | -0.002 | 0.027 |
| Thalamus | 0.030 | 0.034 |
| CSF | 0.034 | 0.140 |
| Pallidum | 0.034 | 0.060 |
| Putamen | 0.047 | 0.033 |
| Caudate | 0.048 | 0.023 |
medians = data.groupby('seg')['qsm'].median().sort_values()
order = medians.index
# plot
fig, ax = plt.subplots(figsize=(5,4))
sns.boxplot(data=data, y='qsm', x='seg', fliersize=0, color='lightblue', order=order, ax=ax)
ax.set_xticks(range(len(ax.get_xticks()))) # Set tick positions explicitly
ax.set_xticklabels(ax.get_xticklabels(), ha='right', rotation=45)
ax.set_ylim(-0.2, 0.3)
ax.axhline(y=0, color='pink', linestyle='-', linewidth=1, zorder=-1)
ax.set_xlabel("Region of interest")
ax.set_ylabel("Susceptibility (ppm)")
ax.set_title("QSM")
plt.show()

Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-10-30T02:24:57.856137+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
nibabel : 5.2.1
numpy : 2.2.6
pandas : 2.3.3
seaborn : 0.13.2
matplotlib: 3.8.4
ipyniivue : 2.3.2
