FreeSurfer#
Author: Steffen Bollmann
Date: 17 Oct 2024
Citation and Resources:#
Tools included in this workflow#
FreeSurfer:
Fischl B. (2012). FreeSurfer. NeuroImage, 62(2), 774–781. https://doi.org/10.1016/j.neuroimage.2012.01.021
Dataset#
MP2RAGE T1-weighted average 7T model (human brain model)
Bollmann, Steffen, Andrew Janke, Lars Marstaller, David Reutens, Kieran O’Brien, and Markus Barth. “MP2RAGE T1-weighted average 7T model” January 1, 2017. doi:10.14264/uql.2017.266
Load FreeSurfer#
# we can use module to load freesurfer in a specific version
import module
await module.load('freesurfer/7.3.2')
await module.list()
['freesurfer/7.3.2']
!recon-all
USAGE: recon-all
Required Arguments:
-subjid <subjid>
-<process directive>
Fully-Automated Directive:
-all : performs all stages of cortical reconstruction
-autorecon-all : same as -all
Manual-Intervention Workflow Directives:
-autorecon1 : process stages 1-5 (see below)
-autorecon2 : process stages 6-23
after autorecon2, check white surfaces:
a. if wm edit was required, then run -autorecon2-wm
b. if control points added, then run -autorecon2-cp
c. proceed to run -autorecon3
-autorecon2-cp : process stages 12-23 (uses -f w/ mri_normalize, -keep w/ mri_seg)
-autorecon2-wm : process stages 15-23
-autorecon2-inflate1 : 6-18
-autorecon2-perhemi : tess, sm1, inf1, q, fix, sm2, inf2, finalsurf, ribbon
-autorecon3 : process stages 24-34
if edits made to correct pial, then run -autorecon-pial
-hemi ?h : just do lh or rh (default is to do both)
Autorecon Processing Stages (see -autorecon# flags above):
1. Motion Correction and Conform
2. NU (Non-Uniform intensity normalization)
3. Talairach transform computation
4. Intensity Normalization 1
5. Skull Strip
6. EM Register (linear volumetric registration)
7. CA Intensity Normalization
8. CA Non-linear Volumetric Registration
9. Remove neck
10. EM Register, with skull
11. CA Label (Aseg: Volumetric Labeling) and Statistics
12. Intensity Normalization 2 (start here for control points)
13. White matter segmentation
14. Edit WM With ASeg
15. Fill (start here for wm edits)
16. Tessellation (begins per-hemisphere operations)
17. Smooth1
18. Inflate1
19. QSphere
20. Automatic Topology Fixer
21. White Surfs (start here for brain edits for pial surf)
22. Smooth2
23. Inflate2
24. Spherical Mapping
25. Spherical Registration
26. Spherical Registration, Contralater hemisphere
27. Map average curvature to subject
28. Cortical Parcellation (Labeling)
29. Cortical Parcellation Statistics
30. Pial Surfs
31. WM/GM Contrast
32. Cortical Ribbon Mask
33. Cortical Parcellation mapped to ASeg
34 Brodmann and exvio EC labels
Step-wise Directives
See -help
Expert Preferences
-pons-crs C R S : col, row, slice of seed point for pons, used in fill
-cc-crs C R S : col, row, slice of seed point for corpus callosum, used in fill
-lh-crs C R S : col, row, slice of seed point for left hemisphere, used in fill
-rh-crs C R S : col, row, slice of seed point for right hemisphere, used in fill
-nofill : do not use the automatic subcort seg to fill
-watershed cmd : control skull stripping/watershed program
-xmask file : custom external brain mask to replace automated skullstripping
-wsless : decrease watershed threshold (leaves less skull, but can strip more brain)
-wsmore : increase watershed threshold (leaves more skull, but can strip less brain)
-wsatlas : use atlas when skull stripping
-no-wsatlas : do not use atlas when skull stripping
-no-wsgcaatlas : do not use GCA atlas when skull stripping
-wsthresh pct : explicity set watershed threshold
-wsseed C R S : identify an index (C, R, S) point in the skull
-norm3diters niters : number of 3d iterations for mri_normalize
-normmaxgrad maxgrad : max grad (-g) for mri_normalize. Default is 1.
-norm1-b N : in the _first_ usage of mri_normalize, use control
point with intensity N below target (default=10.0)
-norm2-b N : in the _second_ usage of mri_normalize, use control
point with intensity N below target (default=10.0)
-norm1-n N : in the _first_ usage of mri_normalize, do N number
of iterations
-norm2-n N : in the _second_ usage of mri_normalize, do N number
of iterations
-cm : conform volumes to the min voxel size
-no-fix-with-ga : do not use genetic algorithm when fixing topology
-fix-diag-only : topology fixer runs until ?h.defect_labels files
are created, then stops
-seg-wlo wlo : set wlo value for mri_segment and mris_make_surfaces
-seg-ghi ghi : set ghi value for mri_segment and mris_make_surfaces
-nothicken : pass '-thicken 0' to mri_segment
-no-ca-align-after : turn off -align-after with mri_ca_register
-no-ca-align : turn off -align with mri_ca_label
-deface : deface subject, written to orig_defaced.mgz
-expert file : read-in expert options file
-xopts-use : use pre-existing expert options file
-xopts-clean : delete pre-existing expert options file
-xopts-overwrite : overwrite pre-existing expert options file
-termscript script : run script before exiting (multiple -termscript flags possible)
This can be good for running custom post-processing after recon-all
The script must be in your path. The subjid is passed as the only argument
The current directory is changed to SUBJECTS_DIR before the script is run
The script should exit with 0 unless there is an error
-mprage : assume scan parameters are MGH MP-RAGE protocol
-washu_mprage : assume scan parameters are Wash.U. MP-RAGE protocol.
both mprage flags affect mri_normalize and mri_segment,
and assumes a darker gm.
-schwartzya3t-atlas : for tal reg, use special young adult 3T atlas
-threads num : set number of threads to use
Notification Files (Optional)
-waitfor file : wait for file to appear before beginning
-notify file : create this file after finishing
Status and Log files (Optional)
-log file : default is scripts/recon-all.log
-status file : default is scripts/recon-all-status.log
-noappend : start new log and status files instead of appending
-no-isrunning : do not check whether this subject is currently being processed
Segmentation of substructures of hippocampus and brainstem
(These deprecated; please see segmentHA_T1.sh, segmentHA_T1.sh, segmentHA_T1_long.sh, segmentBS.sh)
-hippocampal-subfields-T1 : segmentation of hippocampal subfields using input T1 scan
-hippocampal-subfields-T2 file ID : segmentation using an additional scan (given by file);
ID is a user-defined identifier for the analysis
-hippocampal-subfields-T1T2 file ID : segmentation using additional scan (given by file) and input T1
simultaneously; ID is a user-defined identifier for the analysis
-brainstem-structures : segmentation of brainstem structures
Other Arguments (Optional)
-sd subjectsdir : specify subjects dir (default env SUBJECTS_DIR)
-mail username : mail user when done
-umask umask : set unix file permission mask (default 002)
-grp groupid : check that current group is alpha groupid
-onlyversions : print version of each binary and exit
-debug : print out lots of info
-allowcoredump : set coredump limit to unlimited
-dontrun : do everything but execute each command
-version : print version of this script and exit
-help : voluminous bits of wisdom
Download data#
![ -f ./mp2rage.nii ] && echo "$FILE exist." || wget https://imaging.org.au/uploads/Human7T/mp2rageModel_L13_work03-plus-hippocampus-7T-sym-norm-mincanon_v0.8.nii -O ./mp2rage.nii
--2025-06-12 10:28:00-- https://imaging.org.au/uploads/Human7T/mp2rageModel_L13_work03-plus-hippocampus-7T-sym-norm-mincanon_v0.8.nii
Resolving imaging.org.au (imaging.org.au)... 203.101.229.7
Connecting to imaging.org.au (imaging.org.au)|203.101.229.7|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 1536000352 (1.4G) [application/octet-stream]
Saving to: ‘./mp2rage.nii’
./mp2rage.nii 100%[===================>] 1.43G 7.85MB/s in 3m 18s
2025-06-12 10:31:21 (7.38 MB/s) - ‘./mp2rage.nii’ saved [1536000352/1536000352]
!ls
brain_extraction_different_tools.ipynb mp2rage.nii sct_toolbox.ipynb
freesurfer.ipynb qsmxt.ipynb
Run#
!mkdir ./freesurfer_output
!recon-all -subject subjectname -i mp2rage.nii -all -sd ./freesurfer_output
fs-check-version --s subjectname --o /tmp/tmp.yk66v5
Thu Jun 12 10:32:01 UTC 2025
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging
/opt/freesurfer-7.3.2/bin/fs-check-version --s subjectname --o /tmp/tmp.yk66v5
-rwxrwxr-x 1 nobody nobody 18565 Aug 4 2022 /opt/freesurfer-7.3.2/bin/fs-check-version
freesurfer-linux-centos8_x86_64-7.3.2-20220804-6354275
$Id$
Linux jupyter-monidoerig 5.4.0-204-generic #224-Ubuntu SMP Thu Dec 5 13:38:28 UTC 2024 x86_64 x86_64 x86_64 GNU/Linux
pid 3536440
Current FS Version freesurfer-linux-centos8_x86_64-7.3.2-20220804-6354275
Subject does not have a bstampfile, copying /opt/freesurfer-7.3.2/build-stamp.txt
Subject FS Version: freesurfer-linux-centos8_x86_64-7.3.2-20220804-6354275
No constraints on version because REQ=UnSet and FsVerFile=NotThere
#@#% fs-check-version match = 1
fs-check-version Done
INFO: SUBJECTS_DIR is /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
Actual FREESURFER_HOME /usr/local/freesurfer/7.3.2-1
Linux jupyter-monidoerig 5.4.0-204-generic #224-Ubuntu SMP Thu Dec 5 13:38:28 UTC 2024 x86_64 x86_64 x86_64 GNU/Linux
#--------------------------------------------
#@# MotionCor Thu Jun 12 10:34:29 UTC 2025
Found 1 runs
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig/001.mgz
Checking for (invalid) multi-frame inputs...
Only one run found so motion
correction will not be performed. I'll
copy the run to rawavg and continue.
cp /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig/001.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz
mri_info /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz
rawavg.mgz ========================================
Volume information for /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz
type: MGH
dimensions: 640 x 750 x 800
voxel sizes: 0.300000, 0.300000, 0.300000
type: FLOAT (3)
fov: 240.000
dof: 1
xstart: -96.0, xend: 96.0
ystart: -112.5, yend: 112.5
zstart: -120.0, zend: 120.0
TR: 0.00 msec, TE: 0.00 msec, TI: 0.00 msec, flip angle: 0.00 degrees
nframes: 1
PhEncDir: UNKNOWN
FieldStrength: 0.000000
ras xform present
xform info: x_r = 1.0000, y_r = 0.0000, z_r = 0.0000, c_r = 0.3750
: x_a = 0.0000, y_a = 1.0000, z_a = 0.0000, c_a = 0.3750
: x_s = 0.0000, y_s = 0.0000, z_s = 1.0000, c_s = 0.3750
talairach xfm :
Orientation : RAS
Primary Slice Direction: axial
voxel to ras transform:
0.3000 0.0000 0.0000 -95.6250
0.0000 0.3000 0.0000 -112.1250
0.0000 0.0000 0.3000 -119.6250
0.0000 0.0000 0.0000 1.0000
voxel-to-ras determinant 0.027
ras to voxel transform:
3.3333 0.0000 0.0000 318.7500
0.0000 3.3333 0.0000 373.7500
0.0000 0.0000 3.3333 398.7500
0.0000 0.0000 0.0000 1.0000
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname
mri_convert /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz --conform
mri_convert /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz --conform
reading from /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/rawavg.mgz...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (1, 0, 0)
j_ras = (0, 1, 0)
k_ras = (0, 0, 1)
changing data type from float to uchar (noscale = 0)...
MRIchangeType: Building histogram 0 255 1000, flo=0, fhi=0.999, dest_type=0
Reslicing using trilinear interpolation
writing to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz...
mri_add_xform_to_header -c /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/talairach.xfm /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz
INFO: extension is mgz
mri_info /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz
orig.mgz ========================================
Volume information for /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz
type: MGH
dimensions: 256 x 256 x 256
voxel sizes: 1.000000, 1.000000, 1.000000
type: UCHAR (0)
fov: 256.000
dof: 1
xstart: -128.0, xend: 128.0
ystart: -128.0, yend: 128.0
zstart: -128.0, zend: 128.0
TR: 0.00 msec, TE: 0.00 msec, TI: 0.00 msec, flip angle: 0.00 degrees
nframes: 1
PhEncDir: UNKNOWN
FieldStrength: 0.000000
ras xform present
xform info: x_r = -1.0000, y_r = 0.0000, z_r = 0.0000, c_r = 0.3750
: x_a = 0.0000, y_a = 0.0000, z_a = 1.0000, c_a = 0.3750
: x_s = 0.0000, y_s = -1.0000, z_s = 0.0000, c_s = 0.3750
talairach xfm : /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/talairach.xfm
Orientation : LIA
Primary Slice Direction: coronal
voxel to ras transform:
-1.0000 0.0000 0.0000 128.3750
0.0000 0.0000 1.0000 -127.6250
0.0000 -1.0000 0.0000 128.3750
0.0000 0.0000 0.0000 1.0000
voxel-to-ras determinant -1
ras to voxel transform:
-1.0000 -0.0000 -0.0000 128.3750
-0.0000 -0.0000 -1.0000 128.3750
-0.0000 1.0000 -0.0000 127.6250
-0.0000 -0.0000 -0.0000 1.0000
#--------------------------------------------
#@# Talairach Thu Jun 12 10:39:15 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_nu_correct.mni --no-rescale --i orig.mgz --o orig_nu.mgz --ants-n4 --n 1 --proto-iters 1000 --distance 50
/usr/bin/bc
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
/opt/freesurfer-7.3.2/bin/mri_nu_correct.mni
--no-rescale --i orig.mgz --o orig_nu.mgz --ants-n4 --n 1 --proto-iters 1000 --distance 50
nIters 1
mri_nu_correct.mni 7.3.2
Linux jupyter-monidoerig 5.4.0-204-generic #224-Ubuntu SMP Thu Dec 5 13:38:28 UTC 2024 x86_64 x86_64 x86_64 GNU/Linux
Thu Jun 12 10:39:15 UTC 2025
tmpdir is ./tmp.mri_nu_correct.mni.3536918
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
AntsN4BiasFieldCorrectionFs -i orig.mgz -o ./tmp.mri_nu_correct.mni.3536918/nu0.mgz --dtype uchar
AntsN4BiasFieldCorrectionFs done
mri_convert ./tmp.mri_nu_correct.mni.3536918/nu0.mgz orig_nu.mgz --like orig.mgz --conform
mri_convert ./tmp.mri_nu_correct.mni.3536918/nu0.mgz orig_nu.mgz --like orig.mgz --conform
reading from ./tmp.mri_nu_correct.mni.3536918/nu0.mgz...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
INFO: transform src into the like-volume: orig.mgz
writing to orig_nu.mgz...
Thu Jun 12 10:42:36 UTC 2025
mri_nu_correct.mni done
talairach_avi --i orig_nu.mgz --xfm transforms/talairach.auto.xfm
talairach_avi log file is transforms/talairach_avi.log...
mv -f /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/talsrcimg_to_711-2C_as_mni_average_305_t4_vox2vox.txt /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/talsrcimg_to_711-2C_as_mni_average_305_t4_vox2vox.txt
Started at Thu Jun 12 10:42:37 UTC 2025
Ended at Thu Jun 12 10:43:10 UTC 2025
talairach_avi done
cp transforms/talairach.auto.xfm transforms/talairach.xfm
lta_convert --src orig.mgz --trg /opt/freesurfer-7.3.2/average/mni305.cor.mgz --inxfm transforms/talairach.xfm --outlta transforms/talairach.xfm.lta --subject fsaverage --ltavox2vox
7.3.2
--src: orig.mgz src image (geometry).
--trg: /opt/freesurfer-7.3.2/average/mni305.cor.mgz trg image (geometry).
--inmni: transforms/talairach.xfm input MNI/XFM transform.
--outlta: transforms/talairach.xfm.lta output LTA.
--s: fsaverage subject name
--ltavox2vox: output LTA as VOX_TO_VOX transform.
LTA read, type : 1
1.07958 0.00270 -0.01574 0.58327;
0.01399 0.99838 0.31440 -9.30106;
0.00758 -0.40444 1.15055 -29.25244;
0.00000 0.00000 0.00000 1.00000;
setting subject to fsaverage
Writing LTA to file transforms/talairach.xfm.lta...
lta_convert successful.
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms ~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
#--------------------------------------------
#@# Talairach Failure Detection Thu Jun 12 10:43:12 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
talairach_afd -T 0.005 -xfm transforms/talairach.xfm
7.3.2
cwd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
cmdline mri_binarize --i ./tmp.mri_nu_correct.mni.3537203/nu0.mgz --min -1 --o ./tmp.mri_nu_correct.mni.3537203/ones.mgz
sysname Linux
hostname jupyter-monidoerig
machine x86_64
user jovyan
input ./tmp.mri_nu_correct.mni.3537203/nu0.mgz
frame 0
nErode3d 0
nErode2d 0
output ./tmp.mri_nu_correct.mni.3537203/ones.mgz
Binarizing based on threshold
min -1
max +infinity
binval 1
binvalnot 0
fstart = 0, fend = 0, nframes = 1
Starting parallel 1
Found 16777216 values in range
Counting number of voxels in first frame
Found 16777215 voxels in final mask
Writing output to ./tmp.mri_nu_correct.mni.3537203/ones.mgz
Count: 16777215 16777215.000000 16777216 99.999994
mri_binarize done
mri_segstats --id 1 --seg ./tmp.mri_nu_correct.mni.3537203/ones.mgz --i orig.mgz --sum ./tmp.mri_nu_correct.mni.3537203/sum.junk --avgwf ./tmp.mri_nu_correct.mni.3537203/input.mean.dat
7.3.2
cwd
cmdline mri_segstats --id 1 --seg ./tmp.mri_nu_correct.mni.3537203/ones.mgz --i orig.mgz --sum ./tmp.mri_nu_correct.mni.3537203/sum.junk --avgwf ./tmp.mri_nu_correct.mni.3537203/input.mean.dat
sysname Linux
hostname jupyter-monidoerig
machine x86_64
user jovyan
whitesurfname white
UseRobust 0
Loading ./tmp.mri_nu_correct.mni.3537203/ones.mgz
Loading orig.mgz
Voxel Volume is 1 mm^3
Generating list of segmentation ids
Found 1 segmentations
Computing statistics for each segmentation
Reporting on 1 segmentations
Using PrintSegStat
Computing spatial average of each frame
Writing to ./tmp.mri_nu_correct.mni.3537203/input.mean.dat
mri_segstats done
mri_segstats --id 1 --seg ./tmp.mri_nu_correct.mni.3537203/ones.mgz --i ./tmp.mri_nu_correct.mni.3537203/nu0.mgz --sum ./tmp.mri_nu_correct.mni.3537203/sum.junk --avgwf ./tmp.mri_nu_correct.mni.3537203/output.mean.dat
7.3.2
cwd
cmdline mri_segstats --id 1 --seg ./tmp.mri_nu_correct.mni.3537203/ones.mgz --i ./tmp.mri_nu_correct.mni.3537203/nu0.mgz --sum ./tmp.mri_nu_correct.mni.3537203/sum.junk --avgwf ./tmp.mri_nu_correct.mni.3537203/output.mean.dat
sysname Linux
hostname jupyter-monidoerig
machine x86_64
user jovyan
whitesurfname white
UseRobust 0
Loading ./tmp.mri_nu_correct.mni.3537203/ones.mgz
Loading ./tmp.mri_nu_correct.mni.3537203/nu0.mgz
Voxel Volume is 1 mm^3
Generating list of segmentation ids
Found 1 segmentations
Computing statistics for each segmentation
Reporting on 1 segmentations
Using PrintSegStat
Computing spatial average of each frame
Writing to ./tmp.mri_nu_correct.mni.3537203/output.mean.dat
mri_segstats done
mris_calc -o ./tmp.mri_nu_correct.mni.3537203/nu0.mgz ./tmp.mri_nu_correct.mni.3537203/nu0.mgz mul 1.16048835830789136545
Saving result to './tmp.mri_nu_correct.mni.3537203/nu0.mgz' (type = MGH ) [ ok ]
mri_convert ./tmp.mri_nu_correct.mni.3537203/nu0.mgz nu.mgz --like orig.mgz
mri_convert ./tmp.mri_nu_correct.mni.3537203/nu0.mgz nu.mgz --like orig.mgz
reading from ./tmp.mri_nu_correct.mni.3537203/nu0.mgz...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
INFO: transform src into the like-volume: orig.mgz
writing to nu.mgz...
mri_make_uchar nu.mgz transforms/talairach.xfm nu.mgz
type change took 0 minutes and 6 seconds.
FIRST_PERCENTILE 0.010000
WM_PERCENTILE 0.900000
MAX_R 50.000000
i1 = 6, i2 = 82
#mri_make_uchar# mapping 15 211 to 3 110 : b -5.93289 m 0.548893 : thresh 10.8088 maxsat 475.38 : nzero 12342897 nsat 0
Thu Jun 12 10:46:58 UTC 2025
mri_nu_correct.mni done
mri_add_xform_to_header -c /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/talairach.xfm nu.mgz nu.mgz
INFO: extension is mgz
#--------------------------------------------
#@# Intensity Normalization Thu Jun 12 10:46:58 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_normalize -g 1 -seed 1234 -mprage nu.mgz T1.mgz
using max gradient = 1.000
setting seed for random number genererator to 1234
assuming input volume is MGH (Van der Kouwe) MP-RAGE
reading mri_src from nu.mgz...
normalizing image...
NOT doing gentle normalization with control points/label
talairach transform
1.07958 0.00270 -0.01574 0.58327;
0.01399 0.99838 0.31440 -9.30106;
0.00758 -0.40444 1.15055 -29.25244;
0.00000 0.00000 0.00000 1.00000;
processing without aseg, no1d=0
MRInormInit():
INFO: Modifying talairach volume c_(r,a,s) based on average_305
MRInormalize():
MRIsplineNormalize(): npeaks = 19
Starting OpenSpline(): npoints = 19
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
Iterating 2 times
---------------------------------
3d normalization pass 1 of 2
white matter peak found at 110
white matter peak found at 110
gm peak at 62 (62), valley at 51 (51)
csf peak at 32, setting threshold to 52
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
---------------------------------
3d normalization pass 2 of 2
white matter peak found at 110
white matter peak found at 110
gm peak at 65 (65), valley at 53 (53)
csf peak at 33, setting threshold to 54
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
Done iterating ---------------------------------
writing output to T1.mgz
3D bias adjustment took 1 minutes and 45 seconds.
#--------------------------------------------
#@# Skull Stripping Thu Jun 12 10:48:44 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_em_register -skull nu.mgz /opt/freesurfer-7.3.2/average/RB_all_withskull_2020_01_02.gca transforms/talairach_with_skull.lta
aligning to atlas containing skull, setting unknown_nbr_spacing = 5
== Number of threads available to mri_em_register for OpenMP = 1 ==
reading 1 input volumes...
logging results to talairach_with_skull.log
reading '/opt/freesurfer-7.3.2/average/RB_all_withskull_2020_01_02.gca'...
GCAread took 0 minutes and 4 seconds.
average std = 23.0 using min determinant for regularization = 52.8
0 singular and 9205 ill-conditioned covariance matrices regularized
reading 'nu.mgz'...
freeing gibbs priors...done.
accounting for voxel sizes in initial transform
bounding unknown intensity as < 8.9 or > 556.0
total sample mean = 77.3 (1403 zeros)
************************************************
spacing=8, using 3292 sample points, tol=1.00e-05...
************************************************
register_mri: find_optimal_transform
find_optimal_transform: nsamples 3292, passno 0, spacing 8
resetting wm mean[0]: 100 --> 108
resetting gm mean[0]: 61 --> 61
input volume #1 is the most T1-like
using real data threshold=25.0
skull bounding box = (51, 29, 17) --> (205, 241, 220)
finding center of left hemi white matter
using (102, 100, 119) as brain centroid of Right_Cerebral_White_Matter...
MRImask(): AllowDiffGeom = 1
mean wm in atlas = 108, using box (83,74,94) --> (120, 126,144) to find MRI wm
before smoothing, mri peak at 107
robust fit to distribution - 107 +- 3.7
after smoothing, mri peak at 107, scaling input intensities by 1.009
scaling channel 0 by 1.00935
initial log_p = -4.355
************************************************
First Search limited to translation only.
************************************************
max log p = -4.327209 @ (-10.526, 10.526, -10.526)
max log p = -4.238077 @ (5.263, 5.263, 5.263)
max log p = -4.103386 @ (2.632, -2.632, -2.632)
max log p = -4.103386 @ (0.000, 0.000, 0.000)
max log p = -4.087552 @ (-0.658, -0.658, -1.974)
max log p = -4.087552 @ (0.000, 0.000, 0.000)
max log p = -4.087552 @ (0.000, 0.000, 0.000)
Result so far: scale 1.000: max_log_p=-3.772, old_max_log_p =-4.088 (thresh=-4.1)
1.06375 0.00000 0.00000 -11.47446;
0.00000 1.11081 0.29764 -30.89916;
0.00000 -0.27532 1.02750 26.07788;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 26 seconds.
****************************************
Nine parameter search. iteration 1 nscales = 0 ...
****************************************
Result so far: scale 1.000: max_log_p=-3.772, old_max_log_p =-3.772 (thresh=-3.8)
1.06375 0.00000 0.00000 -11.47446;
0.00000 1.11081 0.29764 -30.89916;
0.00000 -0.27532 1.02750 26.07788;
0.00000 0.00000 0.00000 1.00000;
reducing scale to 0.2500
iteration took 1 minutes and 22 seconds.
****************************************
Nine parameter search. iteration 2 nscales = 1 ...
****************************************
Result so far: scale 0.250: max_log_p=-3.654, old_max_log_p =-3.772 (thresh=-3.8)
1.04325 -0.00851 0.03175 -11.59169;
0.00000 1.08921 0.29185 -25.65857;
-0.03480 -0.26485 0.98844 30.03328;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 19 seconds.
****************************************
Nine parameter search. iteration 3 nscales = 1 ...
****************************************
Result so far: scale 0.250: max_log_p=-3.648, old_max_log_p =-3.654 (thresh=-3.7)
1.04325 -0.00851 0.03175 -11.59169;
-0.00116 1.10021 0.33011 -31.41711;
-0.03479 -0.30035 0.97836 35.56596;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 19 seconds.
****************************************
Nine parameter search. iteration 4 nscales = 1 ...
****************************************
Result so far: scale 0.250: max_log_p=-3.647, old_max_log_p =-3.648 (thresh=-3.6)
1.04383 0.00132 -0.00028 -9.04488;
-0.00118 1.12083 0.33630 -32.79573;
-0.00063 -0.30047 0.97888 31.13393;
0.00000 0.00000 0.00000 1.00000;
reducing scale to 0.0625
iteration took 1 minutes and 20 seconds.
****************************************
Nine parameter search. iteration 5 nscales = 2 ...
****************************************
Result so far: scale 0.062: max_log_p=-3.625, old_max_log_p =-3.626 (thresh=-3.6)
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
min search scale 0.025000 reached
***********************************************
Computing MAP estimate using 3292 samples...
***********************************************
dt = 5.00e-06, momentum=0.80, tol=1.00e-05
l_intensity = 1.0000
Aligning input volume to GCA...
Transform matrix
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
nsamples 3292
Quasinewton: input matrix
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
IFLAG= -1 LINE SEARCH FAILED. SEE DOCUMENTATION OF ROUTINE MCSRCH ERROR RETURN OF LINE SEARCH: INFO= 3 POSSIBLE CAUSES: FUNCTION OR GRADIENT ARE INCORRECT OR INCORRECT TOLERANCESoutof QuasiNewtonEMA: 009: -log(p) = -0.0 tol 0.000010
Resulting transform:
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
pass 1, spacing 8: log(p) = -3.625 (old=-4.355)
transform before final EM align:
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
**************************************************
EM alignment process ...
Computing final MAP estimate using 364986 samples.
**************************************************
dt = 5.00e-06, momentum=0.80, tol=1.00e-07
l_intensity = 1.0000
Aligning input volume to GCA...
Transform matrix
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
nsamples 364986
Quasinewton: input matrix
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
IFLAG= -1 LINE SEARCH FAILED. SEE DOCUMENTATION OF ROUTINE MCSRCH ERROR RETURN OF LINE SEARCH: INFO= 6 POSSIBLE CAUSES: FUNCTION OR GRADIENT ARE INCORRECT OR INCORRECT TOLERANCESoutof QuasiNewtonEMA: 011: -log(p) = 4.1 tol 0.000000
final transform:
1.04107 0.01509 0.02101 -12.41482;
-0.01816 1.12024 0.31844 -29.82493;
-0.01776 -0.28240 0.98528 29.92633;
0.00000 0.00000 0.00000 1.00000;
writing output transformation to transforms/talairach_with_skull.lta...
#VMPC# mri_em_register VmPeak 800740
FSRUNTIME@ mri_em_register 0.1757 hours 1 threads
registration took 10 minutes and 33 seconds.
mri_watershed -T1 -brain_atlas /opt/freesurfer-7.3.2/average/RB_all_withskull_2020_01_02.gca transforms/talairach_with_skull.lta T1.mgz brainmask.auto.mgz
Mode: T1 normalized volume
Mode: Use the information of atlas (default parms, --help for details)
*********************************************************
The input file is T1.mgz
The output file is brainmask.auto.mgz
Weighting the input with atlas information before watershed
*************************WATERSHED**************************
Sorting...
first estimation of the COG coord: x=129 y=99 z=114 r=79
first estimation of the main basin volume: 2144899 voxels
Looking for seedpoints
2 found in the cerebellum
16 found in the rest of the brain
global maximum in x=106, y=92, z=76, Imax=255
CSF=12, WM_intensity=110, WM_VARIANCE=5
WM_MIN=110, WM_HALF_MIN=110, WM_HALF_MAX=110, WM_MAX=110
preflooding height equal to 10 percent
done.
Analyze...
main basin size=17403346377 voxels, voxel volume =1.000
= 17403346377 mmm3 = 17403346.944 cm3
done.
PostAnalyze...Basin Prior
8 basins merged thanks to atlas
***** 0 basin(s) merged in 1 iteration(s)
***** 0 voxel(s) added to the main basin
done.
Weighting the input with prior template
****************TEMPLATE DEFORMATION****************
second estimation of the COG coord: x=128,y=106, z=109, r=9955 iterations
^^^^^^^^ couldn't find WM with original limits - expanding ^^^^^^
GLOBAL CSF_MIN=0, CSF_intensity=7, CSF_MAX=21 , nb = 44406
RIGHT_CER CSF_MIN=0, CSF_intensity=4, CSF_MAX=11 , nb = 3024
LEFT_CER CSF_MIN=0, CSF_intensity=4, CSF_MAX=11 , nb = 3060
RIGHT_BRAIN CSF_MIN=0, CSF_intensity=9, CSF_MAX=20 , nb = 18684
LEFT_BRAIN CSF_MIN=0, CSF_intensity=9, CSF_MAX=21 , nb = 18684
OTHER CSF_MIN=0, CSF_intensity=4, CSF_MAX=14 , nb = 954
Problem with the least square interpolation in GM_MIN calculation.
CSF_MAX TRANSITION GM_MIN GM
GLOBAL
before analyzing : 21, 23, 31, 64
after analyzing : 21, 28, 31, 37
RIGHT_CER
before analyzing : 11, 16, 36, 64
after analyzing : 11, 29, 36, 37
LEFT_CER
before analyzing : 11, 16, 37, 61
after analyzing : 11, 30, 37, 37
RIGHT_BRAIN
before analyzing : 20, 22, 29, 64
after analyzing : 20, 26, 29, 35
LEFT_BRAIN
before analyzing : 21, 24, 32, 64
after analyzing : 21, 29, 32, 37
OTHER
before analyzing : 14, 15, 27, 94
after analyzing : 14, 23, 27, 40
mri_strip_skull: done peeling brain
highly tesselated surface with 10242 vertices
matching...65 iterations
*********************VALIDATION*********************
curvature mean = -0.013, std = 0.011
curvature mean = 69.801, std = 8.241
No Rigid alignment: -atlas Mode Off (basic atlas / no registration)
before rotation: sse = 2.23, sigma = 3.29
after rotation: sse = 2.23, sigma = 3.29
Localization of inacurate regions: Erosion-Dilation steps
the sse mean is 2.27, its var is 2.73
before Erosion-Dilatation 0.09% of inacurate vertices
after Erosion-Dilatation 0.00% of inacurate vertices
Validation of the shape of the surface done.
Scaling of atlas fields onto current surface fields
********FINAL ITERATIVE TEMPLATE DEFORMATION********
Compute Local values csf/gray
Fine Segmentation...34 iterations
mri_strip_skull: done peeling brain
Brain Size = 1611792 voxels, voxel volume = 1.000 mm3
= 1611792 mmm3 = 1611.792 cm3
******************************
Saving brainmask.auto.mgz
done
mri_watershed done
cp brainmask.auto.mgz brainmask.mgz
#-------------------------------------
#@# EM Registration Thu Jun 12 10:59:35 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_em_register -uns 3 -mask brainmask.mgz nu.mgz /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca transforms/talairach.lta
setting unknown_nbr_spacing = 3
using MR volume brainmask.mgz to mask input volume...
== Number of threads available to mri_em_register for OpenMP = 1 ==
reading 1 input volumes...
logging results to talairach.log
reading '/opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca'...
GCAread took 0 minutes and 3 seconds.
average std = 7.2 using min determinant for regularization = 5.2
0 singular and 884 ill-conditioned covariance matrices regularized
reading 'nu.mgz'...
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
freeing gibbs priors...done.
accounting for voxel sizes in initial transform
bounding unknown intensity as < 5.9 or > 519.0
total sample mean = 79.1 (1017 zeros)
************************************************
spacing=8, using 2841 sample points, tol=1.00e-05...
************************************************
register_mri: find_optimal_transform
find_optimal_transform: nsamples 2841, passno 0, spacing 8
resetting wm mean[0]: 98 --> 107
resetting gm mean[0]: 61 --> 61
input volume #1 is the most T1-like
using real data threshold=20.9
skull bounding box = (62, 44, 31) --> (194, 181, 196)
finding center of left hemi white matter
using (106, 90, 114) as brain centroid of Right_Cerebral_White_Matter...
MRImask(): AllowDiffGeom = 1
mean wm in atlas = 107, using box (90,73,94) --> (122, 106,134) to find MRI wm
before smoothing, mri peak at 107
robust fit to distribution - 107 +- 3.2
after smoothing, mri peak at 107, scaling input intensities by 1.000
scaling channel 0 by 1
initial log_p = -4.170
************************************************
First Search limited to translation only.
************************************************
max log p = -4.041770 @ (-10.526, 10.526, -10.526)
max log p = -3.763349 @ (5.263, 5.263, 5.263)
max log p = -3.695452 @ (2.632, -2.632, -2.632)
max log p = -3.663278 @ (1.316, 3.947, 1.316)
max log p = -3.653469 @ (-0.658, -0.658, -0.658)
max log p = -3.653469 @ (0.000, 0.000, 0.000)
max log p = -3.653469 @ (0.000, 0.000, 0.000)
max log p = -3.653469 @ (0.000, 0.000, 0.000)
Found translation: (-2.0, 16.4, -7.2): log p = -3.653
****************************************
Nine parameter search. iteration 0 nscales = 0 ...
****************************************
Result so far: scale 1.000: max_log_p=-3.190, old_max_log_p =-3.653 (thresh=-3.6)
1.07500 0.00000 0.00000 -11.63188;
0.00000 1.11081 0.29764 -28.48080;
0.00000 -0.25882 0.96593 30.71957;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 16 seconds.
****************************************
Nine parameter search. iteration 1 nscales = 0 ...
****************************************
Result so far: scale 1.000: max_log_p=-3.190, old_max_log_p =-3.190 (thresh=-3.2)
1.07500 0.00000 0.00000 -11.63188;
0.00000 1.11081 0.29764 -28.48080;
0.00000 -0.25882 0.96593 30.71957;
0.00000 0.00000 0.00000 1.00000;
reducing scale to 0.2500
iteration took 1 minutes and 15 seconds.
****************************************
Nine parameter search. iteration 2 nscales = 1 ...
****************************************
Result so far: scale 0.250: max_log_p=-3.104, old_max_log_p =-3.190 (thresh=-3.2)
1.05409 0.00000 0.00000 -8.93891;
0.00000 1.15247 0.30880 -34.02222;
0.00000 -0.26367 0.98404 27.44393;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 11 seconds.
****************************************
Nine parameter search. iteration 3 nscales = 1 ...
****************************************
Result so far: scale 0.062: max_log_p=-3.052, old_max_log_p =-3.104 (thresh=-3.1)
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
iteration took 1 minutes and 7 seconds.
****************************************
Nine parameter search. iteration 5 nscales = 2 ...
****************************************
Result so far: scale 0.062: max_log_p=-3.052, old_max_log_p =-3.052 (thresh=-3.0)
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
min search scale 0.025000 reached
***********************************************
Computing MAP estimate using 2841 samples...
***********************************************
dt = 5.00e-06, momentum=0.80, tol=1.00e-05
l_intensity = 1.0000
Aligning input volume to GCA...
Transform matrix
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
nsamples 2841
Quasinewton: input matrix
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
IFLAG= -1 LINE SEARCH FAILED. SEE DOCUMENTATION OF ROUTINE MCSRCH ERROR RETURN OF LINE SEARCH: INFO= 3 POSSIBLE CAUSES: FUNCTION OR GRADIENT ARE INCORRECT OR INCORRECT TOLERANCESoutof QuasiNewtonEMA: 008: -log(p) = -0.0 tol 0.000010
Resulting transform:
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
pass 1, spacing 8: log(p) = -3.052 (old=-4.170)
transform before final EM align:
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
**************************************************
EM alignment process ...
Computing final MAP estimate using 315638 samples.
**************************************************
dt = 5.00e-06, momentum=0.80, tol=1.00e-07
l_intensity = 1.0000
Aligning input volume to GCA...
Transform matrix
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
nsamples 315638
Quasinewton: input matrix
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
IFLAG= -1 LINE SEARCH FAILED. SEE DOCUMENTATION OF ROUTINE MCSRCH ERROR RETURN OF LINE SEARCH: INFO= 6 POSSIBLE CAUSES: FUNCTION OR GRADIENT ARE INCORRECT OR INCORRECT TOLERANCESoutof QuasiNewtonEMA: 010: -log(p) = 3.7 tol 0.000000
final transform:
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
writing output transformation to transforms/talairach.lta...
#VMPC# mri_em_register VmPeak 788196
FSRUNTIME@ mri_em_register 0.1346 hours 1 threads
registration took 8 minutes and 5 seconds.
#--------------------------------------
#@# CA Normalize Thu Jun 12 11:07:40 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_ca_normalize -c ctrl_pts.mgz -mask brainmask.mgz nu.mgz /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca transforms/talairach.lta norm.mgz
writing control point volume to ctrl_pts.mgz
using MR volume brainmask.mgz to mask input volume...
reading 1 input volume
reading atlas from '/opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca'...
reading transform from 'transforms/talairach.lta'...
reading input volume from nu.mgz...
resetting wm mean[0]: 98 --> 107
resetting gm mean[0]: 61 --> 61
input volume #1 is the most T1-like
using real data threshold=20.9
skull bounding box = (62, 44, 31) --> (194, 181, 196)
finding center of left hemi white matter
using (106, 90, 114) as brain centroid of Right_Cerebral_White_Matter...
mean wm in atlas = 107, using box (90,73,94) --> (122, 106,134) to find MRI wm
before smoothing, mri peak at 107
robust fit to distribution - 107 +- 3.2
after smoothing, mri peak at 107, scaling input intensities by 1.000
scaling channel 0 by 1
using 246437 sample points...
INFO: compute sample coordinates transform
1.05652 -0.00215 0.00803 -9.89662;
0.00000 1.14977 0.30808 -33.66290;
-0.00863 -0.26274 0.98055 28.36465;
0.00000 0.00000 0.00000 1.00000;
INFO: transform used
finding control points in Left_Cerebral_White_Matter....
found 40230 control points for structure...
bounding box (129, 48, 34) --> (191, 150, 194)
Left_Cerebral_White_Matter: limiting intensities to 96.0 --> 132.0
20 of 9292 (0.2%) samples deleted
finding control points in Right_Cerebral_White_Matter....
found 39478 control points for structure...
bounding box (67, 47, 32) --> (130, 143, 195)
Right_Cerebral_White_Matter: limiting intensities to 97.0 --> 132.0
17 of 9476 (0.2%) samples deleted
finding control points in Left_Cerebellum_White_Matter....
found 3105 control points for structure...
bounding box (130, 124, 62) --> (176, 163, 115)
Left_Cerebellum_White_Matter: limiting intensities to 88.0 --> 132.0
0 of 352 (0.0%) samples deleted
finding control points in Right_Cerebellum_White_Matter....
found 2710 control points for structure...
bounding box (87, 124, 59) --> (128, 162, 115)
Right_Cerebellum_White_Matter: limiting intensities to 88.0 --> 132.0
2 of 393 (0.5%) samples deleted
finding control points in Brain_Stem....
found 3475 control points for structure...
bounding box (113, 114, 97) --> (145, 173, 125)
Brain_Stem: limiting intensities to 88.0 --> 132.0
0 of 523 (0.0%) samples deleted
using 20036 total control points for intensity normalization...
bias field = 0.973 +- 0.051
189 of 19997 control points discarded
finding control points in Left_Cerebral_White_Matter....
found 40230 control points for structure...
bounding box (129, 48, 34) --> (191, 150, 194)
Left_Cerebral_White_Matter: limiting intensities to 90.0 --> 132.0
30 of 9460 (0.3%) samples deleted
finding control points in Right_Cerebral_White_Matter....
found 39478 control points for structure...
bounding box (67, 47, 32) --> (130, 143, 195)
Right_Cerebral_White_Matter: limiting intensities to 91.0 --> 132.0
20 of 9622 (0.2%) samples deleted
finding control points in Left_Cerebellum_White_Matter....
found 3105 control points for structure...
bounding box (130, 124, 62) --> (176, 163, 115)
Left_Cerebellum_White_Matter: limiting intensities to 88.0 --> 132.0
158 of 479 (33.0%) samples deleted
finding control points in Right_Cerebellum_White_Matter....
found 2710 control points for structure...
bounding box (87, 124, 59) --> (128, 162, 115)
Right_Cerebellum_White_Matter: limiting intensities to 88.0 --> 132.0
157 of 462 (34.0%) samples deleted
finding control points in Brain_Stem....
found 3475 control points for structure...
bounding box (113, 114, 97) --> (145, 173, 125)
Brain_Stem: limiting intensities to 88.0 --> 132.0
88 of 638 (13.8%) samples deleted
using 20732 total control points for intensity normalization...
bias field = 1.028 +- 0.043
159 of 19866 control points discarded
writing normalized volume to norm.mgz...
writing control points to ctrl_pts.mgz
freeing GCA...done.
normalization took 1 minutes and 1 seconds.
#--------------------------------------
#@# CA Reg Thu Jun 12 11:08:41 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_ca_register -nobigventricles -T transforms/talairach.lta -align-after -mask brainmask.mgz norm.mgz /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca transforms/talairach.m3z
not handling expanded ventricles...
using previously computed transform transforms/talairach.lta
renormalizing sequences with structure alignment, equivalent to:
-renormalize
-regularize_mean 0.500
-regularize 0.500
using MR volume brainmask.mgz to mask input volume...
== Number of threads available to mri_ca_register for OpenMP = 1 ==
reading 1 input volumes...
logging results to talairach.log
reading input volume 'norm.mgz'...
reading GCA '/opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca'...
label assignment complete, 0 changed (0.00%)
freeing gibbs priors...done.
average std[0] = 5.0
Starting GCAMregister()
label assignment complete, 0 changed (0.00%)
npasses = 1, nlevels = 6
#pass# 1 of 1 ************************
enabling zero nodes
setting smoothness cost coefficient to 0.156
#GCAMreg# pass 0 level1 5 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.16
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.758584
#FOTS# QuadFit found better minimum quadopt=(dt=163.994,rms=0.698519) vs oldopt=(dt=92.48,rms=0.710544)
#GCMRL# 0 dt 163.993517 rms 0.699 7.918% neg 0 invalid 762 tFOTS 17.0700 tGradient 6.7410 tsec 25.0950
#FOTS# QuadFit found better minimum quadopt=(dt=166.748,rms=0.672556) vs oldopt=(dt=92.48,rms=0.674419)
#GCMRL# 2 dt 166.748299 rms 0.673 1.193% neg 0 invalid 762 tFOTS 17.0190 tGradient 6.6850 tsec 24.9640
#FOTS# QuadFit found better minimum quadopt=(dt=144.162,rms=0.668033) vs oldopt=(dt=92.48,rms=0.668858)
#GCMRL# 3 dt 144.161616 rms 0.668 0.673% neg 0 invalid 762 tFOTS 17.1390 tGradient 6.7820 tsec 25.1810
#FOTS# QuadFit found better minimum quadopt=(dt=295.936,rms=0.662801) vs oldopt=(dt=369.92,rms=0.662948)
#GCMRL# 4 dt 295.936000 rms 0.663 0.783% neg 0 invalid 762 tFOTS 16.9760 tGradient 6.6520 tsec 24.8820
#FOTS# QuadFit found better minimum quadopt=(dt=110.976,rms=0.659698) vs oldopt=(dt=92.48,rms=0.659772)
#GCMRL# 5 dt 110.976000 rms 0.660 0.468% neg 0 invalid 762 tFOTS 17.9780 tGradient 6.8620 tsec 26.0940
#FOTS# QuadFit found better minimum quadopt=(dt=517.888,rms=0.654475) vs oldopt=(dt=369.92,rms=0.655164)
#GCMRL# 6 dt 517.888000 rms 0.654 0.792% neg 0 invalid 762 tFOTS 17.0040 tGradient 6.7490 tsec 25.0020
#GCMRL# 7 dt 92.480000 rms 0.652 0.346% neg 0 invalid 762 tFOTS 17.9880 tGradient 6.6850 tsec 25.9240
#GCMRL# 8 dt 1479.680000 rms 0.645 1.041% neg 0 invalid 762 tFOTS 16.9820 tGradient 6.6440 tsec 24.8770
#GCMRL# 9 dt 92.480000 rms 0.643 0.305% neg 0 invalid 762 tFOTS 16.9940 tGradient 6.8600 tsec 25.1110
#FOTS# QuadFit found better minimum quadopt=(dt=517.888,rms=0.641752) vs oldopt=(dt=369.92,rms=0.642094)
#GCMRL# 10 dt 517.888000 rms 0.642 0.264% neg 0 invalid 762 tFOTS 17.9880 tGradient 6.6180 tsec 25.8580
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.641504) vs oldopt=(dt=92.48,rms=0.641536)
#GCMRL# 11 dt 129.472000 rms 0.642 0.000% neg 0 invalid 762 tFOTS 16.9810 tGradient 6.7950 tsec 25.0670
#GCMRL# 12 dt 129.472000 rms 0.641 0.037% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5650 tsec 7.8180
#GCMRL# 13 dt 129.472000 rms 0.641 0.066% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7380 tsec 7.9890
#GCMRL# 16 dt 129.472000 rms 0.638 0.154% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7870 tsec 8.0460
#GCMRL# 17 dt 129.472000 rms 0.637 0.159% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.8120 tsec 8.0630
#GCMRL# 18 dt 129.472000 rms 0.636 0.146% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.8460 tsec 8.0950
#GCMRL# 19 dt 129.472000 rms 0.636 0.139% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7090 tsec 7.9610
#GCMRL# 20 dt 129.472000 rms 0.635 0.145% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.8690 tsec 8.1230
#GCMRL# 21 dt 129.472000 rms 0.634 0.138% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6590 tsec 7.9120
#GCMRL# 22 dt 129.472000 rms 0.633 0.131% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6910 tsec 7.9450
#GCMRL# 23 dt 129.472000 rms 0.632 0.122% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6630 tsec 7.9200
#GCMRL# 24 dt 129.472000 rms 0.631 0.118% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.9610 tsec 8.2520
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.632139
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.630971) vs oldopt=(dt=92.48,rms=0.631031)
#GCMRL# 26 dt 129.472000 rms 0.631 0.185% neg 0 invalid 762 tFOTS 17.0250 tGradient 6.7300 tsec 25.0110
#GCMRL# 27 dt 369.920000 rms 0.630 0.000% neg 0 invalid 762 tFOTS 16.9930 tGradient 6.8050 tsec 25.0980
#GCMRL# 28 dt 369.920000 rms 0.630 0.002% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6470 tsec 7.8970
#GCMRL# 29 dt 369.920000 rms 0.630 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7460 tsec 7.9980
#GCMRL# 30 dt 369.920000 rms 0.630 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6300 tsec 7.8790
setting smoothness cost coefficient to 0.615
#GCAMreg# pass 0 level1 4 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.62
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.638461
#FOTS# QuadFit found better minimum quadopt=(dt=72.716,rms=0.633052) vs oldopt=(dt=103.68,rms=0.633761)
#GCMRL# 32 dt 72.715953 rms 0.633 0.847% neg 0 invalid 762 tFOTS 16.9860 tGradient 5.2830 tsec 23.5260
#FOTS# QuadFit found better minimum quadopt=(dt=217.924,rms=0.626214) vs oldopt=(dt=103.68,rms=0.627862)
#GCMRL# 33 dt 217.924051 rms 0.626 1.080% neg 0 invalid 762 tFOTS 18.0050 tGradient 5.1390 tsec 24.3970
#FOTS# QuadFit found better minimum quadopt=(dt=64.4384,rms=0.622) vs oldopt=(dt=25.92,rms=0.623136)
#GCMRL# 34 dt 64.438356 rms 0.622 0.673% neg 0 invalid 762 tFOTS 17.0220 tGradient 5.2370 tsec 23.5170
#FOTS# QuadFit found better minimum quadopt=(dt=101.517,rms=0.619487) vs oldopt=(dt=103.68,rms=0.619489)
#GCMRL# 35 dt 101.517241 rms 0.619 0.404% neg 0 invalid 762 tFOTS 16.9860 tGradient 5.0750 tsec 23.3140
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.618122) vs oldopt=(dt=25.92,rms=0.618385)
#GCMRL# 36 dt 36.288000 rms 0.618 0.000% neg 0 invalid 762 tFOTS 17.0390 tGradient 5.2200 tsec 23.5350
#GCMRL# 37 dt 36.288000 rms 0.617 0.101% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2040 tsec 6.4370
#GCMRL# 38 dt 36.288000 rms 0.616 0.168% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1020 tsec 6.3440
#GCMRL# 39 dt 36.288000 rms 0.615 0.233% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1060 tsec 6.3460
#GCMRL# 40 dt 36.288000 rms 0.613 0.274% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1310 tsec 6.3720
#GCMRL# 41 dt 36.288000 rms 0.612 0.285% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2020 tsec 6.4460
#GCMRL# 42 dt 36.288000 rms 0.610 0.274% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0700 tsec 6.3100
#GCMRL# 43 dt 36.288000 rms 0.608 0.287% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0730 tsec 6.3160
#GCMRL# 44 dt 36.288000 rms 0.607 0.249% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0790 tsec 6.3120
#GCMRL# 49 dt 36.288000 rms 0.602 0.126% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2370 tsec 6.4920
#GCMRL# 50 dt 36.288000 rms 0.601 0.098% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1980 tsec 6.4530
#GCMRL# 51 dt 36.288000 rms 0.601 0.067% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2180 tsec 6.5140
#GCMRL# 52 dt 0.450000 rms 0.601 0.000% neg 0 invalid 762 tFOTS 16.9280 tGradient 5.1510 tsec 23.3620
#GCAMreg# pass 0 level1 4 level2 1 tsec 249.457 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.62
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=0.5,type=2, relabel=0, neg=no
setting smoothness cost coefficient to 2.353
#GCAMreg# pass 0 level1 3 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.35
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.628375
#GCMRL# 56 dt 0.000000 rms 0.628 0.118% neg 0 invalid 762 tFOTS 14.9970 tGradient 4.5500 tsec 20.8090
#GCAMreg# pass 0 level1 3 level2 1 tsec 47.102 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.35
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=0.5,type=2, relabel=0, neg=no
#FOTS# QuadFit found better minimum quadopt=(dt=0.075,rms=0.627612) vs oldopt=(dt=0.125,rms=0.627613)
#GCMRL# 59 dt 0.075000 rms 0.628 0.000% neg 0 invalid 762 tFOTS 15.9130 tGradient 4.5700 tsec 21.7630
setting smoothness cost coefficient to 8.000
#GCAMreg# pass 0 level1 2 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=8.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.706275
#FOTS# QuadFit found better minimum quadopt=(dt=2.36825,rms=0.68245) vs oldopt=(dt=2.88,rms=0.683533)
#GCMRL# 61 dt 2.368248 rms 0.682 3.373% neg 0 invalid 762 tFOTS 16.1200 tGradient 4.3600 tsec 21.7350
#FOTS# QuadFit found better minimum quadopt=(dt=1.75771,rms=0.679391) vs oldopt=(dt=2.88,rms=0.680485)
#GCMRL# 62 dt 1.757709 rms 0.679 0.448% neg 0 invalid 762 tFOTS 16.0620 tGradient 4.2480 tsec 21.5680
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.679068
#FOTS# QuadFit found better minimum quadopt=(dt=0.252,rms=0.678531) vs oldopt=(dt=0.18,rms=0.678534)
#GCMRL# 66 dt 0.252000 rms 0.679 0.079% neg 0 invalid 762 tFOTS 15.1100 tGradient 4.2680 tsec 20.6440
#FOTS# QuadFit found better minimum quadopt=(dt=0.1875,rms=0.678516) vs oldopt=(dt=0.18,rms=0.678516)
#GCMRL# 67 dt 0.187500 rms 0.679 0.000% neg 0 invalid 762 tFOTS 16.0130 tGradient 4.3370 tsec 21.6380
setting smoothness cost coefficient to 20.000
#GCAMreg# pass 0 level1 1 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=20.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=2.0,type=2, relabel=0, neg=no
#FOTS# QuadFit found better minimum quadopt=(dt=0.384,rms=0.724213) vs oldopt=(dt=0.32,rms=0.724226)
#GCMRL# 70 dt 0.384000 rms 0.724 0.000% neg 0 invalid 762 tFOTS 15.9550 tGradient 4.1070 tsec 21.3690
#GCMRL# 71 dt 0.384000 rms 0.721 0.400% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.0540 tsec 5.3060
#GCMRL# 72 dt 0.384000 rms 0.719 0.358% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.0610 tsec 5.3260
#GCMRL# 73 dt 0.384000 rms 0.717 0.206% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2380 tsec 5.4820
#GCMRL# 74 dt 0.384000 rms 0.716 0.224% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1400 tsec 5.4050
#GCMRL# 75 dt 0.384000 rms 0.714 0.168% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2420 tsec 5.5000
#GCMRL# 76 dt 0.384000 rms 0.714 -0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1010 tsec 6.5340
#FOTS# QuadFit found better minimum quadopt=(dt=0.768,rms=0.713161) vs oldopt=(dt=1.28,rms=0.713384)
#GCMRL# 77 dt 0.768000 rms 0.713 0.176% neg 0 invalid 762 tFOTS 16.0640 tGradient 4.1110 tsec 21.4510
#FOTS# QuadFit found better minimum quadopt=(dt=0.028,rms=0.709522) vs oldopt=(dt=0.02,rms=0.709528)
#GCMRL# 79 dt 0.028000 rms 0.710 0.000% neg 0 invalid 762 tFOTS 16.0370 tGradient 4.2460 tsec 21.5970
#GCMRL# 80 dt 0.028000 rms 0.710 0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2800 tsec 5.5530
#GCAMreg# pass 0 level1 1 level2 1 tsec 157.338 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=20.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.709986
#FOTS# QuadFit found better minimum quadopt=(dt=0.028,rms=0.709517) vs oldopt=(dt=0.02,rms=0.709518)
#GCMRL# 82 dt 0.028000 rms 0.710 0.066% neg 0 invalid 762 tFOTS 16.1210 tGradient 4.2890 tsec 21.6770
#FOTS# QuadFit found better minimum quadopt=(dt=0.024,rms=0.676562) vs oldopt=(dt=0.02,rms=0.676563)
#GCMRL# 85 dt 0.024000 rms 0.677 0.000% neg 0 invalid 762 tFOTS 16.1880 tGradient 3.4010 tsec 20.8930
#GCAMreg# pass 0 level1 0 level2 1 tsec 51.918 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=40.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.677038
#FOTS# QuadFit found better minimum quadopt=(dt=0.006,rms=0.676552) vs oldopt=(dt=0.005,rms=0.676552)
#GCMRL# 87 dt 0.006000 rms 0.677 0.072% neg 0 invalid 762 tFOTS 16.1670 tGradient 3.3650 tsec 20.8350
GCAMregister done in 22.8458 min
Starting GCAmapRenormalizeWithAlignment() without scales
renormalizing by structure alignment....
renormalizing input #0
gca peak = 0.10253 (16)
mri peak = 0.24309 ( 7)
Left_Lateral_Ventricle (4): linear fit = 0.32 x + 0.0 (1137 voxels, overlap=0.004)
Left_Lateral_Ventricle (4): linear fit = 0.40 x + 0.0 (1137 voxels, peak = 5), gca=6.4
gca peak = 0.17690 (16)
mri peak = 0.19113 (10)
Right_Lateral_Ventricle (43): linear fit = 0.44 x + 0.0 (993 voxels, overlap=0.172)
Right_Lateral_Ventricle (43): linear fit = 0.44 x + 0.0 (993 voxels, peak = 7), gca=7.0
gca peak = 0.28275 (96)
mri peak = 0.11886 (106)
Right_Pallidum (52): linear fit = 1.10 x + 0.0 (865 voxels, overlap=0.105)
Right_Pallidum (52): linear fit = 1.10 x + 0.0 (865 voxels, peak = 105), gca=105.1
gca peak = 0.18948 (93)
mri peak = 0.10308 (110)
Left_Pallidum (13): linear fit = 1.13 x + 0.0 (790 voxels, overlap=0.030)
Left_Pallidum (13): linear fit = 1.13 x + 0.0 (790 voxels, peak = 106), gca=105.6
gca peak = 0.20755 (55)
mri peak = 0.09290 (56)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (737 voxels, overlap=1.000)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (737 voxels, peak = 55), gca=55.0
gca peak = 0.31831 (58)
mri peak = 0.11220 (60)
Left_Hippocampus (17): linear fit = 1.02 x + 0.0 (713 voxels, overlap=0.999)
Left_Hippocampus (17): linear fit = 1.02 x + 0.0 (713 voxels, peak = 59), gca=59.4
gca peak = 0.11957 (102)
mri peak = 0.13750 (102)
Right_Cerebral_White_Matter (41): linear fit = 1.02 x + 0.0 (61984 voxels, overlap=0.576)
Right_Cerebral_White_Matter (41): linear fit = 1.02 x + 0.0 (61984 voxels, peak = 105), gca=104.5
gca peak = 0.11429 (102)
mri peak = 0.13708 (104)
Left_Cerebral_White_Matter (2): linear fit = 1.02 x + 0.0 (64475 voxels, overlap=0.597)
Left_Cerebral_White_Matter (2): linear fit = 1.02 x + 0.0 (64475 voxels, peak = 105), gca=104.5
gca peak = 0.14521 (59)
mri peak = 0.03230 (63)
Left_Cerebral_Cortex (3): linear fit = 1.08 x + 0.0 (27092 voxels, overlap=0.559)
Left_Cerebral_Cortex (3): linear fit = 1.08 x + 0.0 (27092 voxels, peak = 63), gca=63.4
gca peak = 0.14336 (58)
mri peak = 0.03626 (60)
Right_Cerebral_Cortex (42): linear fit = 1.08 x + 0.0 (27239 voxels, overlap=0.765)
Right_Cerebral_Cortex (42): linear fit = 1.08 x + 0.0 (27239 voxels, peak = 62), gca=62.4
gca peak = 0.13305 (70)
mri peak = 0.12909 (75)
Right_Caudate (50): linear fit = 1.08 x + 0.0 (602 voxels, overlap=0.560)
Right_Caudate (50): linear fit = 1.08 x + 0.0 (602 voxels, peak = 75), gca=75.2
gca peak = 0.15761 (71)
mri peak = 0.11149 (78)
Left_Caudate (11): linear fit = 1.03 x + 0.0 (884 voxels, overlap=0.757)
Left_Caudate (11): linear fit = 1.03 x + 0.0 (884 voxels, peak = 73), gca=73.5
gca peak = 0.19040 (84)
mri peak = 0.05880 (87)
Left_Cerebellum_White_Matter (7): linear fit = 1.07 x + 0.0 (6844 voxels, overlap=0.799)
Left_Cerebellum_White_Matter (7): linear fit = 1.07 x + 0.0 (6844 voxels, peak = 89), gca=89.5
gca peak = 0.18871 (83)
mri peak = 0.07503 (88)
Right_Cerebellum_White_Matter (46): linear fit = 1.05 x + 0.0 (6722 voxels, overlap=0.828)
Right_Cerebellum_White_Matter (46): linear fit = 1.05 x + 0.0 (6722 voxels, peak = 88), gca=87.6
gca peak = 0.24248 (57)
mri peak = 0.09624 (65)
Left_Amygdala (18): linear fit = 1.10 x + 0.0 (430 voxels, overlap=1.007)
Left_Amygdala (18): linear fit = 1.10 x + 0.0 (430 voxels, peak = 62), gca=62.4
gca peak = 0.35833 (56)
mri peak = 0.10251 (58)
Right_Amygdala (54): linear fit = 1.04 x + 0.0 (501 voxels, overlap=0.958)
Right_Amygdala (54): linear fit = 1.04 x + 0.0 (501 voxels, peak = 59), gca=58.5
gca peak = 0.12897 (85)
mri peak = 0.06635 (91)
Left_Thalamus (10): linear fit = 1.08 x + 0.0 (5328 voxels, overlap=0.630)
Left_Thalamus (10): linear fit = 1.08 x + 0.0 (5328 voxels, peak = 91), gca=91.4
gca peak = 0.13127 (83)
mri peak = 0.06776 (89)
Right_Thalamus (49): linear fit = 1.12 x + 0.0 (4325 voxels, overlap=0.655)
Right_Thalamus (49): linear fit = 1.12 x + 0.0 (4325 voxels, peak = 93), gca=92.5
gca peak = 0.12974 (78)
mri peak = 0.07627 (84)
Left_Putamen (12): linear fit = 1.12 x + 0.0 (2361 voxels, overlap=0.481)
Left_Putamen (12): linear fit = 1.12 x + 0.0 (2361 voxels, peak = 87), gca=87.0
gca peak = 0.17796 (79)
mri peak = 0.09075 (90)
Right_Putamen (51): linear fit = 1.12 x + 0.0 (2325 voxels, overlap=0.594)
Right_Putamen (51): linear fit = 1.12 x + 0.0 (2325 voxels, peak = 89), gca=88.9
gca peak = 0.10999 (80)
mri peak = 0.06213 (91)
Brain_Stem (16): linear fit = 1.21 x + 0.0 (12102 voxels, overlap=0.417)
Brain_Stem (16): linear fit = 1.21 x + 0.0 (12102 voxels, peak = 96), gca=96.4
gca peak = 0.13215 (88)
mri peak = 0.08770 (101)
Right_VentralDC (60): linear fit = 1.21 x + 0.0 (1248 voxels, overlap=0.036)
Right_VentralDC (60): linear fit = 1.21 x + 0.0 (1248 voxels, peak = 106), gca=106.0
gca peak = 0.11941 (89)
mri peak = 0.09670 (101)
Left_VentralDC (28): linear fit = 1.18 x + 0.0 (1470 voxels, overlap=0.014)
Left_VentralDC (28): linear fit = 1.18 x + 0.0 (1470 voxels, peak = 105), gca=105.5
gca peak = 0.20775 (25)
mri peak = 0.25000 ( 7)
gca peak = 0.13297 (21)
mri peak = 0.54545 ( 6)
gca peak Unknown = 0.94777 ( 0)
gca peak Left_Inf_Lat_Vent = 0.19087 (28)
gca peak Third_Ventricle = 0.20775 (25)
gca peak Fourth_Ventricle = 0.13297 (21)
gca peak CSF = 0.16821 (33)
gca peak Left_Accumbens_area = 0.32850 (63)
gca peak Left_undetermined = 0.98480 (28)
gca peak Left_vessel = 0.40887 (53)
gca peak Left_choroid_plexus = 0.10898 (46)
gca peak Right_Inf_Lat_Vent = 0.17798 (26)
gca peak Right_Accumbens_area = 0.30137 (64)
gca peak Right_vessel = 0.47828 (52)
gca peak Right_choroid_plexus = 0.11612 (45)
gca peak Fifth_Ventricle = 0.59466 (35)
gca peak WM_hypointensities = 0.10053 (78)
gca peak non_WM_hypointensities = 0.07253 (60)
gca peak Optic_Chiasm = 0.25330 (73)
not using caudate to estimate GM means
estimating mean gm scale to be 1.05 x + 0.0
estimating mean wm scale to be 1.02 x + 0.0
estimating mean csf scale to be 0.42 x + 0.0
Left_Pallidum too bright - rescaling by 0.961 (from 1.135) to 101.4 (was 105.6)
Right_Pallidum too bright - rescaling by 0.965 (from 1.095) to 101.4 (was 105.1)
saving intensity scales to talairach.label_intensities.txt
GCAmapRenormalizeWithAlignment() took 4.17185 min
noneg pre
Starting GCAMregister()
label assignment complete, 0 changed (0.00%)
npasses = 1, nlevels = 6
#pass# 1 of 1 ************************
enabling zero nodes
setting smoothness cost coefficient to 0.008
#GCAMreg# pass 0 level1 5 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=2.0,type=2, relabel=0, neg=no
#FOTS# QuadFit found better minimum quadopt=(dt=295.936,rms=0.660309) vs oldopt=(dt=369.92,rms=0.660784)
#GCMRL# 91 dt 295.936000 rms 0.660 1.248% neg 0 invalid 762 tFOTS 15.9640 tGradient 6.6800 tsec 23.8960
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.656212) vs oldopt=(dt=92.48,rms=0.656504)
#GCMRL# 92 dt 129.472000 rms 0.656 0.620% neg 0 invalid 762 tFOTS 16.8890 tGradient 6.8300 tsec 24.9680
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.653722) vs oldopt=(dt=92.48,rms=0.654172)
#GCMRL# 93 dt 129.472000 rms 0.654 0.379% neg 0 invalid 762 tFOTS 16.9130 tGradient 6.6120 tsec 24.7670
#GCMRL# 95 dt 92.480000 rms 0.647 0.487% neg 0 invalid 762 tFOTS 17.8740 tGradient 6.6020 tsec 25.7190
#FOTS# QuadFit found better minimum quadopt=(dt=887.808,rms=0.641934) vs oldopt=(dt=1479.68,rms=0.643048)
#GCMRL# 96 dt 887.808000 rms 0.642 0.777% neg 0 invalid 762 tFOTS 16.9270 tGradient 6.6650 tsec 24.8360
#FOTS# QuadFit found better minimum quadopt=(dt=78.9173,rms=0.639691) vs oldopt=(dt=92.48,rms=0.639697)
#GCMRL# 97 dt 78.917293 rms 0.640 0.349% neg 0 invalid 762 tFOTS 17.9570 tGradient 6.5960 tsec 25.8250
#FOTS# QuadFit found better minimum quadopt=(dt=110.976,rms=0.634966) vs oldopt=(dt=92.48,rms=0.635015)
#GCMRL# 99 dt 110.976000 rms 0.635 0.381% neg 0 invalid 762 tFOTS 18.3890 tGradient 7.1700 tsec 26.8140
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.634523) vs oldopt=(dt=92.48,rms=0.634569)
#GCMRL# 100 dt 129.472000 rms 0.635 0.070% neg 0 invalid 762 tFOTS 17.1080 tGradient 6.8190 tsec 25.1930
#FOTS# QuadFit found better minimum quadopt=(dt=73.984,rms=0.633149) vs oldopt=(dt=92.48,rms=0.633153)
#GCMRL# 102 dt 73.984000 rms 0.633 0.090% neg 0 invalid 762 tFOTS 17.0570 tGradient 6.6360 tsec 24.9600
#FOTS# QuadFit found better minimum quadopt=(dt=1183.74,rms=0.631797) vs oldopt=(dt=1479.68,rms=0.631966)
#GCMRL# 103 dt 1183.744000 rms 0.632 0.214% neg 0 invalid 762 tFOTS 18.6300 tGradient 6.7620 tsec 26.6450
#GCMRL# 104 dt 92.480000 rms 0.630 0.249% neg 0 invalid 762 tFOTS 18.5320 tGradient 6.9830 tsec 26.7590
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.629375) vs oldopt=(dt=92.48,rms=0.62943)
#GCMRL# 106 dt 129.472000 rms 0.629 0.083% neg 0 invalid 762 tFOTS 16.9740 tGradient 6.8530 tsec 25.0790
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.629167) vs oldopt=(dt=92.48,rms=0.629196)
#GCMRL# 107 dt 129.472000 rms 0.629 0.000% neg 0 invalid 762 tFOTS 17.9730 tGradient 6.5530 tsec 25.8170
#GCMRL# 108 dt 129.472000 rms 0.629 0.049% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5530 tsec 7.8030
#GCMRL# 109 dt 129.472000 rms 0.628 0.070% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6560 tsec 7.9050
#GCMRL# 110 dt 129.472000 rms 0.628 0.092% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7560 tsec 8.0080
#GCMRL# 114 dt 129.472000 rms 0.624 0.148% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6980 tsec 7.9540
#GCMRL# 115 dt 129.472000 rms 0.624 0.149% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6220 tsec 7.8820
#GCMRL# 116 dt 129.472000 rms 0.623 0.140% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6570 tsec 7.9050
#GCMRL# 117 dt 129.472000 rms 0.622 0.155% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.9790 tsec 8.2280
#GCMRL# 118 dt 129.472000 rms 0.621 0.150% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.9670 tsec 8.2180
#GCMRL# 119 dt 129.472000 rms 0.620 0.129% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7710 tsec 8.0210
#GCMRL# 120 dt 129.472000 rms 0.619 0.139% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7000 tsec 7.9470
#GCMRL# 121 dt 129.472000 rms 0.618 0.141% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5580 tsec 7.8030
#GCMRL# 124 dt 129.472000 rms 0.616 0.120% neg 0 invalid 762 tFOTS 0.0000 tGradient 7.0110 tsec 8.2710
#GCMRL# 125 dt 129.472000 rms 0.615 0.142% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6630 tsec 7.9060
#GCMRL# 126 dt 129.472000 rms 0.614 0.137% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5570 tsec 7.8000
#GCMRL# 127 dt 129.472000 rms 0.613 0.134% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6260 tsec 7.8780
#GCMRL# 128 dt 129.472000 rms 0.613 0.117% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5970 tsec 7.8440
#GCMRL# 129 dt 129.472000 rms 0.612 0.100% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6420 tsec 7.8900
#GCMRL# 130 dt 129.472000 rms 0.611 0.124% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.8710 tsec 8.1210
#GCMRL# 131 dt 129.472000 rms 0.611 0.119% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7460 tsec 7.9900
#GCMRL# 132 dt 129.472000 rms 0.610 0.108% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5330 tsec 7.7790
#GCMRL# 133 dt 129.472000 rms 0.609 0.100% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5260 tsec 7.7760
#GCMRL# 135 dt 129.472000 rms 0.608 0.099% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6590 tsec 7.9040
#GCMRL# 136 dt 129.472000 rms 0.608 0.084% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6990 tsec 7.9460
#GCMRL# 137 dt 129.472000 rms 0.607 0.081% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5700 tsec 7.8090
#GCMRL# 138 dt 129.472000 rms 0.607 0.070% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6020 tsec 7.8420
#GCMRL# 139 dt 129.472000 rms 0.606 0.074% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6570 tsec 7.8990
#GCMRL# 140 dt 129.472000 rms 0.606 0.088% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5970 tsec 7.8360
#GCMRL# 141 dt 129.472000 rms 0.605 0.074% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5370 tsec 7.7780
#GCMRL# 142 dt 129.472000 rms 0.605 0.070% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6640 tsec 7.9070
#GCMRL# 143 dt 129.472000 rms 0.605 0.063% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5020 tsec 7.7410
#GCMRL# 144 dt 129.472000 rms 0.604 0.079% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6200 tsec 7.8630
#GCMRL# 145 dt 129.472000 rms 0.604 0.074% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5390 tsec 7.7760
#GCMRL# 150 dt 129.472000 rms 0.602 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5970 tsec 7.8390
#GCMRL# 151 dt 129.472000 rms 0.602 0.047% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6060 tsec 7.8580
#GCMRL# 152 dt 129.472000 rms 0.601 0.057% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6940 tsec 7.9450
#GCMRL# 153 dt 129.472000 rms 0.601 0.058% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5240 tsec 7.7760
#GCMRL# 154 dt 129.472000 rms 0.601 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7520 tsec 8.0090
#GCMRL# 155 dt 129.472000 rms 0.600 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6730 tsec 7.9250
#GCMRL# 156 dt 129.472000 rms 0.600 0.057% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7440 tsec 7.9970
#GCMRL# 157 dt 129.472000 rms 0.600 0.054% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7600 tsec 8.0130
#GCMRL# 159 dt 129.472000 rms 0.599 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6480 tsec 8.2200
#GCMRL# 160 dt 129.472000 rms 0.599 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7430 tsec 8.0080
#GCMRL# 161 dt 129.472000 rms 0.599 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6420 tsec 7.8840
#GCMRL# 162 dt 129.472000 rms 0.599 0.044% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7830 tsec 8.0250
#GCMRL# 163 dt 129.472000 rms 0.598 0.051% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6660 tsec 7.9250
#GCMRL# 164 dt 129.472000 rms 0.598 0.042% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7260 tsec 7.9790
#GCMRL# 165 dt 129.472000 rms 0.598 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6680 tsec 7.9210
#GCMRL# 166 dt 129.472000 rms 0.598 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6650 tsec 7.9240
#GCMRL# 167 dt 129.472000 rms 0.597 0.028% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6720 tsec 7.9250
#GCMRL# 168 dt 129.472000 rms 0.597 0.040% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6700 tsec 7.9270
#GCMRL# 169 dt 129.472000 rms 0.597 0.047% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6490 tsec 7.9120
#GCMRL# 173 dt 129.472000 rms 0.596 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6790 tsec 7.9380
#GCMRL# 174 dt 129.472000 rms 0.596 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5310 tsec 7.7860
#GCMRL# 175 dt 129.472000 rms 0.596 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7970 tsec 8.0670
#GCMRL# 176 dt 129.472000 rms 0.595 0.033% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6290 tsec 7.8860
#GCMRL# 177 dt 129.472000 rms 0.595 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6200 tsec 7.8630
#GCMRL# 178 dt 129.472000 rms 0.595 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7460 tsec 7.9920
#GCMRL# 179 dt 129.472000 rms 0.595 0.028% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7250 tsec 7.9740
#GCMRL# 180 dt 129.472000 rms 0.595 0.027% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6520 tsec 7.8950
#GCMRL# 183 dt 129.472000 rms 0.594 0.029% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5770 tsec 7.8290
#GCMRL# 184 dt 129.472000 rms 0.594 0.026% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6310 tsec 7.8930
#GCMRL# 185 dt 129.472000 rms 0.594 0.041% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5600 tsec 7.8010
#GCMRL# 186 dt 129.472000 rms 0.594 0.027% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6720 tsec 7.9320
#GCMRL# 187 dt 129.472000 rms 0.594 0.027% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7160 tsec 7.9640
#GCMRL# 188 dt 129.472000 rms 0.593 0.028% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5380 tsec 7.7820
#GCMRL# 189 dt 129.472000 rms 0.593 0.019% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5570 tsec 7.8060
#GCMRL# 190 dt 129.472000 rms 0.593 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6330 tsec 7.8870
#GCMRL# 191 dt 129.472000 rms 0.593 0.024% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5710 tsec 7.8140
#GCMRL# 192 dt 129.472000 rms 0.593 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6500 tsec 7.9130
#GCMRL# 196 dt 129.472000 rms 0.592 0.021% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.5300 tsec 7.8230
#FOTS# QuadFit found better minimum quadopt=(dt=517.888,rms=0.592305) vs oldopt=(dt=369.92,rms=0.592314)
#GCMRL# 197 dt 517.888000 rms 0.592 0.000% neg 0 invalid 762 tFOTS 17.8930 tGradient 6.5410 tsec 25.7320
#GCAMreg# pass 0 level1 5 level2 1 tsec 1201.04 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.592976
#GCMRL# 199 dt 369.920000 rms 0.591 0.362% neg 0 invalid 762 tFOTS 17.1920 tGradient 6.5380 tsec 24.9800
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.590529) vs oldopt=(dt=92.48,rms=0.590554)
#GCMRL# 200 dt 129.472000 rms 0.591 0.051% neg 0 invalid 762 tFOTS 16.9540 tGradient 6.6310 tsec 24.8360
#FOTS# QuadFit found better minimum quadopt=(dt=517.888,rms=0.590044) vs oldopt=(dt=369.92,rms=0.59011)
#GCMRL# 201 dt 517.888000 rms 0.590 0.082% neg 0 invalid 762 tFOTS 17.9300 tGradient 6.5560 tsec 25.7390
#FOTS# QuadFit found better minimum quadopt=(dt=129.472,rms=0.589824) vs oldopt=(dt=92.48,rms=0.589841)
#GCMRL# 202 dt 129.472000 rms 0.590 0.000% neg 0 invalid 762 tFOTS 16.9950 tGradient 6.6410 tsec 24.9370
#GCMRL# 203 dt 129.472000 rms 0.590 0.042% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6130 tsec 7.8680
#GCMRL# 204 dt 129.472000 rms 0.589 0.036% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6690 tsec 7.9200
#GCMRL# 205 dt 129.472000 rms 0.589 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6750 tsec 7.9230
#GCMRL# 206 dt 129.472000 rms 0.589 0.031% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6780 tsec 7.9270
#GCMRL# 207 dt 129.472000 rms 0.589 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6790 tsec 7.9340
#GCMRL# 208 dt 129.472000 rms 0.588 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7360 tsec 7.9860
#GCMRL# 209 dt 129.472000 rms 0.588 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7770 tsec 8.0320
#GCMRL# 210 dt 129.472000 rms 0.588 0.021% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7400 tsec 7.9920
#GCMRL# 211 dt 129.472000 rms 0.588 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6450 tsec 7.8920
#GCMRL# 212 dt 129.472000 rms 0.588 0.036% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7810 tsec 8.0360
#GCMRL# 213 dt 129.472000 rms 0.588 0.024% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7120 tsec 7.9590
#GCMRL# 214 dt 129.472000 rms 0.587 0.012% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6810 tsec 7.9730
#GCMRL# 215 dt 369.920000 rms 0.587 0.023% neg 0 invalid 762 tFOTS 17.8600 tGradient 6.7670 tsec 25.8680
#FOTS# QuadFit found better minimum quadopt=(dt=32.368,rms=0.587309) vs oldopt=(dt=23.12,rms=0.587313)
#GCMRL# 216 dt 32.368000 rms 0.587 0.000% neg 0 invalid 762 tFOTS 18.7680 tGradient 6.7270 tsec 26.7760
#GCMRL# 217 dt 32.368000 rms 0.587 0.002% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.7400 tsec 7.9760
#GCMRL# 218 dt 32.368000 rms 0.587 0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6390 tsec 7.8790
setting smoothness cost coefficient to 0.031
#GCAMreg# pass 0 level1 4 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.58983
#FOTS# QuadFit found better minimum quadopt=(dt=156.781,rms=0.585278) vs oldopt=(dt=103.68,rms=0.585659)
#GCMRL# 220 dt 156.781065 rms 0.585 0.772% neg 0 invalid 762 tFOTS 16.8100 tGradient 5.1960 tsec 23.2480
#FOTS# QuadFit found better minimum quadopt=(dt=207.515,rms=0.577415) vs oldopt=(dt=103.68,rms=0.579197)
#GCMRL# 221 dt 207.515152 rms 0.577 1.344% neg 0 invalid 762 tFOTS 16.8240 tGradient 5.0800 tsec 23.1450
#FOTS# QuadFit found better minimum quadopt=(dt=64,rms=0.573321) vs oldopt=(dt=25.92,rms=0.574334)
#GCMRL# 222 dt 64.000000 rms 0.573 0.709% neg 0 invalid 762 tFOTS 16.8120 tGradient 5.2430 tsec 23.2910
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.570234) vs oldopt=(dt=103.68,rms=0.570602)
#GCMRL# 223 dt 145.152000 rms 0.570 0.538% neg 0 invalid 762 tFOTS 15.8190 tGradient 5.1590 tsec 22.2150
#FOTS# QuadFit found better minimum quadopt=(dt=62.208,rms=0.567687) vs oldopt=(dt=103.68,rms=0.568579)
#GCMRL# 224 dt 62.208000 rms 0.568 0.447% neg 0 invalid 762 tFOTS 16.8080 tGradient 5.1310 tsec 23.1780
#FOTS# QuadFit found better minimum quadopt=(dt=113.672,rms=0.565925) vs oldopt=(dt=103.68,rms=0.565929)
#GCMRL# 225 dt 113.671642 rms 0.566 0.310% neg 0 invalid 762 tFOTS 16.8350 tGradient 5.1700 tsec 23.2500
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.564047) vs oldopt=(dt=25.92,rms=0.564412)
#GCMRL# 226 dt 36.288000 rms 0.564 0.332% neg 0 invalid 762 tFOTS 16.9020 tGradient 5.2770 tsec 23.4230
#FOTS# QuadFit found better minimum quadopt=(dt=580.608,rms=0.554882) vs oldopt=(dt=414.72,rms=0.556925)
#GCMRL# 227 dt 580.608000 rms 0.555 1.625% neg 0 invalid 762 tFOTS 15.9520 tGradient 5.2270 tsec 22.4400
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.55334) vs oldopt=(dt=25.92,rms=0.553672)
#GCMRL# 228 dt 36.288000 rms 0.553 0.278% neg 0 invalid 762 tFOTS 17.0050 tGradient 5.3410 tsec 23.5940
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.551974) vs oldopt=(dt=103.68,rms=0.552189)
#GCMRL# 229 dt 145.152000 rms 0.552 0.247% neg 0 invalid 762 tFOTS 17.0310 tGradient 5.2430 tsec 23.5290
#FOTS# QuadFit found better minimum quadopt=(dt=77.848,rms=0.550892) vs oldopt=(dt=103.68,rms=0.550996)
#GCMRL# 230 dt 77.847953 rms 0.551 0.196% neg 0 invalid 762 tFOTS 18.0110 tGradient 5.3220 tsec 24.5920
#FOTS# QuadFit found better minimum quadopt=(dt=82.944,rms=0.550122) vs oldopt=(dt=103.68,rms=0.550172)
#GCMRL# 231 dt 82.944000 rms 0.550 0.140% neg 0 invalid 762 tFOTS 17.0410 tGradient 5.3260 tsec 23.6170
#FOTS# QuadFit found better minimum quadopt=(dt=79.4161,rms=0.549216) vs oldopt=(dt=103.68,rms=0.549297)
#GCMRL# 232 dt 79.416058 rms 0.549 0.165% neg 0 invalid 762 tFOTS 18.0380 tGradient 5.2380 tsec 24.5270
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.548731) vs oldopt=(dt=25.92,rms=0.548831)
#GCMRL# 233 dt 36.288000 rms 0.549 0.088% neg 0 invalid 762 tFOTS 16.0120 tGradient 5.2680 tsec 22.5290
#FOTS# QuadFit found better minimum quadopt=(dt=580.608,rms=0.545387) vs oldopt=(dt=414.72,rms=0.546)
#GCMRL# 234 dt 580.608000 rms 0.545 0.609% neg 0 invalid 762 tFOTS 17.0190 tGradient 5.2010 tsec 23.4840
#FOTS# QuadFit found better minimum quadopt=(dt=67.2593,rms=0.543626) vs oldopt=(dt=103.68,rms=0.544153)
#GCMRL# 235 dt 67.259259 rms 0.544 0.323% neg 0 invalid 762 tFOTS 16.9810 tGradient 5.3070 tsec 23.5470
#FOTS# QuadFit found better minimum quadopt=(dt=82.944,rms=0.543101) vs oldopt=(dt=103.68,rms=0.543167)
#GCMRL# 236 dt 82.944000 rms 0.543 0.097% neg 0 invalid 762 tFOTS 17.0900 tGradient 5.2170 tsec 23.5620
#FOTS# QuadFit found better minimum quadopt=(dt=82.944,rms=0.542423) vs oldopt=(dt=103.68,rms=0.542478)
#GCMRL# 237 dt 82.944000 rms 0.542 0.125% neg 0 invalid 762 tFOTS 17.0720 tGradient 5.3040 tsec 23.6330
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.542141) vs oldopt=(dt=25.92,rms=0.542193)
#GCMRL# 238 dt 36.288000 rms 0.542 0.052% neg 0 invalid 762 tFOTS 17.1160 tGradient 5.2970 tsec 23.6820
#GCMRL# 239 dt 1658.880000 rms 0.537 0.923% neg 0 invalid 762 tFOTS 17.0360 tGradient 5.2470 tsec 23.5370
#FOTS# QuadFit found better minimum quadopt=(dt=69.8182,rms=0.534) vs oldopt=(dt=103.68,rms=0.534844)
#GCMRL# 240 dt 69.818182 rms 0.534 0.584% neg 0 invalid 762 tFOTS 17.1380 tGradient 5.2920 tsec 23.6920
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.533615) vs oldopt=(dt=25.92,rms=0.533669)
#GCMRL# 241 dt 36.288000 rms 0.534 0.072% neg 0 invalid 762 tFOTS 17.0320 tGradient 5.3050 tsec 23.5870
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.533199) vs oldopt=(dt=103.68,rms=0.533246)
#GCMRL# 242 dt 145.152000 rms 0.533 0.078% neg 0 invalid 762 tFOTS 16.9750 tGradient 5.2070 tsec 23.4430
#FOTS# QuadFit found better minimum quadopt=(dt=82.944,rms=0.532682) vs oldopt=(dt=103.68,rms=0.532732)
#GCMRL# 243 dt 82.944000 rms 0.533 0.097% neg 0 invalid 762 tFOTS 17.0070 tGradient 5.1780 tsec 23.4490
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.532493) vs oldopt=(dt=25.92,rms=0.532519)
#GCMRL# 244 dt 36.288000 rms 0.532 0.000% neg 0 invalid 762 tFOTS 17.0200 tGradient 5.2140 tsec 23.5340
#GCMRL# 245 dt 36.288000 rms 0.532 0.026% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2000 tsec 6.4450
#GCMRL# 246 dt 36.288000 rms 0.532 0.033% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2040 tsec 6.4560
#GCMRL# 247 dt 36.288000 rms 0.532 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2070 tsec 6.4580
#GCMRL# 248 dt 36.288000 rms 0.532 0.069% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2440 tsec 6.5100
#GCMRL# 249 dt 36.288000 rms 0.531 0.079% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2200 tsec 6.4730
#GCMRL# 250 dt 36.288000 rms 0.531 0.081% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3700 tsec 6.6230
#GCMRL# 251 dt 36.288000 rms 0.530 0.074% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0920 tsec 6.3430
#GCMRL# 252 dt 36.288000 rms 0.530 0.082% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1240 tsec 6.3710
#GCMRL# 253 dt 36.288000 rms 0.529 0.094% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2990 tsec 6.5550
#GCMRL# 254 dt 36.288000 rms 0.529 0.118% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2300 tsec 6.4890
#GCMRL# 255 dt 36.288000 rms 0.528 0.116% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1170 tsec 6.3750
#GCMRL# 256 dt 36.288000 rms 0.527 0.121% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2280 tsec 6.4870
#GCMRL# 257 dt 36.288000 rms 0.527 0.121% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2460 tsec 6.4890
#GCMRL# 258 dt 36.288000 rms 0.526 0.106% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2530 tsec 6.5140
#GCMRL# 259 dt 36.288000 rms 0.526 0.110% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2150 tsec 6.4610
#GCMRL# 260 dt 36.288000 rms 0.525 0.107% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1870 tsec 6.4320
#GCMRL# 261 dt 36.288000 rms 0.525 0.108% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2190 tsec 6.4860
#GCMRL# 262 dt 36.288000 rms 0.524 0.094% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3120 tsec 6.5560
#GCMRL# 263 dt 36.288000 rms 0.524 0.088% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2230 tsec 6.4760
#GCMRL# 264 dt 36.288000 rms 0.523 0.098% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2930 tsec 6.5710
#GCMRL# 265 dt 36.288000 rms 0.523 0.092% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2700 tsec 6.5540
#GCMRL# 266 dt 36.288000 rms 0.522 0.090% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1240 tsec 6.3790
#GCMRL# 267 dt 36.288000 rms 0.522 0.093% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1400 tsec 6.4090
#GCMRL# 268 dt 36.288000 rms 0.521 0.096% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2660 tsec 6.5390
#GCMRL# 269 dt 36.288000 rms 0.521 0.084% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2610 tsec 6.5060
#GCMRL# 270 dt 36.288000 rms 0.520 0.075% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2880 tsec 6.5330
#GCMRL# 271 dt 36.288000 rms 0.520 0.060% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1180 tsec 6.3630
#GCMRL# 272 dt 36.288000 rms 0.520 0.076% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2570 tsec 6.4980
#GCMRL# 273 dt 36.288000 rms 0.519 0.083% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0510 tsec 6.2900
#GCMRL# 274 dt 36.288000 rms 0.519 0.080% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0920 tsec 6.3270
#GCMRL# 275 dt 36.288000 rms 0.518 0.068% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1890 tsec 6.4280
#GCMRL# 276 dt 36.288000 rms 0.518 0.059% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2790 tsec 6.5140
#GCMRL# 277 dt 36.288000 rms 0.518 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1830 tsec 6.4210
#GCMRL# 278 dt 36.288000 rms 0.518 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1860 tsec 6.4210
#GCMRL# 279 dt 36.288000 rms 0.517 0.066% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1420 tsec 6.3830
#GCMRL# 280 dt 36.288000 rms 0.517 0.073% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1940 tsec 6.4320
#GCMRL# 281 dt 36.288000 rms 0.517 0.058% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2340 tsec 6.4740
#GCMRL# 282 dt 36.288000 rms 0.516 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1810 tsec 6.4150
#GCMRL# 283 dt 36.288000 rms 0.516 0.059% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2640 tsec 6.5140
#GCMRL# 284 dt 36.288000 rms 0.516 0.064% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1840 tsec 6.4240
#GCMRL# 285 dt 36.288000 rms 0.515 0.051% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2100 tsec 6.4500
#GCMRL# 286 dt 36.288000 rms 0.515 0.042% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2020 tsec 6.4380
#GCMRL# 287 dt 36.288000 rms 0.515 0.033% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1470 tsec 6.3880
#GCMRL# 288 dt 36.288000 rms 0.515 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1400 tsec 6.3920
#GCMRL# 289 dt 36.288000 rms 0.515 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2460 tsec 6.5020
#GCMRL# 290 dt 36.288000 rms 0.514 0.040% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1530 tsec 6.4050
#GCMRL# 291 dt 36.288000 rms 0.514 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1840 tsec 6.4300
#GCMRL# 292 dt 36.288000 rms 0.514 0.040% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2350 tsec 6.4930
#GCMRL# 293 dt 36.288000 rms 0.514 0.047% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2000 tsec 6.4550
#GCMRL# 294 dt 36.288000 rms 0.514 0.048% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1920 tsec 6.4440
#GCMRL# 295 dt 36.288000 rms 0.513 0.040% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2650 tsec 6.5300
#GCMRL# 296 dt 36.288000 rms 0.513 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2420 tsec 6.4920
#GCMRL# 297 dt 36.288000 rms 0.513 0.040% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2320 tsec 6.4830
#GCMRL# 298 dt 36.288000 rms 0.513 0.041% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1760 tsec 6.4350
#GCMRL# 299 dt 36.288000 rms 0.513 0.027% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2140 tsec 6.4680
#GCMRL# 300 dt 36.288000 rms 0.512 0.038% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2380 tsec 6.4840
#GCMRL# 301 dt 36.288000 rms 0.512 0.033% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2180 tsec 6.4870
#GCMRL# 302 dt 36.288000 rms 0.512 0.011% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1990 tsec 6.4530
#GCMRL# 303 dt 36.288000 rms 0.512 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3500 tsec 6.6190
#GCMRL# 304 dt 36.288000 rms 0.512 0.029% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1310 tsec 6.3840
#GCMRL# 305 dt 36.288000 rms 0.512 0.024% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3390 tsec 6.6010
#GCMRL# 306 dt 36.288000 rms 0.512 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3030 tsec 6.5550
#GCMRL# 307 dt 36.288000 rms 0.511 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1680 tsec 6.4290
#GCMRL# 308 dt 36.288000 rms 0.511 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2780 tsec 6.5290
#GCMRL# 309 dt 36.288000 rms 0.511 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2090 tsec 6.4700
#GCMRL# 310 dt 36.288000 rms 0.511 0.027% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3220 tsec 6.6110
#GCMRL# 311 dt 36.288000 rms 0.511 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2620 tsec 6.5470
#GCMRL# 312 dt 36.288000 rms 0.511 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3210 tsec 6.6220
#GCMRL# 313 dt 36.288000 rms 0.511 0.006% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2030 tsec 6.4670
#GCMRL# 314 dt 36.288000 rms 0.510 0.018% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1940 tsec 6.4390
#GCMRL# 315 dt 36.288000 rms 0.510 0.012% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1750 tsec 6.4550
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.510125) vs oldopt=(dt=103.68,rms=0.510161)
#GCMRL# 316 dt 145.152000 rms 0.510 0.055% neg 0 invalid 762 tFOTS 16.9560 tGradient 5.1030 tsec 23.3040
#FOTS# QuadFit found better minimum quadopt=(dt=9.072,rms=0.510125) vs oldopt=(dt=6.48,rms=0.510128)
#GCMRL# 317 dt 9.072000 rms 0.510 0.000% neg 0 invalid 762 tFOTS 17.2150 tGradient 5.1320 tsec 24.7680
#GCMRL# 318 dt 9.072000 rms 0.510 0.002% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1100 tsec 6.3720
#GCAMreg# pass 0 level1 4 level2 1 tsec 1111.53 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.510875
#FOTS# QuadFit found better minimum quadopt=(dt=115.469,rms=0.508374) vs oldopt=(dt=103.68,rms=0.508378)
#GCMRL# 320 dt 115.468531 rms 0.508 0.490% neg 0 invalid 762 tFOTS 18.0300 tGradient 5.2090 tsec 24.4970
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.507497) vs oldopt=(dt=25.92,rms=0.507669)
#GCMRL# 321 dt 36.288000 rms 0.507 0.172% neg 0 invalid 762 tFOTS 16.1030 tGradient 5.2060 tsec 22.5740
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.506808) vs oldopt=(dt=103.68,rms=0.506945)
#GCMRL# 322 dt 145.152000 rms 0.507 0.136% neg 0 invalid 762 tFOTS 16.9980 tGradient 5.1270 tsec 23.3760
#FOTS# QuadFit found better minimum quadopt=(dt=82.944,rms=0.506307) vs oldopt=(dt=103.68,rms=0.506332)
#GCMRL# 323 dt 82.944000 rms 0.506 0.099% neg 0 invalid 762 tFOTS 17.0170 tGradient 5.0920 tsec 23.3670
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.506184) vs oldopt=(dt=25.92,rms=0.506193)
#GCMRL# 324 dt 36.288000 rms 0.506 0.000% neg 0 invalid 762 tFOTS 18.0020 tGradient 5.2680 tsec 24.5590
#GCMRL# 325 dt 36.288000 rms 0.506 0.026% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2190 tsec 6.4710
#GCMRL# 326 dt 36.288000 rms 0.506 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0950 tsec 6.3480
#GCMRL# 327 dt 36.288000 rms 0.506 0.038% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0900 tsec 6.3430
#GCMRL# 328 dt 36.288000 rms 0.505 0.054% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1420 tsec 6.3890
#GCMRL# 329 dt 36.288000 rms 0.505 0.069% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1170 tsec 6.3560
#GCMRL# 330 dt 36.288000 rms 0.505 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0270 tsec 6.2740
#GCMRL# 331 dt 36.288000 rms 0.504 0.057% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1210 tsec 6.3590
#GCMRL# 332 dt 36.288000 rms 0.504 0.049% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1020 tsec 6.3440
#GCMRL# 333 dt 36.288000 rms 0.504 0.051% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0180 tsec 6.2630
#GCMRL# 334 dt 36.288000 rms 0.504 0.050% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1380 tsec 6.3940
#GCMRL# 335 dt 36.288000 rms 0.503 0.051% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0460 tsec 6.2830
#GCMRL# 336 dt 36.288000 rms 0.503 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1390 tsec 6.3850
#GCMRL# 337 dt 36.288000 rms 0.503 0.048% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0900 tsec 6.3330
#GCMRL# 338 dt 36.288000 rms 0.503 0.049% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1740 tsec 6.4130
#GCMRL# 339 dt 36.288000 rms 0.502 0.054% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1820 tsec 6.4150
#GCMRL# 340 dt 36.288000 rms 0.502 0.055% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1740 tsec 6.4190
#GCMRL# 341 dt 36.288000 rms 0.502 0.038% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2000 tsec 6.4410
#GCMRL# 342 dt 36.288000 rms 0.502 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1750 tsec 6.4220
#GCMRL# 343 dt 36.288000 rms 0.501 0.044% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1320 tsec 6.3740
#GCMRL# 344 dt 36.288000 rms 0.501 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1380 tsec 6.3790
#GCMRL# 345 dt 36.288000 rms 0.501 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1740 tsec 6.4150
#GCMRL# 346 dt 36.288000 rms 0.501 0.045% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1360 tsec 6.3770
#GCMRL# 347 dt 36.288000 rms 0.501 0.031% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1420 tsec 6.3750
#GCMRL# 348 dt 36.288000 rms 0.500 0.043% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1420 tsec 6.3820
#GCMRL# 349 dt 36.288000 rms 0.500 0.025% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1350 tsec 6.3790
#GCMRL# 350 dt 36.288000 rms 0.500 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1440 tsec 6.3970
#GCMRL# 351 dt 36.288000 rms 0.500 0.033% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1110 tsec 6.3480
#GCMRL# 352 dt 36.288000 rms 0.500 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1390 tsec 6.3780
#GCMRL# 353 dt 36.288000 rms 0.500 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1810 tsec 6.4190
#GCMRL# 354 dt 36.288000 rms 0.500 0.032% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1710 tsec 6.4060
#GCMRL# 355 dt 36.288000 rms 0.499 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1370 tsec 6.3720
#GCMRL# 356 dt 36.288000 rms 0.499 0.031% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2120 tsec 6.4460
#GCMRL# 357 dt 36.288000 rms 0.499 0.031% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2560 tsec 6.4950
#GCMRL# 358 dt 36.288000 rms 0.499 0.037% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1280 tsec 6.3700
#GCMRL# 359 dt 36.288000 rms 0.499 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1180 tsec 6.3550
#GCMRL# 360 dt 36.288000 rms 0.499 0.026% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.3010 tsec 6.5440
#GCMRL# 361 dt 36.288000 rms 0.499 0.029% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1180 tsec 6.3590
#GCMRL# 362 dt 36.288000 rms 0.498 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1180 tsec 6.3640
#GCMRL# 363 dt 36.288000 rms 0.498 0.018% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1570 tsec 6.4350
#FOTS# QuadFit found better minimum quadopt=(dt=62.208,rms=0.498249) vs oldopt=(dt=103.68,rms=0.498259)
#GCMRL# 364 dt 62.208000 rms 0.498 0.000% neg 0 invalid 762 tFOTS 16.8580 tGradient 5.1780 tsec 23.3220
#GCMRL# 365 dt 62.208000 rms 0.498 0.003% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1810 tsec 6.4240
#GCMRL# 366 dt 62.208000 rms 0.498 0.007% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1510 tsec 6.3900
#GCMRL# 367 dt 62.208000 rms 0.498 0.018% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1860 tsec 6.4190
setting smoothness cost coefficient to 0.118
#GCAMreg# pass 0 level1 3 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.509209
#FOTS# QuadFit found better minimum quadopt=(dt=23,rms=0.507552) vs oldopt=(dt=32,rms=0.50775)
#GCMRL# 369 dt 23.000000 rms 0.508 0.325% neg 0 invalid 762 tFOTS 16.8350 tGradient 4.6430 tsec 22.7200
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.506084) vs oldopt=(dt=32,rms=0.506238)
#GCMRL# 370 dt 44.800000 rms 0.506 0.289% neg 0 invalid 762 tFOTS 15.8830 tGradient 4.5410 tsec 21.6610
#FOTS# QuadFit found better minimum quadopt=(dt=94.6087,rms=0.503948) vs oldopt=(dt=32,rms=0.50453)
#GCMRL# 371 dt 94.608696 rms 0.504 0.422% neg 0 invalid 762 tFOTS 16.8400 tGradient 4.5440 tsec 22.6270
#FOTS# QuadFit found better minimum quadopt=(dt=35.2711,rms=0.500346) vs oldopt=(dt=32,rms=0.500346)
#GCMRL# 372 dt 35.271111 rms 0.500 0.715% neg 0 invalid 762 tFOTS 15.8460 tGradient 4.7770 tsec 21.8740
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.498996) vs oldopt=(dt=32,rms=0.499012)
#GCMRL# 373 dt 38.400000 rms 0.499 0.270% neg 0 invalid 762 tFOTS 15.9220 tGradient 4.7070 tsec 21.8750
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.497041) vs oldopt=(dt=32,rms=0.497145)
#GCMRL# 374 dt 38.400000 rms 0.497 0.392% neg 0 invalid 762 tFOTS 15.9870 tGradient 4.6880 tsec 21.9260
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.496371) vs oldopt=(dt=32,rms=0.496441)
#GCMRL# 375 dt 25.600000 rms 0.496 0.135% neg 0 invalid 762 tFOTS 15.9940 tGradient 4.5370 tsec 21.7790
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.49469) vs oldopt=(dt=32,rms=0.495015)
#GCMRL# 376 dt 44.800000 rms 0.495 0.339% neg 0 invalid 762 tFOTS 15.9940 tGradient 4.5450 tsec 21.7900
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.494218) vs oldopt=(dt=32,rms=0.494242)
#GCMRL# 377 dt 25.600000 rms 0.494 0.095% neg 0 invalid 762 tFOTS 15.9980 tGradient 4.7510 tsec 22.0000
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.49293) vs oldopt=(dt=32,rms=0.493188)
#GCMRL# 378 dt 44.800000 rms 0.493 0.261% neg 0 invalid 762 tFOTS 15.9990 tGradient 4.5520 tsec 21.8100
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.492541) vs oldopt=(dt=32,rms=0.492576)
#GCMRL# 379 dt 25.600000 rms 0.493 0.079% neg 0 invalid 762 tFOTS 16.0300 tGradient 4.6090 tsec 21.8920
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.491364) vs oldopt=(dt=32,rms=0.491524)
#GCMRL# 380 dt 44.800000 rms 0.491 0.239% neg 0 invalid 762 tFOTS 17.0100 tGradient 4.5450 tsec 22.8060
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.491054) vs oldopt=(dt=8,rms=0.491117)
#GCMRL# 381 dt 11.200000 rms 0.491 0.063% neg 0 invalid 762 tFOTS 16.0480 tGradient 4.5800 tsec 21.8820
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.490527) vs oldopt=(dt=32,rms=0.490592)
#GCMRL# 382 dt 44.800000 rms 0.491 0.107% neg 0 invalid 762 tFOTS 16.1930 tGradient 4.6170 tsec 22.0620
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.489446) vs oldopt=(dt=32,rms=0.489571)
#GCMRL# 383 dt 44.800000 rms 0.489 0.221% neg 0 invalid 762 tFOTS 17.0670 tGradient 4.6150 tsec 22.9370
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.489184) vs oldopt=(dt=8,rms=0.489243)
#GCMRL# 384 dt 11.200000 rms 0.489 0.053% neg 0 invalid 762 tFOTS 15.8710 tGradient 4.5190 tsec 21.6300
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.488658) vs oldopt=(dt=32,rms=0.48874)
#GCMRL# 385 dt 44.800000 rms 0.489 0.108% neg 0 invalid 762 tFOTS 15.9110 tGradient 4.5060 tsec 21.6580
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.487978) vs oldopt=(dt=32,rms=0.487989)
#GCMRL# 386 dt 38.400000 rms 0.488 0.139% neg 0 invalid 762 tFOTS 16.8680 tGradient 4.5560 tsec 22.6630
#FOTS# QuadFit found better minimum quadopt=(dt=19.2,rms=0.487699) vs oldopt=(dt=32,rms=0.487782)
#GCMRL# 387 dt 19.200000 rms 0.488 0.057% neg 0 invalid 762 tFOTS 15.8600 tGradient 4.4460 tsec 21.5480
#FOTS# QuadFit found better minimum quadopt=(dt=409.6,rms=0.485709) vs oldopt=(dt=512,rms=0.485764)
#GCMRL# 388 dt 409.600000 rms 0.486 0.408% neg 0 invalid 762 tFOTS 16.8680 tGradient 4.5420 tsec 22.6490
#GCMRL# 389 dt 32.000000 rms 0.484 0.317% neg 0 invalid 762 tFOTS 15.8540 tGradient 5.2530 tsec 22.3560
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.483185) vs oldopt=(dt=32,rms=0.483254)
#GCMRL# 390 dt 38.400000 rms 0.483 0.204% neg 0 invalid 762 tFOTS 16.8540 tGradient 5.1720 tsec 23.2750
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.482337) vs oldopt=(dt=32,rms=0.482434)
#GCMRL# 391 dt 44.800000 rms 0.482 0.176% neg 0 invalid 762 tFOTS 16.0880 tGradient 4.6370 tsec 21.9810
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.481884) vs oldopt=(dt=32,rms=0.481886)
#GCMRL# 392 dt 25.600000 rms 0.482 0.094% neg 0 invalid 762 tFOTS 16.1050 tGradient 5.0780 tsec 22.4350
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.481628) vs oldopt=(dt=32,rms=0.481662)
#GCMRL# 393 dt 44.800000 rms 0.482 0.053% neg 0 invalid 762 tFOTS 16.0620 tGradient 4.4960 tsec 21.8050
#GCMRL# 394 dt 32.000000 rms 0.481 0.053% neg 0 invalid 762 tFOTS 16.9060 tGradient 4.4190 tsec 22.5770
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.481192) vs oldopt=(dt=32,rms=0.481214)
#GCMRL# 395 dt 25.600000 rms 0.481 0.000% neg 0 invalid 762 tFOTS 17.0160 tGradient 4.5510 tsec 22.8630
#GCMRL# 396 dt 25.600000 rms 0.481 0.015% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.9020 tsec 6.1520
#GCMRL# 397 dt 25.600000 rms 0.481 0.073% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4190 tsec 5.6710
#GCMRL# 398 dt 25.600000 rms 0.480 0.098% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4640 tsec 5.7160
#GCMRL# 399 dt 25.600000 rms 0.480 0.081% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5390 tsec 5.7920
#GCMRL# 400 dt 25.600000 rms 0.479 0.108% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5690 tsec 5.8200
#GCMRL# 401 dt 25.600000 rms 0.479 0.116% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.9840 tsec 6.2500
#GCMRL# 402 dt 25.600000 rms 0.478 0.131% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5760 tsec 5.8290
#GCMRL# 403 dt 25.600000 rms 0.477 0.155% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5380 tsec 5.7990
#GCMRL# 404 dt 25.600000 rms 0.477 0.112% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5280 tsec 5.7750
#GCMRL# 405 dt 25.600000 rms 0.476 0.146% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.8900 tsec 6.1420
#GCMRL# 406 dt 25.600000 rms 0.476 0.091% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.8950 tsec 6.1450
#GCMRL# 407 dt 25.600000 rms 0.475 0.081% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6620 tsec 5.9170
#GCMRL# 408 dt 25.600000 rms 0.475 0.115% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5470 tsec 5.7960
#GCMRL# 409 dt 25.600000 rms 0.474 0.142% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4480 tsec 5.7010
#GCMRL# 410 dt 25.600000 rms 0.474 0.130% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4030 tsec 5.6620
#GCMRL# 411 dt 25.600000 rms 0.473 0.087% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4620 tsec 5.7530
#GCMRL# 412 dt 25.600000 rms 0.473 -0.021% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.8120 tsec 7.2450
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.473133) vs oldopt=(dt=8,rms=0.47314)
#GCMRL# 413 dt 11.200000 rms 0.473 0.009% neg 0 invalid 762 tFOTS 17.0130 tGradient 4.8010 tsec 23.0980
#GCMRL# 414 dt 32.000000 rms 0.473 0.029% neg 0 invalid 762 tFOTS 16.1290 tGradient 4.5550 tsec 21.9460
#FOTS# QuadFit found better minimum quadopt=(dt=19.2,rms=0.472969) vs oldopt=(dt=32,rms=0.472976)
#GCAMreg# pass 0 level1 3 level2 1 tsec 771.575 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.473628
#FOTS# QuadFit found better minimum quadopt=(dt=80.8989,rms=0.469444) vs oldopt=(dt=32,rms=0.470339)
#GCMRL# 416 dt 80.898876 rms 0.469 0.883% neg 0 invalid 762 tFOTS 16.9880 tGradient 4.6490 tsec 22.8940
#FOTS# QuadFit found better minimum quadopt=(dt=24.1717,rms=0.468538) vs oldopt=(dt=32,rms=0.46869)
#GCMRL# 417 dt 24.171674 rms 0.469 0.193% neg 0 invalid 762 tFOTS 16.0400 tGradient 4.5440 tsec 21.8370
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.468213) vs oldopt=(dt=32,rms=0.468216)
#GCMRL# 418 dt 38.400000 rms 0.468 0.069% neg 0 invalid 762 tFOTS 16.0460 tGradient 4.5460 tsec 21.8410
#FOTS# QuadFit found better minimum quadopt=(dt=19.2,rms=0.468077) vs oldopt=(dt=32,rms=0.468108)
#GCMRL# 419 dt 19.200000 rms 0.468 0.000% neg 0 invalid 762 tFOTS 16.0520 tGradient 4.6760 tsec 22.0290
#GCMRL# 420 dt 19.200000 rms 0.468 0.066% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5230 tsec 5.7730
#GCMRL# 421 dt 19.200000 rms 0.468 0.055% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6550 tsec 5.9050
#GCMRL# 422 dt 19.200000 rms 0.467 0.076% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6700 tsec 5.9220
#GCMRL# 423 dt 19.200000 rms 0.467 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6890 tsec 5.9400
#GCMRL# 424 dt 19.200000 rms 0.467 0.086% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4540 tsec 5.7100
#GCMRL# 425 dt 19.200000 rms 0.466 0.081% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4520 tsec 5.6950
#GCMRL# 426 dt 19.200000 rms 0.466 0.068% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4430 tsec 5.7030
#GCMRL# 427 dt 19.200000 rms 0.466 0.064% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5270 tsec 5.7760
#GCMRL# 428 dt 19.200000 rms 0.465 0.080% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4290 tsec 5.6740
#GCMRL# 429 dt 19.200000 rms 0.465 0.065% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5150 tsec 5.7670
#GCMRL# 430 dt 19.200000 rms 0.465 0.059% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4380 tsec 5.7090
#GCMRL# 431 dt 19.200000 rms 0.464 0.035% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4430 tsec 5.6960
#GCMRL# 432 dt 19.200000 rms 0.464 0.063% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4260 tsec 5.6870
#GCMRL# 433 dt 19.200000 rms 0.464 0.053% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6690 tsec 5.9710
#GCMRL# 434 dt 19.200000 rms 0.464 0.050% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.9880 tsec 6.2820
#GCMRL# 435 dt 19.200000 rms 0.463 0.064% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0140 tsec 6.3860
#GCMRL# 436 dt 19.200000 rms 0.463 0.045% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6620 tsec 5.9860
#GCMRL# 437 dt 19.200000 rms 0.463 0.018% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.7010 tsec 5.9660
#GCMRL# 438 dt 19.200000 rms 0.463 0.048% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4840 tsec 5.7340
#GCMRL# 439 dt 19.200000 rms 0.463 0.060% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4590 tsec 5.7110
#GCMRL# 440 dt 19.200000 rms 0.463 0.005% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4710 tsec 5.7260
#GCMRL# 441 dt 19.200000 rms 0.462 0.034% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5180 tsec 5.7870
#GCMRL# 442 dt 19.200000 rms 0.462 0.055% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.6210 tsec 5.8870
#GCMRL# 443 dt 19.200000 rms 0.462 -0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5770 tsec 6.9870
#FOTS# QuadFit found better minimum quadopt=(dt=38.4,rms=0.462012) vs oldopt=(dt=32,rms=0.462015)
#GCMRL# 444 dt 38.400000 rms 0.462 0.023% neg 0 invalid 762 tFOTS 16.0280 tGradient 4.5390 tsec 21.8160
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.461955) vs oldopt=(dt=8,rms=0.46197)
setting smoothness cost coefficient to 0.400
#GCAMreg# pass 0 level1 2 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.485295
#GCMRL# 446 dt 0.000000 rms 0.485 0.128% neg 0 invalid 762 tFOTS 16.0560 tGradient 4.1540 tsec 21.4660
#GCMRL# 447 dt 0.150000 rms 0.485 0.000% neg 0 invalid 762 tFOTS 16.0400 tGradient 4.1550 tsec 22.6310
#GCAMreg# pass 0 level1 2 level2 1 tsec 54.96 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.485295
#GCMRL# 449 dt 0.000000 rms 0.485 0.128% neg 0 invalid 762 tFOTS 16.0400 tGradient 4.1080 tsec 21.4000
#GCMRL# 450 dt 0.150000 rms 0.485 0.000% neg 0 invalid 762 tFOTS 15.9780 tGradient 4.1330 tsec 22.5260
setting smoothness cost coefficient to 1.000
#GCAMreg# pass 0 level1 1 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.530295
#GCMRL# 452 dt 1.280000 rms 0.527 0.634% neg 0 invalid 762 tFOTS 16.0150 tGradient 3.9350 tsec 21.2060
#FOTS# QuadFit found better minimum quadopt=(dt=0.064,rms=0.526903) vs oldopt=(dt=0.08,rms=0.526903)
#GCMRL# 453 dt 0.064000 rms 0.527 0.000% neg 0 invalid 762 tFOTS 16.0080 tGradient 3.9900 tsec 21.2850
#GCAMreg# pass 0 level1 1 level2 1 tsec 53.214 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.527486
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.526684) vs oldopt=(dt=0.32,rms=0.526705)
#GCMRL# 455 dt 0.448000 rms 0.527 0.152% neg 0 invalid 762 tFOTS 16.0540 tGradient 4.0190 tsec 21.3280
#GCMRL# 456 dt 0.080000 rms 0.527 0.000% neg 0 invalid 762 tFOTS 16.0290 tGradient 3.9670 tsec 21.2910
#GCMRL# 457 dt 0.080000 rms 0.527 0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.9410 tsec 5.2000
resetting metric properties...
setting smoothness cost coefficient to 2.000
#GCAMreg# pass 0 level1 0 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=2.0,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.470002
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.456551) vs oldopt=(dt=0.32,rms=0.46005)
#GCMRL# 459 dt 0.448000 rms 0.457 2.862% neg 0 invalid 762 tFOTS 15.1080 tGradient 3.1710 tsec 19.5400
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.454157) vs oldopt=(dt=0.32,rms=0.45483)
#GCMRL# 460 dt 0.448000 rms 0.454 0.524% neg 0 invalid 762 tFOTS 15.0300 tGradient 3.2370 tsec 19.5230
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.452642) vs oldopt=(dt=0.32,rms=0.453058)
#GCMRL# 461 dt 0.448000 rms 0.453 0.334% neg 0 invalid 762 tFOTS 14.9980 tGradient 3.1180 tsec 19.3680
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.45174) vs oldopt=(dt=0.32,rms=0.451997)
#GCMRL# 462 dt 0.448000 rms 0.452 0.199% neg 0 invalid 762 tFOTS 15.0070 tGradient 3.1940 tsec 19.4530
#FOTS# QuadFit found better minimum quadopt=(dt=0.458333,rms=0.45102) vs oldopt=(dt=0.32,rms=0.45123)
#GCMRL# 463 dt 0.458333 rms 0.451 0.159% neg 0 invalid 762 tFOTS 15.0430 tGradient 3.2130 tsec 19.5100
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.450522) vs oldopt=(dt=0.32,rms=0.450659)
#GCMRL# 464 dt 0.448000 rms 0.451 0.111% neg 0 invalid 762 tFOTS 15.0180 tGradient 3.3810 tsec 19.6560
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.450085) vs oldopt=(dt=0.32,rms=0.450201)
#GCMRL# 465 dt 0.448000 rms 0.450 0.097% neg 0 invalid 762 tFOTS 15.0320 tGradient 3.1710 tsec 19.4530
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.449786) vs oldopt=(dt=0.32,rms=0.44987)
#GCMRL# 466 dt 0.448000 rms 0.450 0.066% neg 0 invalid 762 tFOTS 15.0060 tGradient 3.3610 tsec 19.6270
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.449478) vs oldopt=(dt=0.32,rms=0.449555)
#GCMRL# 467 dt 0.448000 rms 0.449 0.068% neg 0 invalid 762 tFOTS 15.0110 tGradient 3.2150 tsec 19.4840
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.449275) vs oldopt=(dt=0.32,rms=0.449329)
#GCMRL# 468 dt 0.448000 rms 0.449 0.000% neg 0 invalid 762 tFOTS 15.0190 tGradient 3.3520 tsec 19.6610
#GCMRL# 469 dt 0.448000 rms 0.449 0.051% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.1800 tsec 4.4360
#GCMRL# 470 dt 0.448000 rms 0.449 0.074% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.3890 tsec 4.6470
#GCMRL# 471 dt 0.448000 rms 0.448 0.094% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.3840 tsec 4.6390
#GCMRL# 472 dt 0.448000 rms 0.448 0.096% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.1730 tsec 4.4360
#GCMRL# 473 dt 0.448000 rms 0.447 0.085% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2040 tsec 4.4610
#GCMRL# 474 dt 0.448000 rms 0.447 0.070% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.1670 tsec 4.4220
#GCMRL# 475 dt 0.448000 rms 0.447 0.041% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.1690 tsec 4.4230
#GCMRL# 476 dt 0.448000 rms 0.447 0.030% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.3900 tsec 4.6430
#GCMRL# 477 dt 0.448000 rms 0.447 -0.002% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2930 tsec 6.2580
#FOTS# QuadFit found better minimum quadopt=(dt=0.112,rms=0.446836) vs oldopt=(dt=0.08,rms=0.446836)
#GCMRL# 478 dt 0.112000 rms 0.447 0.001% neg 0 invalid 762 tFOTS 14.9840 tGradient 3.2970 tsec 19.5280
#FOTS# QuadFit found better minimum quadopt=(dt=0.064,rms=0.44684) vs oldopt=(dt=0.08,rms=0.44684)
#GCAMreg# pass 0 level1 0 level2 1 tsec 282.369 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=5.00e-02, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=0.5,type=2, relabel=0, neg=no
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.447587
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.443597) vs oldopt=(dt=0.32,rms=0.444475)
#GCMRL# 480 dt 0.448000 rms 0.444 0.891% neg 0 invalid 762 tFOTS 14.9910 tGradient 3.3770 tsec 19.6110
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.443144) vs oldopt=(dt=0.32,rms=0.443265)
#GCMRL# 481 dt 0.448000 rms 0.443 0.102% neg 0 invalid 762 tFOTS 15.0030 tGradient 3.1590 tsec 19.4090
#FOTS# QuadFit found better minimum quadopt=(dt=0.448,rms=0.442984) vs oldopt=(dt=0.32,rms=0.443023)
#GCMRL# 482 dt 0.448000 rms 0.443 0.000% neg 0 invalid 762 tFOTS 15.1110 tGradient 3.1660 tsec 19.5830
#GCMRL# 483 dt 0.448000 rms 0.443 0.006% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2460 tsec 4.5170
#GCMRL# 484 dt 0.448000 rms 0.443 0.018% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2540 tsec 4.5090
#GCMRL# 485 dt 0.448000 rms 0.443 0.000% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2530 tsec 5.0630
GCAMregister done in 77.4366 min
********************* ALLOWING NEGATIVE NODES IN DEFORMATION********************************
noneg post
Starting GCAMregister()
label assignment complete, 0 changed (0.00%)
npasses = 1, nlevels = 6
#pass# 1 of 1 ************************
enabling zero nodes
setting smoothness cost coefficient to 0.008
#GCAMreg# pass 0 level1 5 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.441636
#GCMRL# 487 dt 0.000000 rms 0.441 0.164% neg 0 invalid 762 tFOTS 17.9880 tGradient 6.3500 tsec 25.5880
#GCAMreg# pass 0 level1 5 level2 1 tsec 56.615 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.441636
#FOTS# QuadFit found better minimum quadopt=(dt=295.936,rms=0.440537) vs oldopt=(dt=369.92,rms=0.440559)
#GCMRL# 489 dt 295.936000 rms 0.441 0.249% neg 0 invalid 762 tFOTS 18.0200 tGradient 6.3580 tsec 25.6270
#FOTS# QuadFit found better minimum quadopt=(dt=32.368,rms=0.440489) vs oldopt=(dt=23.12,rms=0.440497)
#GCMRL# 490 dt 32.368000 rms 0.440 0.000% neg 0 invalid 762 tFOTS 18.9530 tGradient 6.5050 tsec 26.7520
#GCMRL# 491 dt 32.368000 rms 0.440 0.007% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.4930 tsec 7.7360
#GCMRL# 492 dt 32.368000 rms 0.440 0.005% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6220 tsec 7.8740
#GCMRL# 493 dt 32.368000 rms 0.440 0.007% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6270 tsec 7.8730
#GCMRL# 494 dt 32.368000 rms 0.440 0.009% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6360 tsec 7.8860
#GCMRL# 495 dt 32.368000 rms 0.440 0.017% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6020 tsec 7.8570
#GCMRL# 496 dt 32.368000 rms 0.440 0.019% neg 0 invalid 762 tFOTS 0.0000 tGradient 6.6080 tsec 7.8560
setting smoothness cost coefficient to 0.031
#GCAMreg# pass 0 level1 4 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.440894
#FOTS# QuadFit found better minimum quadopt=(dt=9.072,rms=0.440119) vs oldopt=(dt=6.48,rms=0.440123)
#GCMRL# 498 dt 9.072000 rms 0.440 0.176% neg 0 invalid 762 tFOTS 19.1320 tGradient 5.2920 tsec 25.6740
#FOTS# QuadFit found better minimum quadopt=(dt=9.072,rms=0.440099) vs oldopt=(dt=6.48,rms=0.440104)
#GCMRL# 499 dt 9.072000 rms 0.440 0.000% neg 0 invalid 762 tFOTS 18.9800 tGradient 4.9940 tsec 25.2710
#GCMRL# 500 dt 9.072000 rms 0.440 0.006% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.2110 tsec 6.4680
#GCAMreg# pass 0 level1 4 level2 1 tsec 69.255 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.440843
#FOTS# QuadFit found better minimum quadopt=(dt=145.152,rms=0.437637) vs oldopt=(dt=103.68,rms=0.438117)
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 502 dt 145.152000 rms 0.438 0.727% neg 0 invalid 762 tFOTS 18.1170 tGradient 5.0720 tsec 25.7960
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.437455) vs oldopt=(dt=25.92,rms=0.437473)
#GCMRL# 503 dt 36.288000 rms 0.437 0.000% neg 0 invalid 762 tFOTS 18.1720 tGradient 5.0020 tsec 24.4660
#GCMRL# 504 dt 36.288000 rms 0.437 0.041% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1550 tsec 6.4250
#GCMRL# 505 dt 36.288000 rms 0.437 0.068% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.1350 tsec 6.3930
#GCMRL# 506 dt 36.288000 rms 0.437 0.096% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.9940 tsec 6.2390
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 507 dt 36.288000 rms 0.436 0.128% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0000 tsec 7.4580
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 508 dt 36.288000 rms 0.435 0.119% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0010 tsec 7.4470
#GCMRL# 509 dt 36.288000 rms 0.435 0.123% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0390 tsec 6.2830
iter 0, gcam->neg = 2
after 7 iterations, nbhd size=1, neg = 0
#GCMRL# 510 dt 36.288000 rms 0.435 0.093% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.0460 tsec 11.6420
#FOTS# QuadFit found better minimum quadopt=(dt=36.288,rms=0.434557) vs oldopt=(dt=25.92,rms=0.43456)
setting smoothness cost coefficient to 0.118
#GCAMreg# pass 0 level1 3 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.435641
#FOTS# QuadFit found better minimum quadopt=(dt=30.2959,rms=0.433782) vs oldopt=(dt=32,rms=0.433795)
iter 0, gcam->neg = 4
after 7 iterations, nbhd size=1, neg = 0
#GCMRL# 512 dt 30.295858 rms 0.434 0.420% neg 0 invalid 762 tFOTS 19.0540 tGradient 4.4610 tsec 30.2220
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.432413) vs oldopt=(dt=32,rms=0.432535)
iter 0, gcam->neg = 4
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 513 dt 44.800000 rms 0.432 0.323% neg 0 invalid 762 tFOTS 19.0190 tGradient 4.4530 tsec 25.9740
#FOTS# QuadFit found better minimum quadopt=(dt=22.2762,rms=0.431624) vs oldopt=(dt=32,rms=0.431815)
#GCMRL# 514 dt 22.276243 rms 0.432 0.000% neg 0 invalid 762 tFOTS 18.9160 tGradient 4.3800 tsec 24.5830
#GCMRL# 515 dt 22.276243 rms 0.431 0.138% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3780 tsec 5.6270
iter 0, gcam->neg = 2
after 7 iterations, nbhd size=1, neg = 0
#GCMRL# 516 dt 22.276243 rms 0.430 0.194% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4620 tsec 11.0430
#GCMRL# 517 dt 22.276243 rms 0.429 0.192% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3800 tsec 5.6350
iter 0, gcam->neg = 3
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 518 dt 22.276243 rms 0.429 0.199% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3810 tsec 6.8520
iter 0, gcam->neg = 1
after 6 iterations, nbhd size=1, neg = 0
#GCMRL# 519 dt 22.276243 rms 0.428 0.141% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2920 tsec 10.2740
iter 0, gcam->neg = 6
after 10 iterations, nbhd size=1, neg = 0
#GCMRL# 520 dt 22.276243 rms 0.427 0.176% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3300 tsec 12.6500
#GCMRL# 521 dt 22.276243 rms 0.427 0.143% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3520 tsec 5.6020
iter 0, gcam->neg = 6
after 11 iterations, nbhd size=1, neg = 0
#GCMRL# 522 dt 22.276243 rms 0.426 0.088% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4360 tsec 13.4730
iter 0, gcam->neg = 2
after 2 iterations, nbhd size=0, neg = 0
#GCMRL# 523 dt 22.276243 rms 0.426 0.141% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5200 tsec 8.2050
#GCMRL# 524 dt 22.276243 rms 0.425 0.130% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4580 tsec 5.7310
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 525 dt 22.276243 rms 0.425 0.100% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.5220 tsec 6.9650
iter 0, gcam->neg = 3
after 8 iterations, nbhd size=1, neg = 0
#GCMRL# 526 dt 22.276243 rms 0.424 0.058% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3870 tsec 11.5100
#FOTS# QuadFit found better minimum quadopt=(dt=25.6,rms=0.42397) vs oldopt=(dt=32,rms=0.423973)
#GCMRL# 527 dt 25.600000 rms 0.424 0.089% neg 0 invalid 762 tFOTS 18.9140 tGradient 4.4410 tsec 24.5990
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.423915) vs oldopt=(dt=8,rms=0.423917)
#GCMRL# 528 dt 11.200000 rms 0.424 0.000% neg 0 invalid 762 tFOTS 18.9980 tGradient 4.2820 tsec 24.5630
#GCMRL# 529 dt 11.200000 rms 0.424 0.023% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3390 tsec 5.5840
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 530 dt 11.200000 rms 0.424 0.026% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3740 tsec 6.8810
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 531 dt 11.200000 rms 0.424 0.025% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3200 tsec 6.7780
iter 0, gcam->neg = 1
after 6 iterations, nbhd size=1, neg = 0
#GCAMreg# pass 0 level1 3 level2 1 tsec 267.456 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.42429
#FOTS# QuadFit found better minimum quadopt=(dt=66.3358,rms=0.419519) vs oldopt=(dt=32,rms=0.420377)
#GCMRL# 533 dt 66.335766 rms 0.420 1.124% neg 0 invalid 762 tFOTS 18.9430 tGradient 4.3080 tsec 24.5020
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.418766) vs oldopt=(dt=8,rms=0.418914)
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 534 dt 11.200000 rms 0.419 0.000% neg 0 invalid 762 tFOTS 18.9390 tGradient 4.3760 tsec 25.8510
#GCMRL# 535 dt 11.200000 rms 0.419 0.061% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4420 tsec 5.6970
#GCMRL# 536 dt 11.200000 rms 0.418 0.057% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.4100 tsec 5.6690
#GCMRL# 537 dt 11.200000 rms 0.418 0.056% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.3050 tsec 5.5990
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.417494) vs oldopt=(dt=32,rms=0.417535)
#GCMRL# 538 dt 44.800000 rms 0.417 0.129% neg 0 invalid 762 tFOTS 19.0650 tGradient 4.3430 tsec 24.6670
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.417241) vs oldopt=(dt=8,rms=0.417279)
setting smoothness cost coefficient to 0.400
#GCAMreg# pass 0 level1 2 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.423798
#FOTS# QuadFit found better minimum quadopt=(dt=2.304,rms=0.422956) vs oldopt=(dt=2.88,rms=0.422956)
#GCMRL# 540 dt 2.304000 rms 0.423 0.199% neg 0 invalid 762 tFOTS 19.0150 tGradient 4.0850 tsec 24.3510
#FOTS# QuadFit found better minimum quadopt=(dt=1.008,rms=0.422955) vs oldopt=(dt=0.72,rms=0.422958)
#GCMRL# 541 dt 1.008000 rms 0.423 0.000% neg 0 invalid 762 tFOTS 19.0300 tGradient 4.0540 tsec 24.3900
#GCMRL# 542 dt 1.008000 rms 0.423 0.003% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1710 tsec 5.4170
#GCAMreg# pass 0 level1 2 level2 1 tsec 64.839 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.423656
#FOTS# QuadFit found better minimum quadopt=(dt=8.67257,rms=0.421995) vs oldopt=(dt=11.52,rms=0.422103)
#GCMRL# 544 dt 8.672566 rms 0.422 0.392% neg 0 invalid 762 tFOTS 19.0400 tGradient 4.0250 tsec 24.3200
#FOTS# QuadFit found better minimum quadopt=(dt=13.9429,rms=0.421202) vs oldopt=(dt=11.52,rms=0.421229)
#GCMRL# 545 dt 13.942857 rms 0.421 0.000% neg 0 invalid 762 tFOTS 19.0830 tGradient 4.0220 tsec 24.3960
#GCMRL# 546 dt 13.942857 rms 0.420 0.273% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.0640 tsec 5.3130
iter 0, gcam->neg = 2
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 547 dt 13.942857 rms 0.419 0.356% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1910 tsec 6.6840
iter 0, gcam->neg = 2
after 1 iterations, nbhd size=0, neg = 0
#GCMRL# 548 dt 13.942857 rms 0.417 0.301% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2250 tsec 7.2950
iter 0, gcam->neg = 2
after 1 iterations, nbhd size=0, neg = 0
#GCMRL# 549 dt 13.942857 rms 0.415 0.468% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.0270 tsec 7.1030
iter 0, gcam->neg = 2
after 3 iterations, nbhd size=0, neg = 0
#GCMRL# 550 dt 13.942857 rms 0.413 0.469% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1830 tsec 8.3760
iter 0, gcam->neg = 6
after 9 iterations, nbhd size=1, neg = 0
#GCMRL# 551 dt 13.942857 rms 0.412 0.342% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1240 tsec 11.8660
iter 0, gcam->neg = 3
after 1 iterations, nbhd size=0, neg = 0
#GCMRL# 552 dt 13.942857 rms 0.411 0.129% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1000 tsec 7.1460
iter 0, gcam->neg = 2
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 553 dt 13.942857 rms 0.411 -0.010% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.2280 tsec 7.8580
#FOTS# QuadFit found better minimum quadopt=(dt=4.032,rms=0.411389) vs oldopt=(dt=2.88,rms=0.411395)
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 554 dt 4.032000 rms 0.411 0.012% neg 0 invalid 762 tFOTS 18.8340 tGradient 4.1960 tsec 25.4650
#FOTS# QuadFit found better minimum quadopt=(dt=0.0016875,rms=0.411365) vs oldopt=(dt=0.0028125,rms=0.411365)
setting smoothness cost coefficient to 1.000
#GCAMreg# pass 0 level1 1 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.417255
#GCMRL# 556 dt 0.000000 rms 0.416 0.193% neg 0 invalid 762 tFOTS 18.0740 tGradient 3.9380 tsec 23.2580
#GCAMreg# pass 0 level1 1 level2 1 tsec 51.94 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.417255
#GCMRL# 558 dt 0.000000 rms 0.416 0.193% neg 0 invalid 762 tFOTS 18.0360 tGradient 3.9540 tsec 23.2440
resetting metric properties...
setting smoothness cost coefficient to 2.000
#GCAMreg# pass 0 level1 0 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.408762
#FOTS# QuadFit found better minimum quadopt=(dt=1.88843,rms=0.385579) vs oldopt=(dt=1.28,rms=0.388123)
iter 0, gcam->neg = 624
after 13 iterations, nbhd size=1, neg = 0
#GCMRL# 560 dt 1.888427 rms 0.386 5.618% neg 0 invalid 762 tFOTS 19.1240 tGradient 3.2010 tsec 32.5150
#GCMRL# 561 dt 0.000013 rms 0.386 0.000% neg 0 invalid 762 tFOTS 24.1650 tGradient 3.2570 tsec 28.7100
#GCAMreg# pass 0 level1 0 level2 1 tsec 71.174 sigma 0.5
l_jacobian=1.00 l_label=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.386748
#FOTS# QuadFit found better minimum quadopt=(dt=0.028,rms=0.385744) vs oldopt=(dt=0.02,rms=0.385752)
#GCMRL# 563 dt 0.028000 rms 0.386 0.259% neg 0 invalid 762 tFOTS 19.0890 tGradient 3.2290 tsec 23.5650
#FOTS# QuadFit found better minimum quadopt=(dt=0.007,rms=0.385783) vs oldopt=(dt=0.005,rms=0.385784)
#GCMRL# 564 dt 0.007000 rms 0.386 0.000% neg 0 invalid 762 tFOTS 19.1000 tGradient 3.2780 tsec 24.8060
#GCMRL# 565 dt 0.007000 rms 0.386 0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.3310 tsec 4.5920
#GCMRL# 566 dt 0.007000 rms 0.386 0.006% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2150 tsec 4.4670
label assignment complete, 0 changed (0.00%)
GCAMregister done in 20.459 min
Starting GCAMcomputeMaxPriorLabels()
Morphing with label term set to 0 *******************************
Starting GCAMregister()
label assignment complete, 0 changed (0.00%)
npasses = 1, nlevels = 6
#pass# 1 of 1 ************************
enabling zero nodes
setting smoothness cost coefficient to 0.008
#GCAMreg# pass 0 level1 5 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.375653
#GCAMreg# pass 0 level1 5 level2 1 tsec 29.715 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.01
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=256, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.375653
#FOTS# QuadFit found better minimum quadopt=(dt=8.092,rms=0.375651) vs oldopt=(dt=5.78,rms=0.375651)
#GCMRL# 569 dt 8.092000 rms 0.376 0.001% neg 0 invalid 762 tFOTS 18.5480 tGradient 5.6610 tsec 25.4340
#FOTS# QuadFit found better minimum quadopt=(dt=2.023,rms=0.375651) vs oldopt=(dt=1.445,rms=0.375651)
#GCMRL# 570 dt 2.023000 rms 0.376 0.000% neg 0 invalid 762 tFOTS 18.4940 tGradient 5.6670 tsec 25.4230
#GCMRL# 571 dt 2.023000 rms 0.376 0.000% neg 0 invalid 762 tFOTS 0.0000 tGradient 5.7100 tsec 6.9370
setting smoothness cost coefficient to 0.031
#GCAMreg# pass 0 level1 4 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.37578
#FOTS# QuadFit found better minimum quadopt=(dt=2.268,rms=0.375776) vs oldopt=(dt=1.62,rms=0.375777)
#GCMRL# 573 dt 2.268000 rms 0.376 0.001% neg 0 invalid 762 tFOTS 18.5980 tGradient 4.2000 tsec 24.0240
#FOTS# QuadFit found better minimum quadopt=(dt=1.944,rms=0.375775) vs oldopt=(dt=1.62,rms=0.375775)
#GCMRL# 574 dt 1.944000 rms 0.376 0.000% neg 0 invalid 762 tFOTS 18.4010 tGradient 4.2210 tsec 23.8700
#GCAMreg# pass 0 level1 4 level2 1 tsec 58.629 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.03
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=64, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.375775
#GCMRL# 576 dt 25.920000 rms 0.376 0.015% neg 0 invalid 762 tFOTS 17.3170 tGradient 4.1390 tsec 22.6710
#FOTS# QuadFit found better minimum quadopt=(dt=20.8,rms=0.375706) vs oldopt=(dt=25.92,rms=0.375708)
#GCMRL# 577 dt 20.800000 rms 0.376 0.000% neg 0 invalid 762 tFOTS 18.3320 tGradient 4.1540 tsec 23.7380
#GCMRL# 578 dt 20.800000 rms 0.376 0.002% neg 0 invalid 762 tFOTS 0.0000 tGradient 4.1520 tsec 5.3630
setting smoothness cost coefficient to 0.118
#GCAMreg# pass 0 level1 3 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.376188
#FOTS# QuadFit found better minimum quadopt=(dt=11.2,rms=0.375971) vs oldopt=(dt=8,rms=0.375989)
#GCMRL# 580 dt 11.200000 rms 0.376 0.058% neg 0 invalid 762 tFOTS 18.4540 tGradient 3.5500 tsec 23.2300
#FOTS# QuadFit found better minimum quadopt=(dt=6.4,rms=0.37594) vs oldopt=(dt=8,rms=0.375943)
#GCMRL# 581 dt 6.400000 rms 0.376 0.000% neg 0 invalid 762 tFOTS 18.5110 tGradient 3.5560 tsec 23.3410
#GCAMreg# pass 0 level1 3 level2 1 tsec 56.677 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.12
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=16, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.37594
#FOTS# QuadFit found better minimum quadopt=(dt=86.9565,rms=0.374039) vs oldopt=(dt=32,rms=0.374485)
iter 0, gcam->neg = 22
after 9 iterations, nbhd size=1, neg = 0
#GCMRL# 583 dt 86.956522 rms 0.374 0.497% neg 0 invalid 762 tFOTS 18.4170 tGradient 3.5490 tsec 29.7040
#FOTS# QuadFit found better minimum quadopt=(dt=25.4316,rms=0.373277) vs oldopt=(dt=32,rms=0.373375)
iter 0, gcam->neg = 7
after 3 iterations, nbhd size=0, neg = 0
#GCMRL# 584 dt 25.431579 rms 0.373 0.000% neg 0 invalid 762 tFOTS 18.2710 tGradient 3.5660 tsec 26.0110
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
#GCMRL# 585 dt 25.431579 rms 0.373 0.083% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.5450 tsec 5.9450
iter 0, gcam->neg = 7
after 2 iterations, nbhd size=0, neg = 0
#GCMRL# 586 dt 25.431579 rms 0.373 0.095% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.5370 tsec 7.1120
iter 0, gcam->neg = 9
after 12 iterations, nbhd size=1, neg = 0
#GCMRL# 587 dt 25.431579 rms 0.372 0.083% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.5400 tsec 12.9440
iter 0, gcam->neg = 10
after 5 iterations, nbhd size=0, neg = 0
#GCMRL# 588 dt 25.431579 rms 0.372 0.108% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.5410 tsec 8.8930
iter 0, gcam->neg = 15
after 11 iterations, nbhd size=1, neg = 0
#GCMRL# 589 dt 25.431579 rms 0.372 0.096% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.5540 tsec 12.5700
#FOTS# QuadFit found better minimum quadopt=(dt=44.8,rms=0.371393) vs oldopt=(dt=32,rms=0.371426)
iter 0, gcam->neg = 1
after 0 iterations, nbhd size=0, neg = 0
setting smoothness cost coefficient to 0.400
#GCAMreg# pass 0 level1 2 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.374087
#FOTS# QuadFit found better minimum quadopt=(dt=3.456,rms=0.373984) vs oldopt=(dt=2.88,rms=0.373988)
#GCMRL# 591 dt 3.456000 rms 0.374 0.028% neg 0 invalid 762 tFOTS 18.4720 tGradient 3.2850 tsec 22.9790
#FOTS# QuadFit found better minimum quadopt=(dt=1.008,rms=0.373976) vs oldopt=(dt=0.72,rms=0.373977)
#GCMRL# 592 dt 1.008000 rms 0.374 0.000% neg 0 invalid 762 tFOTS 17.4710 tGradient 3.2810 tsec 22.0240
#GCMRL# 593 dt 1.008000 rms 0.374 0.001% neg 0 invalid 762 tFOTS 0.0000 tGradient 3.2810 tsec 4.5030
iter 0, gcam->neg = 1
after 1 iterations, nbhd size=0, neg = 0
#GCAMreg# pass 0 level1 2 level2 1 tsec 61.177 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=0.40
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=4, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.373974
#FOTS# QuadFit found better minimum quadopt=(dt=21.7358,rms=0.372501) vs oldopt=(dt=11.52,rms=0.372842)
iter 0, gcam->neg = 11
after 3 iterations, nbhd size=0, neg = 0
#GCMRL# 595 dt 21.735849 rms 0.372 0.394% neg 0 invalid 762 tFOTS 18.4650 tGradient 3.2850 tsec 25.9860
#FOTS# QuadFit found better minimum quadopt=(dt=22.8571,rms=0.37164) vs oldopt=(dt=11.52,rms=0.371845)
iter 0, gcam->neg = 17
after 10 iterations, nbhd size=1, neg = 0
#GCMRL# 596 dt 22.857143 rms 0.372 0.000% neg 0 invalid 762 tFOTS 18.4650 tGradient 3.2770 tsec 30.0950
iter 0, gcam->neg = 27
after 15 iterations, nbhd size=1, neg = 0
setting smoothness cost coefficient to 1.000
#GCAMreg# pass 0 level1 1 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.376315
#FOTS# QuadFit found better minimum quadopt=(dt=-0.000160217,rms=0.376315) vs oldopt=(dt=7.8125e-05,rms=0.376315)
#GCMRL# 598 dt -0.000160 rms 0.376 0.000% neg 0 invalid 762 tFOTS 23.9490 tGradient 3.1070 tsec 28.2820
#GCAMreg# pass 0 level1 1 level2 1 tsec 55.45 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=1.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=1, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.376315
resetting metric properties...
setting smoothness cost coefficient to 2.000
#GCAMreg# pass 0 level1 0 level2 0 tsec 0 sigma 2
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=2.0,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=2.000...
GCAMRegisterLevel(): init RMS 0.368515
#FOTS# QuadFit found better minimum quadopt=(dt=1.11117,rms=0.361078) vs oldopt=(dt=1.28,rms=0.361224)
iter 0, gcam->neg = 480
after 16 iterations, nbhd size=1, neg = 0
#GCMRL# 601 dt 1.111171 rms 0.362 1.841% neg 0 invalid 762 tFOTS 18.4190 tGradient 2.3440 tsec 32.7700
#FOTS# QuadFit found better minimum quadopt=(dt=2.34375e-05,rms=0.361732) vs oldopt=(dt=1.95313e-05,rms=0.361732)
#GCMRL# 602 dt 0.000023 rms 0.362 0.000% neg 0 invalid 762 tFOTS 23.2410 tGradient 2.3370 tsec 26.8400
#GCAMreg# pass 0 level1 0 level2 1 tsec 68.481 sigma 0.5
l_jacobian=1.00 l_log_likelihood=0.20 l_smoothness=2.00
tol=2.50e-01, dt=5.00e-02, exp_k=20.0, momentum=0.90, levels=6, niter=500, lbl_dist=10.00, avgs=0, sigma=0.5,type=2, relabel=0, neg=yes
blurring input image with Gaussian with sigma=0.500...
GCAMRegisterLevel(): init RMS 0.361732
#FOTS# QuadFit found better minimum quadopt=(dt=0.064,rms=0.361659) vs oldopt=(dt=0.08,rms=0.361659)
#GCMRL# 604 dt 0.064000 rms 0.362 0.020% neg 0 invalid 762 tFOTS 18.4540 tGradient 2.3380 tsec 22.0240
#FOTS# QuadFit found better minimum quadopt=(dt=0.028,rms=0.361639) vs oldopt=(dt=0.02,rms=0.361643)
#GCMRL# 605 dt 0.028000 rms 0.362 0.000% neg 0 invalid 762 tFOTS 18.2940 tGradient 2.3300 tsec 21.8820
#GCMRL# 606 dt 0.028000 rms 0.362 0.004% neg 0 invalid 762 tFOTS 0.0000 tGradient 2.3230 tsec 3.5330
GCAMregister done in 12.4903 min
writing output transformation to transforms/talairach.m3z...
GCAMwrite
Calls to gcamLogLikelihoodEnergy 4499 tmin = 18.5523
Calls to gcamLabelEnergy 3971 tmin = 2.07455
Calls to gcamJacobianEnergy 4499 tmin = 14.8373
Calls to gcamSmoothnessEnergy 4499 tmin = 14.6877
Calls to gcamLogLikelihoodTerm 608 tmin = 5.47622
Calls to gcamLabelTerm 568 tmin = 9.66203
Calls to gcamJacobianTerm 608 tmin = 11.415
Calls to gcamSmoothnessTerm 608 tmin = 3.24693
Calls to gcamComputeGradient 608 tmin = 52.2942
Calls to gcamComputeMetricProperties 6257 tmin = 20.5359
mri_ca_register took 2 hours, 17 minutes and 45 seconds.
#VMPC# mri_ca_register VmPeak 2023712
FSRUNTIME@ mri_ca_register 2.2959 hours 1 threads
#--------------------------------------
#@# SubCort Seg Thu Jun 12 13:26:28 UTC 2025
mri_ca_label -relabel_unlikely 9 .3 -prior 0.5 -align norm.mgz transforms/talairach.m3z /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca aseg.auto_noCCseg.mgz
sysname Linux
hostname jupyter-monidoerig
machine x86_64
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_ca_label -relabel_unlikely 9 .3 -prior 0.5 -align norm.mgz transforms/talairach.m3z /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca aseg.auto_noCCseg.mgz
relabeling unlikely voxels with window_size = 9 and prior threshold 0.30
using Gibbs prior factor = 0.500
renormalizing sequences with structure alignment, equivalent to:
-renormalize
-renormalize_mean 0.500
-regularize 0.500
== Number of threads available to for OpenMP = 1 ==
reading 1 input volumes
reading classifier array from /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca
reading input volume from norm.mgz
average std[0] = 7.2
reading transform from transforms/talairach.m3z
setting orig areas to linear transform determinant scaled 6.27
Atlas used for the 3D morph was /opt/freesurfer-7.3.2/average/RB_all_2020-01-02.gca
average std = 7.2 using min determinant for regularization = 5.2
0 singular and 0 ill-conditioned covariance matrices regularized
labeling volume...
renormalizing by structure alignment....
renormalizing input #0
gca peak = 0.15521 (20)
mri peak = 0.17945 ( 7)
Left_Lateral_Ventricle (4): linear fit = 0.35 x + 0.0 (1560 voxels, overlap=0.004)
Left_Lateral_Ventricle (4): linear fit = 0.40 x + 0.0 (1560 voxels, peak = 7), gca=8.0
gca peak = 0.20380 (13)
mri peak = 0.17272 ( 7)
Right_Lateral_Ventricle (43): linear fit = 0.43 x + 0.0 (1589 voxels, overlap=0.049)
Right_Lateral_Ventricle (43): linear fit = 0.43 x + 0.0 (1589 voxels, peak = 6), gca=5.5
gca peak = 0.26283 (96)
mri peak = 0.16332 (106)
Right_Pallidum (52): linear fit = 1.10 x + 0.0 (819 voxels, overlap=0.128)
Right_Pallidum (52): linear fit = 1.10 x + 0.0 (819 voxels, peak = 105), gca=105.1
gca peak = 0.15814 (97)
mri peak = 0.11072 (106)
Left_Pallidum (13): linear fit = 1.12 x + 0.0 (858 voxels, overlap=0.026)
Left_Pallidum (13): linear fit = 1.12 x + 0.0 (858 voxels, peak = 109), gca=109.1
gca peak = 0.27624 (56)
mri peak = 0.10696 (56)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (782 voxels, overlap=1.011)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (782 voxels, peak = 56), gca=56.0
gca peak = 0.28723 (59)
mri peak = 0.09347 (60)
Left_Hippocampus (17): linear fit = 1.02 x + 0.0 (838 voxels, overlap=1.011)
Left_Hippocampus (17): linear fit = 1.02 x + 0.0 (838 voxels, peak = 60), gca=60.5
gca peak = 0.07623 (103)
mri peak = 0.13857 (102)
Right_Cerebral_White_Matter (41): linear fit = 1.00 x + 0.0 (41973 voxels, overlap=0.615)
Right_Cerebral_White_Matter (41): linear fit = 1.00 x + 0.0 (41973 voxels, peak = 103), gca=103.0
gca peak = 0.07837 (105)
mri peak = 0.13751 (104)
Left_Cerebral_White_Matter (2): linear fit = 1.00 x + 0.0 (43265 voxels, overlap=0.620)
Left_Cerebral_White_Matter (2): linear fit = 1.00 x + 0.0 (43265 voxels, peak = 105), gca=105.0
gca peak = 0.10165 (58)
mri peak = 0.04398 (66)
Left_Cerebral_Cortex (3): linear fit = 1.10 x + 0.0 (29448 voxels, overlap=0.783)
Left_Cerebral_Cortex (3): linear fit = 1.10 x + 0.0 (29448 voxels, peak = 64), gca=63.5
gca peak = 0.11113 (58)
mri peak = 0.04406 (64)
Right_Cerebral_Cortex (42): linear fit = 1.08 x + 0.0 (28388 voxels, overlap=0.735)
Right_Cerebral_Cortex (42): linear fit = 1.08 x + 0.0 (28388 voxels, peak = 62), gca=62.4
gca peak = 0.27796 (67)
mri peak = 0.18231 (76)
Right_Caudate (50): linear fit = 1.12 x + 0.0 (935 voxels, overlap=0.231)
Right_Caudate (50): linear fit = 1.12 x + 0.0 (935 voxels, peak = 75), gca=74.7
gca peak = 0.14473 (69)
mri peak = 0.13884 (77)
Left_Caudate (11): linear fit = 1.00 x + 0.0 (901 voxels, overlap=0.937)
Left_Caudate (11): linear fit = 1.00 x + 0.0 (901 voxels, peak = 69), gca=69.0
gca peak = 0.14301 (56)
mri peak = 0.05310 (48)
Left_Cerebellum_Cortex (8): linear fit = 0.86 x + 0.0 (14929 voxels, overlap=0.755)
Left_Cerebellum_Cortex (8): linear fit = 0.86 x + 0.0 (14929 voxels, peak = 48), gca=47.9
gca peak = 0.14610 (55)
mri peak = 0.04965 (46)
Right_Cerebellum_Cortex (47): linear fit = 0.83 x + 0.0 (15407 voxels, overlap=0.643)
Right_Cerebellum_Cortex (47): linear fit = 0.83 x + 0.0 (15407 voxels, peak = 46), gca=45.9
gca peak = 0.16309 (85)
mri peak = 0.07994 (89)
Left_Cerebellum_White_Matter (7): linear fit = 1.07 x + 0.0 (5075 voxels, overlap=0.821)
Left_Cerebellum_White_Matter (7): linear fit = 1.07 x + 0.0 (5075 voxels, peak = 91), gca=90.5
gca peak = 0.15172 (84)
mri peak = 0.08530 (88)
Right_Cerebellum_White_Matter (46): linear fit = 1.07 x + 0.0 (5192 voxels, overlap=0.827)
Right_Cerebellum_White_Matter (46): linear fit = 1.07 x + 0.0 (5192 voxels, peak = 89), gca=89.5
gca peak = 0.30461 (58)
mri peak = 0.10000 (63)
Left_Amygdala (18): linear fit = 1.10 x + 0.0 (469 voxels, overlap=0.856)
Left_Amygdala (18): linear fit = 1.10 x + 0.0 (469 voxels, peak = 64), gca=63.5
gca peak = 0.32293 (57)
mri peak = 0.11073 (62)
Right_Amygdala (54): linear fit = 1.10 x + 0.0 (449 voxels, overlap=0.625)
Right_Amygdala (54): linear fit = 1.10 x + 0.0 (449 voxels, peak = 62), gca=62.4
gca peak = 0.11083 (90)
mri peak = 0.08778 (91)
Left_Thalamus (10): linear fit = 1.05 x + 0.0 (4105 voxels, overlap=0.842)
Left_Thalamus (10): linear fit = 1.05 x + 0.0 (4105 voxels, peak = 95), gca=94.9
gca peak = 0.11393 (83)
mri peak = 0.08291 (89)
Right_Thalamus (49): linear fit = 1.10 x + 0.0 (4093 voxels, overlap=0.697)
Right_Thalamus (49): linear fit = 1.10 x + 0.0 (4093 voxels, peak = 91), gca=90.9
gca peak = 0.08575 (81)
mri peak = 0.09072 (84)
Left_Putamen (12): linear fit = 1.10 x + 0.0 (1968 voxels, overlap=0.477)
Left_Putamen (12): linear fit = 1.10 x + 0.0 (1968 voxels, peak = 89), gca=88.7
gca peak = 0.08618 (78)
mri peak = 0.09372 (88)
Right_Putamen (51): linear fit = 1.10 x + 0.0 (2102 voxels, overlap=0.522)
Right_Putamen (51): linear fit = 1.10 x + 0.0 (2102 voxels, peak = 85), gca=85.4
gca peak = 0.08005 (78)
mri peak = 0.06509 (96)
Brain_Stem (16): linear fit = 1.18 x + 0.0 (10329 voxels, overlap=0.363)
Brain_Stem (16): linear fit = 1.18 x + 0.0 (10329 voxels, peak = 92), gca=92.4
gca peak = 0.12854 (88)
mri peak = 0.08940 (101)
Right_VentralDC (60): linear fit = 1.20 x + 0.0 (1512 voxels, overlap=0.017)
Right_VentralDC (60): linear fit = 1.20 x + 0.0 (1512 voxels, peak = 105), gca=105.2
gca peak = 0.15703 (87)
mri peak = 0.10143 (101)
Left_VentralDC (28): linear fit = 1.20 x + 0.0 (1493 voxels, overlap=0.012)
Left_VentralDC (28): linear fit = 1.20 x + 0.0 (1493 voxels, peak = 104), gca=104.0
gca peak = 0.17522 (25)
mri peak = 0.42105 ( 9)
gca peak = 0.17113 (14)
mri peak = 0.35667 ( 3)
Fourth_Ventricle (15): linear fit = 0.12 x + 0.0 (291 voxels, overlap=0.016)
Fourth_Ventricle (15): linear fit = 0.12 x + 0.0 (291 voxels, peak = 2), gca=1.8
gca peak Unknown = 0.94777 ( 0)
gca peak Left_Inf_Lat_Vent = 0.16627 (28)
gca peak Third_Ventricle = 0.17522 (25)
gca peak Fourth_Ventricle = 0.17113 (14)
gca peak CSF = 0.20346 (36)
gca peak Left_Accumbens_area = 0.70646 (62)
gca peak Left_undetermined = 1.00000 (28)
gca peak Left_vessel = 0.89917 (53)
gca peak Left_choroid_plexus = 0.11689 (35)
gca peak Right_Inf_Lat_Vent = 0.25504 (23)
gca peak Right_Accumbens_area = 0.31650 (65)
gca peak Right_vessel = 0.77268 (52)
gca peak Right_choroid_plexus = 0.13275 (38)
gca peak Fifth_Ventricle = 0.60973 (33)
gca peak WM_hypointensities = 0.11013 (77)
gca peak non_WM_hypointensities = 0.11354 (41)
gca peak Optic_Chiasm = 0.51646 (76)
not using caudate to estimate GM means
estimating mean gm scale to be 1.06 x + 0.0
estimating mean wm scale to be 1.00 x + 0.0
estimating mean csf scale to be 0.41 x + 0.0
Left_Pallidum too bright - rescaling by 0.924 (from 1.125) to 100.9 (was 109.1)
Right_Pallidum too bright - rescaling by 0.960 (from 1.095) to 100.9 (was 105.1)
saving intensity scales to aseg.auto_noCCseg.label_intensities.txt
renormalizing by structure alignment....
renormalizing input #0
gca peak = 0.31706 ( 7)
mri peak = 0.17945 ( 7)
Left_Lateral_Ventricle (4): linear fit = 0.82 x + 0.0 (1560 voxels, overlap=0.926)
Left_Lateral_Ventricle (4): linear fit = 0.82 x + 0.0 (1560 voxels, peak = 6), gca=5.8
gca peak = 0.29738 ( 6)
mri peak = 0.17272 ( 7)
Right_Lateral_Ventricle (43): linear fit = 1.25 x + 0.0 (1589 voxels, overlap=0.924)
Right_Lateral_Ventricle (43): linear fit = 1.25 x + 0.0 (1589 voxels, peak = 7), gca=7.5
gca peak = 0.28156 (100)
mri peak = 0.16332 (106)
Right_Pallidum (52): linear fit = 1.04 x + 0.0 (819 voxels, overlap=0.821)
Right_Pallidum (52): linear fit = 1.04 x + 0.0 (819 voxels, peak = 104), gca=104.5
gca peak = 0.15478 (99)
mri peak = 0.11072 (106)
Left_Pallidum (13): linear fit = 1.08 x + 0.0 (858 voxels, overlap=0.466)
Left_Pallidum (13): linear fit = 1.08 x + 0.0 (858 voxels, peak = 106), gca=106.4
gca peak = 0.26416 (56)
mri peak = 0.10696 (56)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (782 voxels, overlap=1.011)
Right_Hippocampus (53): linear fit = 1.00 x + 0.0 (782 voxels, peak = 56), gca=56.0
gca peak = 0.33569 (58)
mri peak = 0.09347 (60)
Left_Hippocampus (17): linear fit = 0.99 x + 0.0 (838 voxels, overlap=1.009)
Left_Hippocampus (17): linear fit = 0.99 x + 0.0 (838 voxels, peak = 57), gca=57.1
gca peak = 0.07621 (103)
mri peak = 0.13857 (102)
Right_Cerebral_White_Matter (41): linear fit = 1.00 x + 0.0 (41973 voxels, overlap=0.615)
Right_Cerebral_White_Matter (41): linear fit = 1.00 x + 0.0 (41973 voxels, peak = 103), gca=103.0
gca peak = 0.07845 (105)
mri peak = 0.13751 (104)
Left_Cerebral_White_Matter (2): linear fit = 1.00 x + 0.0 (43265 voxels, overlap=0.620)
Left_Cerebral_White_Matter (2): linear fit = 1.00 x + 0.0 (43265 voxels, peak = 105), gca=105.0
gca peak = 0.08782 (63)
mri peak = 0.04398 (66)
Left_Cerebral_Cortex (3): linear fit = 1.00 x + 0.0 (29448 voxels, overlap=0.960)
Left_Cerebral_Cortex (3): linear fit = 1.00 x + 0.0 (29448 voxels, peak = 63), gca=63.0
gca peak = 0.10681 (63)
mri peak = 0.04406 (64)
Right_Cerebral_Cortex (42): linear fit = 1.02 x + 0.0 (28388 voxels, overlap=0.926)
Right_Cerebral_Cortex (42): linear fit = 1.02 x + 0.0 (28388 voxels, peak = 65), gca=64.6
gca peak = 0.22478 (74)
mri peak = 0.18231 (76)
Right_Caudate (50): linear fit = 1.00 x + 0.0 (935 voxels, overlap=1.000)
Right_Caudate (50): linear fit = 1.00 x + 0.0 (935 voxels, peak = 74), gca=74.0
gca peak = 0.14473 (69)
mri peak = 0.13884 (77)
Left_Caudate (11): linear fit = 1.00 x + 0.0 (901 voxels, overlap=0.937)
Left_Caudate (11): linear fit = 1.00 x + 0.0 (901 voxels, peak = 69), gca=69.0
gca peak = 0.16470 (48)
mri peak = 0.05310 (48)
Left_Cerebellum_Cortex (8): linear fit = 1.00 x + 0.0 (14929 voxels, overlap=1.000)
Left_Cerebellum_Cortex (8): linear fit = 1.00 x + 0.0 (14929 voxels, peak = 48), gca=48.0
gca peak = 0.18229 (46)
mri peak = 0.04965 (46)
Right_Cerebellum_Cortex (47): linear fit = 1.05 x + 0.0 (15407 voxels, overlap=0.996)
Right_Cerebellum_Cortex (47): linear fit = 1.05 x + 0.0 (15407 voxels, peak = 49), gca=48.5
gca peak = 0.15119 (90)
mri peak = 0.07994 (89)
Left_Cerebellum_White_Matter (7): linear fit = 1.00 x + 0.0 (5075 voxels, overlap=0.982)
Left_Cerebellum_White_Matter (7): linear fit = 1.00 x + 0.0 (5075 voxels, peak = 90), gca=89.6
gca peak = 0.16700 (90)
mri peak = 0.08530 (88)
Right_Cerebellum_White_Matter (46): linear fit = 0.99 x + 0.0 (5192 voxels, overlap=0.982)
Right_Cerebellum_White_Matter (46): linear fit = 0.99 x + 0.0 (5192 voxels, peak = 89), gca=88.7
gca peak = 0.23301 (63)
mri peak = 0.10000 (63)
Left_Amygdala (18): linear fit = 1.01 x + 0.0 (469 voxels, overlap=1.028)
Left_Amygdala (18): linear fit = 1.01 x + 0.0 (469 voxels, peak = 64), gca=63.9
gca peak = 0.29567 (63)
mri peak = 0.11073 (62)
Right_Amygdala (54): linear fit = 1.00 x + 0.0 (449 voxels, overlap=1.021)
Right_Amygdala (54): linear fit = 1.00 x + 0.0 (449 voxels, peak = 63), gca=63.0
gca peak = 0.10834 (95)
mri peak = 0.08778 (91)
Left_Thalamus (10): linear fit = 0.99 x + 0.0 (4105 voxels, overlap=0.971)
Left_Thalamus (10): linear fit = 0.99 x + 0.0 (4105 voxels, peak = 94), gca=93.6
gca peak = 0.11610 (91)
mri peak = 0.08291 (89)
Right_Thalamus (49): linear fit = 1.00 x + 0.0 (4093 voxels, overlap=0.955)
Right_Thalamus (49): linear fit = 1.00 x + 0.0 (4093 voxels, peak = 91), gca=91.0
gca peak = 0.07778 (89)
mri peak = 0.09072 (84)
Left_Putamen (12): linear fit = 1.00 x + 0.0 (1968 voxels, overlap=0.718)
Left_Putamen (12): linear fit = 1.00 x + 0.0 (1968 voxels, peak = 89), gca=89.0
gca peak = 0.08743 (83)
mri peak = 0.09372 (88)
Right_Putamen (51): linear fit = 1.00 x + 0.0 (2102 voxels, overlap=0.840)
Right_Putamen (51): linear fit = 1.00 x + 0.0 (2102 voxels, peak = 83), gca=83.0
gca peak = 0.07618 (93)
mri peak = 0.06509 (96)
Brain_Stem (16): linear fit = 0.98 x + 0.0 (10329 voxels, overlap=0.855)
Brain_Stem (16): linear fit = 0.98 x + 0.0 (10329 voxels, peak = 91), gca=90.7
gca peak = 0.10886 (105)
mri peak = 0.08940 (101)
Right_VentralDC (60): linear fit = 1.00 x + 0.0 (1512 voxels, overlap=0.835)
Right_VentralDC (60): linear fit = 1.00 x + 0.0 (1512 voxels, peak = 106), gca=105.5
gca peak = 0.16520 (102)
mri peak = 0.10143 (101)
Left_VentralDC (28): linear fit = 0.99 x + 0.0 (1493 voxels, overlap=0.880)
Left_VentralDC (28): linear fit = 0.99 x + 0.0 (1493 voxels, peak = 100), gca=100.5
gca peak = 0.33670 (11)
mri peak = 0.42105 ( 9)
gca peak = 0.38155 ( 6)
mri peak = 0.35667 ( 3)
Fourth_Ventricle (15): linear fit = 0.32 x + 0.0 (291 voxels, overlap=0.435)
Fourth_Ventricle (15): linear fit = 0.32 x + 0.0 (291 voxels, peak = 2), gca=1.9
gca peak Unknown = 0.94777 ( 0)
gca peak Left_Inf_Lat_Vent = 0.20560 (29)
gca peak Third_Ventricle = 0.33670 (11)
gca peak Fourth_Ventricle = 0.38155 ( 6)
gca peak CSF = 0.34330 (15)
gca peak Left_Accumbens_area = 0.61521 (62)
gca peak Left_undetermined = 1.00000 (28)
gca peak Left_vessel = 0.89917 (53)
gca peak Left_choroid_plexus = 0.11689 (35)
gca peak Right_Inf_Lat_Vent = 0.25695 (23)
gca peak Right_Accumbens_area = 0.37254 (72)
gca peak Right_vessel = 0.77268 (52)
gca peak Right_choroid_plexus = 0.13275 (38)
gca peak Fifth_Ventricle = 0.59846 (14)
gca peak WM_hypointensities = 0.11042 (77)
gca peak non_WM_hypointensities = 0.11354 (41)
gca peak Optic_Chiasm = 0.54524 (76)
not using caudate to estimate GM means
estimating mean gm scale to be 1.00 x + 0.0
estimating mean wm scale to be 1.00 x + 0.0
estimating mean csf scale to be 1.03 x + 0.0
Left_Pallidum too bright - rescaling by 0.948 (from 1.075) to 100.9 (was 106.4)
Right_Pallidum too bright - rescaling by 0.965 (from 1.045) to 100.9 (was 104.5)
saving intensity scales to aseg.auto_noCCseg.label_intensities.txt
saving sequentially combined intensity scales to aseg.auto_noCCseg.label_intensities.txt
47186 voxels changed in iteration 0 of unlikely voxel relabeling
187 voxels changed in iteration 1 of unlikely voxel relabeling
4 voxels changed in iteration 2 of unlikely voxel relabeling
0 voxels changed in iteration 3 of unlikely voxel relabeling
27342 gm and wm labels changed (%33 to gray, %67 to white out of all changed labels)
283 hippocampal voxels changed.
0 amygdala voxels changed.
Reclassifying using Gibbs Priors
pass 1: 60355 changed. image ll: -2.078, PF=0.500
pass 2: 16145 changed. image ll: -2.078, PF=0.500
pass 3: 5268 changed.
pass 4: 1940 changed.
35968 voxels changed in iteration 0 of unlikely voxel relabeling
451 voxels changed in iteration 1 of unlikely voxel relabeling
81 voxels changed in iteration 2 of unlikely voxel relabeling
0 voxels changed in iteration 3 of unlikely voxel relabeling
5829 voxels changed in iteration 0 of unlikely voxel relabeling
91 voxels changed in iteration 1 of unlikely voxel relabeling
0 voxels changed in iteration 2 of unlikely voxel relabeling
4774 voxels changed in iteration 0 of unlikely voxel relabeling
31 voxels changed in iteration 1 of unlikely voxel relabeling
2 voxels changed in iteration 2 of unlikely voxel relabeling
0 voxels changed in iteration 3 of unlikely voxel relabeling
4560 voxels changed in iteration 0 of unlikely voxel relabeling
29 voxels changed in iteration 1 of unlikely voxel relabeling
2 voxels changed in iteration 2 of unlikely voxel relabeling
1 voxels changed in iteration 3 of unlikely voxel relabeling
0 voxels changed in iteration 4 of unlikely voxel relabeling
!!!!!!!!! ventricle segment 1 with volume 7087 above threshold 100 - not erasing !!!!!!!!!!
!!!!!!!!! ventricle segment 0 with volume 561 above threshold 100 - not erasing !!!!!!!!!!
!!!!!!!!! ventricle segment 1 with volume 7108 above threshold 100 - not erasing !!!!!!!!!!
!!!!!!!!! ventricle segment 0 with volume 504 above threshold 100 - not erasing !!!!!!!!!!
writing labeled volume to aseg.auto_noCCseg.mgz
mri_ca_label utimesec 1947.324402
mri_ca_label stimesec 8.425987
mri_ca_label ru_maxrss 2103700
mri_ca_label ru_ixrss 0
mri_ca_label ru_idrss 0
mri_ca_label ru_isrss 0
mri_ca_label ru_minflt 1522708
mri_ca_label ru_majflt 167
mri_ca_label ru_nswap 0
mri_ca_label ru_inblock 181432
mri_ca_label ru_oublock 672
mri_ca_label ru_msgsnd 0
mri_ca_label ru_msgrcv 0
mri_ca_label ru_nsignals 0
mri_ca_label ru_nvcsw 514
mri_ca_label ru_nivcsw 20713
auto-labeling took 32 minutes and 37 seconds.
#--------------------------------------
#@# CC Seg Thu Jun 12 13:59:06 UTC 2025
mri_cc -aseg aseg.auto_noCCseg.mgz -o aseg.auto.mgz -lta /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/cc_up.lta subjectname
will read input aseg from aseg.auto_noCCseg.mgz
writing aseg with cc labels to aseg.auto.mgz
will write lta as /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/transforms/cc_up.lta
reading aseg from /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/aseg.auto_noCCseg.mgz
reading norm from /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/norm.mgz
17460 voxels in left wm, 19637 in right wm, xrange [126, 131]
searching rotation angles z=[-7 7], y=[-7 7]
searching scale 1 Z rot 6.8 global minimum found at slice 128.0, rotations (-0.13, 0.03)
final transformation (x=128.0, yr=-0.129, zr=0.034):
1.00000 -0.00059 -0.00224 0.20948;
0.00059 1.00000 -0.00000 35.92439;
0.00224 0.00000 1.00000 36.71302;
0.00000 0.00000 0.00000 1.00000;
updating x range to be [127, 131] in xformed coordinates
best xformed slice 128
min_x_fornix = 150
min_x_fornix = 156
min_x_fornix = 158
min_x_fornix = 156
min_x_fornix = 152
cc center is found at 128 92 91
eigenvectors:
-0.00013 -0.00000 1.00000;
-0.16866 -0.98567 -0.00002;
0.98567 -0.16866 0.00012;
error in mid anterior detected - correcting...
writing aseg with callosum to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/aseg.auto.mgz...
corpus callosum segmentation took 0.5 minutes
#VMPC# mri_cc VmPeak 450060
mri_cc done
#--------------------------------------
#@# Merge ASeg Thu Jun 12 13:59:36 UTC 2025
cp aseg.auto.mgz aseg.presurf.mgz
#--------------------------------------------
#@# Intensity Normalization2 Thu Jun 12 13:59:36 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_normalize -seed 1234 -mprage -aseg aseg.presurf.mgz -mask brainmask.mgz norm.mgz brain.mgz
setting seed for random number genererator to 1234
assuming input volume is MGH (Van der Kouwe) MP-RAGE
using segmentation for initial intensity normalization
using MR volume brainmask.mgz to mask input volume...
reading mri_src from norm.mgz...
Reading aseg aseg.presurf.mgz
normalizing image...
NOT doing gentle normalization with control points/label
processing with aseg
removing outliers in the aseg WM...
308 control points removed
Building bias image
building Voronoi diagram...
performing soap bubble smoothing, sigma = 0...
Smoothing with sigma 8
Applying bias correction
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
Iterating 2 times
---------------------------------
3d normalization pass 1 of 2
white matter peak found at 110
white matter peak found at 110
gm peak at 63 (63), valley at 29 (29)
csf peak at 32, setting threshold to 52
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
---------------------------------
3d normalization pass 2 of 2
white matter peak found at 110
white matter peak found at 110
gm peak at 64 (64), valley at 25 (25)
csf peak at 33, setting threshold to 53
building Voronoi diagram...
performing soap bubble smoothing, sigma = 8...
Done iterating ---------------------------------
writing output to brain.mgz
3D bias adjustment took 2 minutes and 30 seconds.
#--------------------------------------------
#@# Mask BFS Thu Jun 12 14:02:08 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_mask -T 5 brain.mgz brainmask.mgz brain.finalsurfs.mgz
threshold mask volume at 5
DoAbs = 0
Found 1563568 voxels in mask (pct= 9.32)
Writing masked volume to brain.finalsurfs.mgz...done.
#--------------------------------------------
#@# WM Segmentation Thu Jun 12 14:02:09 UTC 2025
AntsDenoiseImageFs -i brain.mgz -o antsdn.brain.mgz
mri_segment -wsizemm 13 -mprage antsdn.brain.mgz wm.seg.mgz
wsizemm = 13, voxres = 1, wsize = 13
Widening wm low from 89 to 79
assuming input volume is MGH (Van der Kouwe) MP-RAGE
wm mean: 110
wsize: 13
wm low: 79
wm hi: 125
gray low: 30
gray hi: 99
Doing initial trinary intensity segmentation
Using local statistics to label ambiguous voxels
Autodetecting stats
Computing class statistics for intensity windows...
CCS WM (103.0): 102.6 +- 6.3 [79.0 --> 125.0]
CCS GM (74.0) : 72.3 +- 10.5 [30.0 --> 95.0]
white_mean 102.629
white_sigma 6.3106
gray_mean 72.3238
gray_sigma 10.4832
setting bottom of white matter range wm_low to 82.8
setting top of gray matter range gray_hi to 93.3
wm_low 82.807
wm_hi 125
gray_low 30
gray_hi 93.2901
Redoing initial intensity segmentation...
Recomputing local statistics to label ambiguous voxels...
wm_low 82.807
wm_hi 125
gray_low 30
gray_hi 93.2901
using local geometry to label remaining ambiguous voxels...
polvwsize = 5, polvlen = 3, gray_hi = 93.2901, wm_low = 82.807
MRIcpolvMedianCurveSegment(): wsize=5, len=3, gmhi=93.2901, wmlow=82.807
116645 voxels processed (0.70%)
54510 voxels white (0.32%)
62135 voxels non-white (0.37%)
Reclassifying voxels using Gaussian border classifier niter=1
MRIreclassify(): wm_low=77.807, gray_hi=93.2901, wsize=13
207002 voxels tested (1.23%)
40536 voxels changed (0.24%)
41711 multi-scale searches (0.25%)
Recovering bright white
MRIrecoverBrightWhite()
wm_low 82.807
wm_hi 125
slack 6.3106
pct_thresh 0.33
intensity_thresh 131.311
nvox_thresh 8.58
181 voxels tested (0.00%)
37 voxels changed (0.00%)
removing voxels with positive offset direction...
MRIremoveWrongDirection() wsize=3, lowthr=77.807, hithr=93.2901
smoothing input volume with sigma = 0.250
78157 voxels tested (0.47%)
14991 voxels changed (0.09%)
thicken = 1
removing 1-dimensional structures...
MRIremove1dStructures(): max_iter=10000, thresh=2, WM_MIN_VAL=5
2692 sparsely connected voxels removed in 1 iterations
thickening thin strands....
thickness 4
nsegments 20
wm_hi 125
948 diagonally connected voxels added...
MRIthickenThinWMStrands(): thickness=4, nsegments=20
20 segments, 4508 filled
MRIfindBrightNonWM(): 1452 bright non-wm voxels segmented.
MRIfilterMorphology() WM_MIN_VAL=5, DIAGONAL_FILL=230
white matter segmentation took 1.0 minutes
writing output to wm.seg.mgz...
mri_edit_wm_with_aseg -keep-in wm.seg.mgz brain.mgz aseg.presurf.mgz wm.asegedit.mgz
preserving editing changes in input volume...
auto filling took 0.51 minutes
reading wm segmentation from wm.seg.mgz...
0 voxels added to wm to prevent paths from MTL structures to cortex
2913 additional wm voxels added
0 additional wm voxels added
SEG EDIT: 44278 voxels turned on, 31168 voxels turned off.
propagating editing to output volume from wm.seg.mgz
writing edited volume to wm.asegedit.mgz....
mri_pretess wm.asegedit.mgz wm norm.mgz wm.mgz
Iteration Number : 1
pass 1 (xy+): 10 found - 10 modified | TOTAL: 10
pass 2 (xy+): 0 found - 10 modified | TOTAL: 10
pass 1 (xy-): 9 found - 9 modified | TOTAL: 19
pass 2 (xy-): 0 found - 9 modified | TOTAL: 19
pass 1 (yz+): 18 found - 18 modified | TOTAL: 37
pass 2 (yz+): 0 found - 18 modified | TOTAL: 37
pass 1 (yz-): 19 found - 19 modified | TOTAL: 56
pass 2 (yz-): 0 found - 19 modified | TOTAL: 56
pass 1 (xz+): 17 found - 17 modified | TOTAL: 73
pass 2 (xz+): 0 found - 17 modified | TOTAL: 73
pass 1 (xz-): 9 found - 9 modified | TOTAL: 82
pass 2 (xz-): 0 found - 9 modified | TOTAL: 82
Iteration Number : 1
pass 1 (+++): 12 found - 12 modified | TOTAL: 12
pass 2 (+++): 0 found - 12 modified | TOTAL: 12
pass 1 (+++): 12 found - 12 modified | TOTAL: 24
pass 2 (+++): 0 found - 12 modified | TOTAL: 24
pass 1 (+++): 16 found - 16 modified | TOTAL: 40
pass 2 (+++): 0 found - 16 modified | TOTAL: 40
pass 1 (+++): 3 found - 3 modified | TOTAL: 43
pass 2 (+++): 0 found - 3 modified | TOTAL: 43
Iteration Number : 1
pass 1 (++): 43 found - 43 modified | TOTAL: 43
pass 2 (++): 0 found - 43 modified | TOTAL: 43
pass 1 (+-): 18 found - 18 modified | TOTAL: 61
pass 2 (+-): 0 found - 18 modified | TOTAL: 61
pass 1 (--): 44 found - 44 modified | TOTAL: 105
pass 2 (--): 0 found - 44 modified | TOTAL: 105
pass 1 (-+): 15 found - 15 modified | TOTAL: 120
pass 2 (-+): 0 found - 15 modified | TOTAL: 120
Iteration Number : 2
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 1 found - 1 modified | TOTAL: 1
pass 2 (xy-): 0 found - 1 modified | TOTAL: 1
pass 1 (yz+): 6 found - 6 modified | TOTAL: 7
pass 2 (yz+): 0 found - 6 modified | TOTAL: 7
pass 1 (yz-): 0 found - 0 modified | TOTAL: 7
pass 1 (xz+): 0 found - 0 modified | TOTAL: 7
pass 1 (xz-): 2 found - 2 modified | TOTAL: 9
pass 2 (xz-): 0 found - 2 modified | TOTAL: 9
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 1 found - 1 modified | TOTAL: 1
pass 2 (+-): 0 found - 1 modified | TOTAL: 1
pass 1 (--): 1 found - 1 modified | TOTAL: 2
pass 2 (--): 0 found - 1 modified | TOTAL: 2
pass 1 (-+): 3 found - 3 modified | TOTAL: 5
pass 2 (-+): 0 found - 3 modified | TOTAL: 5
Iteration Number : 3
pass 1 (xy+): 1 found - 1 modified | TOTAL: 1
pass 2 (xy+): 0 found - 1 modified | TOTAL: 1
pass 1 (xy-): 0 found - 0 modified | TOTAL: 1
pass 1 (yz+): 0 found - 0 modified | TOTAL: 1
pass 1 (yz-): 0 found - 0 modified | TOTAL: 1
pass 1 (xz+): 0 found - 0 modified | TOTAL: 1
pass 1 (xz-): 0 found - 0 modified | TOTAL: 1
Iteration Number : 3
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Iteration Number : 4
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 4
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 4
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 260 (out of 560962: 0.046349)
binarizing input wm segmentation...
Ambiguous edge configurations...
mri_pretess done
#--------------------------------------------
#@# Fill Thu Jun 12 14:04:28 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_fill -a ../scripts/ponscc.cut.log -xform transforms/talairach.lta -segmentation aseg.presurf.mgz -ctab /opt/freesurfer-7.3.2/SubCorticalMassLUT.txt wm.mgz filled.mgz
logging cutting plane coordinates to ../scripts/ponscc.cut.log...
INFO: Using transforms/talairach.lta and its offset for Talairach volume ...
using segmentation aseg.presurf.mgz...
reading input volume...done.
searching for cutting planes...voxel to talairach voxel transform
1.05652 -0.00215 0.00803 -9.89665;
0.00000 1.14977 0.30808 -33.66292;
-0.00863 -0.26274 0.98055 28.36463;
0.00000 0.00000 0.00000 1.00000;
voxel to talairach voxel transform
1.05652 -0.00215 0.00803 -9.89665;
0.00000 1.14977 0.30808 -33.66292;
-0.00863 -0.26274 0.98055 28.36463;
0.00000 0.00000 0.00000 1.00000;
reading segmented volume aseg.presurf.mgz
removing CC from segmentation
Looking for area (min, max) = (350, 1400)
area[0] = 1280 (min = 350, max = 1400), aspect = 0.50 (min = 0.10, max = 0.75)
no need to search
using seed (126, 121, 150), TAL = (2.0, 22.0, 7.0)
talairach voxel to voxel transform
0.94644 -0.00000 -0.00775 9.58643;
-0.00208 0.81148 -0.25494 34.52746;
0.00778 0.21743 0.95146 -19.59134;
0.00000 0.00000 0.00000 1.00000;
segmentation indicates cc at (126, 121, 150) --> (2.0, 22.0, 7.0)
done.
filling took 1.2 minutes
talairach cc position changed to (2.00, 22.00, 7.00)
Erasing brainstem...done.
seed_search_size = 9, min_neighbors = 5
search rh wm seed point around talairach space:(20.00, 22.00, 7.00) SRC: (110.64, 94.25, 150.28)
search lh wm seed point around talairach space (-16.00, 22.00, 7.00), SRC: (144.71, 94.17, 150.56)
compute mri_fill using aseg
Erasing Brain Stem and Cerebellum ...
Define left and right masks using aseg:
Building Voronoi diagram ...
Using the Voronoi diagram for separating WM into two hemispheres ...
Find the largest connected component for each hemisphere ...
Embedding colortable
mri_fill done, writing output to filled.mgz...
cp filled.mgz filled.auto.mgz
#--------------------------------------------
#@# Tessellate lh Thu Jun 12 14:05:40 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mri_pretess ../mri/filled.mgz 255 ../mri/norm.mgz ../mri/filled-pretess255.mgz
Iteration Number : 1
pass 1 (xy+): 1 found - 1 modified | TOTAL: 1
pass 2 (xy+): 0 found - 1 modified | TOTAL: 1
pass 1 (xy-): 0 found - 0 modified | TOTAL: 1
pass 1 (yz+): 16 found - 16 modified | TOTAL: 17
pass 2 (yz+): 0 found - 16 modified | TOTAL: 17
pass 1 (yz-): 9 found - 9 modified | TOTAL: 26
pass 2 (yz-): 0 found - 9 modified | TOTAL: 26
pass 1 (xz+): 1 found - 1 modified | TOTAL: 27
pass 2 (xz+): 0 found - 1 modified | TOTAL: 27
pass 1 (xz-): 0 found - 0 modified | TOTAL: 27
Iteration Number : 1
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 2 found - 2 modified | TOTAL: 2
pass 2 (+++): 0 found - 2 modified | TOTAL: 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 2
Iteration Number : 1
pass 1 (++): 1 found - 1 modified | TOTAL: 1
pass 2 (++): 0 found - 1 modified | TOTAL: 1
pass 1 (+-): 1 found - 1 modified | TOTAL: 2
pass 2 (+-): 0 found - 1 modified | TOTAL: 2
pass 1 (--): 1 found - 1 modified | TOTAL: 3
pass 2 (--): 0 found - 1 modified | TOTAL: 3
pass 1 (-+): 0 found - 0 modified | TOTAL: 3
Iteration Number : 2
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 1 found - 1 modified | TOTAL: 1
pass 2 (xy-): 0 found - 1 modified | TOTAL: 1
pass 1 (yz+): 2 found - 2 modified | TOTAL: 3
pass 2 (yz+): 0 found - 2 modified | TOTAL: 3
pass 1 (yz-): 1 found - 1 modified | TOTAL: 4
pass 2 (yz-): 0 found - 1 modified | TOTAL: 4
pass 1 (xz+): 0 found - 0 modified | TOTAL: 4
pass 1 (xz-): 0 found - 0 modified | TOTAL: 4
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 3
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 36 (out of 271467: 0.013261)
Ambiguous edge configurations...
mri_pretess done
mri_tessellate ../mri/filled-pretess255.mgz 255 ../surf/lh.orig.nofix
7.3.2
7.3.2
slice 40: 1563 vertices, 1694 faces
slice 50: 7479 vertices, 7726 faces
slice 60: 16728 vertices, 17090 faces
slice 70: 27379 vertices, 27754 faces
slice 80: 37574 vertices, 37917 faces
slice 90: 47458 vertices, 47807 faces
slice 100: 57721 vertices, 58081 faces
slice 110: 67494 vertices, 67836 faces
slice 120: 76761 vertices, 77125 faces
slice 130: 86317 vertices, 86691 faces
slice 140: 94986 vertices, 95319 faces
slice 150: 102605 vertices, 102883 faces
slice 160: 109181 vertices, 109419 faces
slice 170: 114787 vertices, 114997 faces
slice 180: 119729 vertices, 119898 faces
slice 190: 122912 vertices, 123024 faces
slice 200: 123586 vertices, 123606 faces
slice 210: 123586 vertices, 123606 faces
slice 220: 123586 vertices, 123606 faces
slice 230: 123586 vertices, 123606 faces
slice 240: 123586 vertices, 123606 faces
slice 250: 123586 vertices, 123606 faces
using the conformed surface RAS to save vertex points...
writing ../surf/lh.orig.nofix
using vox2ras matrix:
-1.00000 0.00000 0.00000 128.00000;
0.00000 0.00000 1.00000 -128.00000;
0.00000 -1.00000 0.00000 128.00000;
0.00000 0.00000 0.00000 1.00000;
rm -f ../mri/filled-pretess255.mgz
mris_extract_main_component ../surf/lh.orig.nofix ../surf/lh.orig.nofix
counting number of connected components...
123586 voxel in cpt #1: X=-20 [v=123586,e=370818,f=247212] located at (-27.235659, -22.251646, 30.684002)
For the whole surface: X=-20 [v=123586,e=370818,f=247212]
One single component has been found
nothing to do
done
#--------------------------------------------
#@# Tessellate rh Thu Jun 12 14:05:46 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mri_pretess ../mri/filled.mgz 127 ../mri/norm.mgz ../mri/filled-pretess127.mgz
Iteration Number : 1
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 1 found - 1 modified | TOTAL: 1
pass 2 (xy-): 0 found - 1 modified | TOTAL: 1
pass 1 (yz+): 9 found - 9 modified | TOTAL: 10
pass 2 (yz+): 0 found - 9 modified | TOTAL: 10
pass 1 (yz-): 16 found - 16 modified | TOTAL: 26
pass 2 (yz-): 0 found - 16 modified | TOTAL: 26
pass 1 (xz+): 0 found - 0 modified | TOTAL: 26
pass 1 (xz-): 0 found - 0 modified | TOTAL: 26
Iteration Number : 1
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 1
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (xy+): 0 found - 0 modified | TOTAL: 0
pass 1 (xy-): 0 found - 0 modified | TOTAL: 0
pass 1 (yz+): 0 found - 0 modified | TOTAL: 0
pass 1 (yz-): 0 found - 0 modified | TOTAL: 0
pass 1 (xz+): 0 found - 0 modified | TOTAL: 0
pass 1 (xz-): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
pass 1 (+++): 0 found - 0 modified | TOTAL: 0
Iteration Number : 2
pass 1 (++): 0 found - 0 modified | TOTAL: 0
pass 1 (+-): 0 found - 0 modified | TOTAL: 0
pass 1 (--): 0 found - 0 modified | TOTAL: 0
pass 1 (-+): 0 found - 0 modified | TOTAL: 0
Total Number of Modified Voxels = 26 (out of 270470: 0.009613)
Ambiguous edge configurations...
mri_pretess done
mri_tessellate ../mri/filled-pretess127.mgz 127 ../surf/rh.orig.nofix
7.3.2
7.3.2
slice 40: 1562 vertices, 1687 faces
slice 50: 7406 vertices, 7648 faces
slice 60: 16583 vertices, 16949 faces
slice 70: 27176 vertices, 27548 faces
slice 80: 37330 vertices, 37673 faces
slice 90: 47228 vertices, 47575 faces
slice 100: 57505 vertices, 57869 faces
slice 110: 67438 vertices, 67789 faces
slice 120: 76827 vertices, 77194 faces
slice 130: 86268 vertices, 86647 faces
slice 140: 94765 vertices, 95086 faces
slice 150: 102254 vertices, 102537 faces
slice 160: 108810 vertices, 109053 faces
slice 170: 114390 vertices, 114613 faces
slice 180: 119252 vertices, 119429 faces
slice 190: 122404 vertices, 122520 faces
slice 200: 123084 vertices, 123108 faces
slice 210: 123084 vertices, 123108 faces
slice 220: 123084 vertices, 123108 faces
slice 230: 123084 vertices, 123108 faces
slice 240: 123084 vertices, 123108 faces
slice 250: 123084 vertices, 123108 faces
using the conformed surface RAS to save vertex points...
writing ../surf/rh.orig.nofix
using vox2ras matrix:
-1.00000 0.00000 0.00000 128.00000;
0.00000 0.00000 1.00000 -128.00000;
0.00000 -1.00000 0.00000 128.00000;
0.00000 0.00000 0.00000 1.00000;
rm -f ../mri/filled-pretess127.mgz
mris_extract_main_component ../surf/rh.orig.nofix ../surf/rh.orig.nofix
counting number of connected components...
123084 voxel in cpt #1: X=-24 [v=123084,e=369324,f=246216] located at (26.963943, -22.324535, 30.661808)
For the whole surface: X=-24 [v=123084,e=369324,f=246216]
One single component has been found
nothing to do
done
#--------------------------------------------
#@# Smooth1 lh Thu Jun 12 14:05:51 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_smooth -nw -seed 1234 ../surf/lh.orig.nofix ../surf/lh.smoothwm.nofix
setting seed for random number generator to 1234
smoothing surface tessellation for 10 iterations...
smoothing complete - recomputing first and second fundamental forms...
#--------------------------------------------
#@# Smooth1 rh Thu Jun 12 14:05:54 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_smooth -nw -seed 1234 ../surf/rh.orig.nofix ../surf/rh.smoothwm.nofix
setting seed for random number generator to 1234
smoothing surface tessellation for 10 iterations...
smoothing complete - recomputing first and second fundamental forms...
#--------------------------------------------
#@# Inflation1 lh Thu Jun 12 14:05:57 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_inflate -no-save-sulc ../surf/lh.smoothwm.nofix ../surf/lh.inflated.nofix
Not saving sulc
Reading ../surf/lh.smoothwm.nofix
avg radius = 47.1 mm, total surface area = 69045 mm^2
step 060: RMS=0.032 (target=0.015) writing inflated surface to ../surf/lh.inflated.nofix
inflation took 0.4 minutes
inflation complete.
Not saving sulc
mris_inflate utimesec 19.815110
mris_inflate stimesec 1.292207
mris_inflate ru_maxrss 195840
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 534457
mris_inflate ru_majflt 15
mris_inflate ru_nswap 0
mris_inflate ru_inblock 3696
mris_inflate ru_oublock 8696
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 276
mris_inflate ru_nivcsw 91
#--------------------------------------------
#@# Inflation1 rh Thu Jun 12 14:06:19 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_inflate -no-save-sulc ../surf/rh.smoothwm.nofix ../surf/rh.inflated.nofix
Not saving sulc
Reading ../surf/rh.smoothwm.nofix
avg radius = 47.0 mm, total surface area = 68801 mm^2
step 060: RMS=0.029 (target=0.015) writing inflated surface to ../surf/rh.inflated.nofix
inflation took 0.4 minutes
inflation complete.
Not saving sulc
mris_inflate utimesec 19.937156
mris_inflate stimesec 1.216313
mris_inflate ru_maxrss 195080
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 522054
mris_inflate ru_majflt 0
mris_inflate ru_nswap 0
mris_inflate ru_inblock 0
mris_inflate ru_oublock 8664
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 266
mris_inflate ru_nivcsw 271
#--------------------------------------------
#@# QSphere lh Thu Jun 12 14:06:40 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_sphere -q -p 6 -a 128 -seed 1234 ../surf/lh.inflated.nofix ../surf/lh.qsphere.nofix
doing quick spherical unfolding.
limitting unfolding to 6 passes
using n_averages = 128
setting seed for random number genererator to 1234
version: 7.3.2
available threads: 1
scaling brain by 0.321...
inflating...
projecting onto sphere...
surface projected - minimizing metric distortion...
vertex spacing 1.04 +- 0.60 (0.00-->6.25) (max @ vno 94710 --> 94732)
face area 0.03 +- 0.03 (-0.15-->0.61)
Entering MRISinflateToSphere()
inflating to sphere (rms error < 2.00)
000: dt: 0.0000, rms radial error=176.279, avgs=0
005/300: dt: 0.9000, rms radial error=176.018, avgs=0
010/300: dt: 0.9000, rms radial error=175.458, avgs=0
015/300: dt: 0.9000, rms radial error=174.720, avgs=0
020/300: dt: 0.9000, rms radial error=173.881, avgs=0
025/300: dt: 0.9000, rms radial error=172.989, avgs=0
030/300: dt: 0.9000, rms radial error=172.071, avgs=0
035/300: dt: 0.9000, rms radial error=171.136, avgs=0
040/300: dt: 0.9000, rms radial error=170.192, avgs=0
045/300: dt: 0.9000, rms radial error=169.246, avgs=0
050/300: dt: 0.9000, rms radial error=168.300, avgs=0
055/300: dt: 0.9000, rms radial error=167.357, avgs=0
060/300: dt: 0.9000, rms radial error=166.417, avgs=0
065/300: dt: 0.9000, rms radial error=165.482, avgs=0
070/300: dt: 0.9000, rms radial error=164.551, avgs=0
075/300: dt: 0.9000, rms radial error=163.625, avgs=0
080/300: dt: 0.9000, rms radial error=162.705, avgs=0
085/300: dt: 0.9000, rms radial error=161.790, avgs=0
090/300: dt: 0.9000, rms radial error=160.880, avgs=0
095/300: dt: 0.9000, rms radial error=159.976, avgs=0
100/300: dt: 0.9000, rms radial error=159.077, avgs=0
105/300: dt: 0.9000, rms radial error=158.183, avgs=0
110/300: dt: 0.9000, rms radial error=157.294, avgs=0
115/300: dt: 0.9000, rms radial error=156.409, avgs=0
120/300: dt: 0.9000, rms radial error=155.530, avgs=0
125/300: dt: 0.9000, rms radial error=154.656, avgs=0
130/300: dt: 0.9000, rms radial error=153.787, avgs=0
135/300: dt: 0.9000, rms radial error=152.923, avgs=0
140/300: dt: 0.9000, rms radial error=152.064, avgs=0
145/300: dt: 0.9000, rms radial error=151.209, avgs=0
150/300: dt: 0.9000, rms radial error=150.359, avgs=0
155/300: dt: 0.9000, rms radial error=149.514, avgs=0
160/300: dt: 0.9000, rms radial error=148.674, avgs=0
165/300: dt: 0.9000, rms radial error=147.838, avgs=0
170/300: dt: 0.9000, rms radial error=147.007, avgs=0
175/300: dt: 0.9000, rms radial error=146.180, avgs=0
180/300: dt: 0.9000, rms radial error=145.358, avgs=0
185/300: dt: 0.9000, rms radial error=144.541, avgs=0
190/300: dt: 0.9000, rms radial error=143.728, avgs=0
195/300: dt: 0.9000, rms radial error=142.920, avgs=0
200/300: dt: 0.9000, rms radial error=142.116, avgs=0
205/300: dt: 0.9000, rms radial error=141.316, avgs=0
210/300: dt: 0.9000, rms radial error=140.521, avgs=0
215/300: dt: 0.9000, rms radial error=139.730, avgs=0
220/300: dt: 0.9000, rms radial error=138.944, avgs=0
225/300: dt: 0.9000, rms radial error=138.162, avgs=0
230/300: dt: 0.9000, rms radial error=137.384, avgs=0
235/300: dt: 0.9000, rms radial error=136.610, avgs=0
240/300: dt: 0.9000, rms radial error=135.841, avgs=0
245/300: dt: 0.9000, rms radial error=135.076, avgs=0
250/300: dt: 0.9000, rms radial error=134.316, avgs=0
255/300: dt: 0.9000, rms radial error=133.559, avgs=0
260/300: dt: 0.9000, rms radial error=132.807, avgs=0
265/300: dt: 0.9000, rms radial error=132.059, avgs=0
270/300: dt: 0.9000, rms radial error=131.315, avgs=0
275/300: dt: 0.9000, rms radial error=130.575, avgs=0
280/300: dt: 0.9000, rms radial error=129.840, avgs=0
285/300: dt: 0.9000, rms radial error=129.108, avgs=0
290/300: dt: 0.9000, rms radial error=128.381, avgs=0
295/300: dt: 0.9000, rms radial error=127.658, avgs=0
300/300: dt: 0.9000, rms radial error=126.939, avgs=0
spherical inflation complete.
epoch 1 (K=10.0), pass 1, starting sse = 14165.20
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00042
epoch 2 (K=40.0), pass 1, starting sse = 2219.93
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00043
epoch 3 (K=160.0), pass 1, starting sse = 204.14
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.24/20 = 0.01191
epoch 4 (K=640.0), pass 1, starting sse = 7.62
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.15/24 = 0.00630
final distance error %100000.00
writing spherical brain to ../surf/lh.qsphere.nofix
spherical transformation took 0.0466 hours
FSRUNTIME@ mris_sphere 0.0466 hours 1 threads
#VMPC# mris_sphere VmPeak 473784
mris_sphere done
#--------------------------------------------
#@# QSphere rh Thu Jun 12 14:09:28 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_sphere -q -p 6 -a 128 -seed 1234 ../surf/rh.inflated.nofix ../surf/rh.qsphere.nofix
doing quick spherical unfolding.
limitting unfolding to 6 passes
using n_averages = 128
setting seed for random number genererator to 1234
version: 7.3.2
available threads: 1
scaling brain by 0.321...
inflating...
projecting onto sphere...
surface projected - minimizing metric distortion...
vertex spacing 1.04 +- 0.59 (0.00-->6.15) (max @ vno 56154 --> 56161)
face area 0.03 +- 0.03 (-0.11-->0.63)
Entering MRISinflateToSphere()
inflating to sphere (rms error < 2.00)
000: dt: 0.0000, rms radial error=176.325, avgs=0
005/300: dt: 0.9000, rms radial error=176.064, avgs=0
010/300: dt: 0.9000, rms radial error=175.503, avgs=0
015/300: dt: 0.9000, rms radial error=174.765, avgs=0
020/300: dt: 0.9000, rms radial error=173.924, avgs=0
025/300: dt: 0.9000, rms radial error=173.023, avgs=0
030/300: dt: 0.9000, rms radial error=172.100, avgs=0
035/300: dt: 0.9000, rms radial error=171.166, avgs=0
040/300: dt: 0.9000, rms radial error=170.223, avgs=0
045/300: dt: 0.9000, rms radial error=169.277, avgs=0
050/300: dt: 0.9000, rms radial error=168.332, avgs=0
055/300: dt: 0.9000, rms radial error=167.390, avgs=0
060/300: dt: 0.9000, rms radial error=166.450, avgs=0
065/300: dt: 0.9000, rms radial error=165.515, avgs=0
070/300: dt: 0.9000, rms radial error=164.584, avgs=0
075/300: dt: 0.9000, rms radial error=163.658, avgs=0
080/300: dt: 0.9000, rms radial error=162.739, avgs=0
085/300: dt: 0.9000, rms radial error=161.825, avgs=0
090/300: dt: 0.9000, rms radial error=160.916, avgs=0
095/300: dt: 0.9000, rms radial error=160.013, avgs=0
100/300: dt: 0.9000, rms radial error=159.115, avgs=0
105/300: dt: 0.9000, rms radial error=158.222, avgs=0
110/300: dt: 0.9000, rms radial error=157.334, avgs=0
115/300: dt: 0.9000, rms radial error=156.451, avgs=0
120/300: dt: 0.9000, rms radial error=155.573, avgs=0
125/300: dt: 0.9000, rms radial error=154.699, avgs=0
130/300: dt: 0.9000, rms radial error=153.830, avgs=0
135/300: dt: 0.9000, rms radial error=152.965, avgs=0
140/300: dt: 0.9000, rms radial error=152.106, avgs=0
145/300: dt: 0.9000, rms radial error=151.251, avgs=0
150/300: dt: 0.9000, rms radial error=150.402, avgs=0
155/300: dt: 0.9000, rms radial error=149.556, avgs=0
160/300: dt: 0.9000, rms radial error=148.716, avgs=0
165/300: dt: 0.9000, rms radial error=147.880, avgs=0
170/300: dt: 0.9000, rms radial error=147.049, avgs=0
175/300: dt: 0.9000, rms radial error=146.223, avgs=0
180/300: dt: 0.9000, rms radial error=145.401, avgs=0
185/300: dt: 0.9000, rms radial error=144.584, avgs=0
190/300: dt: 0.9000, rms radial error=143.771, avgs=0
195/300: dt: 0.9000, rms radial error=142.963, avgs=0
200/300: dt: 0.9000, rms radial error=142.159, avgs=0
205/300: dt: 0.9000, rms radial error=141.359, avgs=0
210/300: dt: 0.9000, rms radial error=140.564, avgs=0
215/300: dt: 0.9000, rms radial error=139.774, avgs=0
220/300: dt: 0.9000, rms radial error=138.987, avgs=0
225/300: dt: 0.9000, rms radial error=138.205, avgs=0
230/300: dt: 0.9000, rms radial error=137.427, avgs=0
235/300: dt: 0.9000, rms radial error=136.654, avgs=0
240/300: dt: 0.9000, rms radial error=135.885, avgs=0
245/300: dt: 0.9000, rms radial error=135.120, avgs=0
250/300: dt: 0.9000, rms radial error=134.359, avgs=0
255/300: dt: 0.9000, rms radial error=133.603, avgs=0
260/300: dt: 0.9000, rms radial error=132.851, avgs=0
265/300: dt: 0.9000, rms radial error=132.103, avgs=0
270/300: dt: 0.9000, rms radial error=131.359, avgs=0
275/300: dt: 0.9000, rms radial error=130.619, avgs=0
280/300: dt: 0.9000, rms radial error=129.884, avgs=0
285/300: dt: 0.9000, rms radial error=129.152, avgs=0
290/300: dt: 0.9000, rms radial error=128.425, avgs=0
295/300: dt: 0.9000, rms radial error=127.701, avgs=0
300/300: dt: 0.9000, rms radial error=126.982, avgs=0
spherical inflation complete.
epoch 1 (K=10.0), pass 1, starting sse = 14093.30
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00039
epoch 2 (K=40.0), pass 1, starting sse = 2207.83
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.01/13 = 0.00049
epoch 3 (K=160.0), pass 1, starting sse = 199.57
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.20/18 = 0.01128
epoch 4 (K=640.0), pass 1, starting sse = 7.76
taking momentum steps...
taking momentum steps...
taking momentum steps...
taking momentum steps...
pass 1 complete, delta sse/iter = 0.20/22 = 0.00914
final distance error %100000.00
writing spherical brain to ../surf/rh.qsphere.nofix
spherical transformation took 0.0437 hours
FSRUNTIME@ mris_sphere 0.0437 hours 1 threads
#VMPC# mris_sphere VmPeak 473020
mris_sphere done
#@# Fix Topology lh Thu Jun 12 14:12:05 UTC 2025
mris_fix_topology -mgz -sphere qsphere.nofix -inflated inflated.nofix -orig orig.nofix -out orig.premesh -ga -seed 1234 subjectname lh
reading spherical homeomorphism from 'qsphere.nofix'
reading inflated coordinates from 'inflated.nofix'
reading original coordinates from 'orig.nofix'
using genetic algorithm with optimized parameters
setting seed for random number genererator to 1234
*************************************************************
Topology Correction Parameters
retessellation mode: genetic search
number of patches/generation : 10
number of generations : 10
surface mri loglikelihood coefficient : 1.0
volume mri loglikelihood coefficient : 10.0
normal dot loglikelihood coefficient : 1.0
quadratic curvature loglikelihood coefficient : 1.0
volume resolution : 2
eliminate vertices during search : 1
initial patch selection : 1
select all defect vertices : 0
ordering dependant retessellation: 0
use precomputed edge table : 0
smooth retessellated patch : 2
match retessellated patch : 1
verbose mode : 0
*************************************************************
INFO: assuming .mgz format
writing corrected surface to 'orig.premesh'
7.3.2
7.3.2
before topology correction, eno=-20 (nv=123586, nf=247212, ne=370818, g=11)
using quasi-homeomorphic spherical map to tessellate cortical surface...
Correction of the Topology
Finding true center and radius of Spherical Surface...done
Surface centered at (0,0,0) with radius 100.0 in 9 iterations
marking ambiguous vertices...
2265 ambiguous faces found in tessellation
segmenting defects...
20 defects found, arbitrating ambiguous regions...
analyzing neighboring defects...
20 defects to be corrected
0 vertices coincident
reading input surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.qsphere.nofix...
reading brain volume from brain...
reading wm segmentation from wm...
Reading original properties of orig.nofix
Reading vertex positions of inflated.nofix
Computing Initial Surface Statistics
-face loglikelihood: -9.3632 (-4.6816)
-vertex loglikelihood: -6.3223 (-3.1612)
-normal dot loglikelihood: -3.6319 (-3.6319)
-quad curv loglikelihood: -6.0087 (-3.0044)
Total Loglikelihood : -25.3261
CORRECTING DEFECT 0 (vertices=64, convex hull=114, v0=46)
After retessellation of defect 0 (v0=46), euler #=-17 (122208,366038,243813) : difference with theory (-17) = 0
CORRECTING DEFECT 1 (vertices=146, convex hull=139, v0=198)
After retessellation of defect 1 (v0=198), euler #=-16 (122276,366315,244023) : difference with theory (-16) = 0
CORRECTING DEFECT 2 (vertices=112, convex hull=131, v0=1074)
After retessellation of defect 2 (v0=1074), euler #=-15 (122341,366575,244219) : difference with theory (-15) = 0
CORRECTING DEFECT 3 (vertices=277, convex hull=137, v0=11411)
After retessellation of defect 3 (v0=11411), euler #=-14 (122405,366829,244410) : difference with theory (-14) = 0
CORRECTING DEFECT 4 (vertices=7, convex hull=31, v0=16234)
After retessellation of defect 4 (v0=16234), euler #=-13 (122407,366845,244425) : difference with theory (-13) = 0
CORRECTING DEFECT 5 (vertices=93, convex hull=125, v0=25452)
After retessellation of defect 5 (v0=25452), euler #=-12 (122438,366998,244548) : difference with theory (-12) = 0
CORRECTING DEFECT 6 (vertices=8, convex hull=26, v0=30341)
After retessellation of defect 6 (v0=30341), euler #=-11 (122440,367012,244561) : difference with theory (-11) = 0
CORRECTING DEFECT 7 (vertices=58, convex hull=90, v0=37517)
After retessellation of defect 7 (v0=37517), euler #=-10 (122471,367145,244664) : difference with theory (-10) = 0
CORRECTING DEFECT 8 (vertices=142, convex hull=134, v0=51389)
After retessellation of defect 8 (v0=51389), euler #=-9 (122530,367392,244853) : difference with theory (-9) = 0
CORRECTING DEFECT 9 (vertices=14, convex hull=18, v0=55197)
After retessellation of defect 9 (v0=55197), euler #=-8 (122534,367409,244867) : difference with theory (-8) = 0
CORRECTING DEFECT 10 (vertices=5, convex hull=28, v0=55480)
After retessellation of defect 10 (v0=55480), euler #=-7 (122535,367419,244877) : difference with theory (-7) = 0
CORRECTING DEFECT 11 (vertices=27, convex hull=39, v0=64579)
After retessellation of defect 11 (v0=64579), euler #=-6 (122547,367470,244917) : difference with theory (-6) = 0
CORRECTING DEFECT 12 (vertices=42, convex hull=63, v0=67279)
After retessellation of defect 12 (v0=67279), euler #=-5 (122564,367548,244979) : difference with theory (-5) = 0
CORRECTING DEFECT 13 (vertices=20, convex hull=26, v0=78184)
After retessellation of defect 13 (v0=78184), euler #=-4 (122569,367571,244998) : difference with theory (-4) = 0
CORRECTING DEFECT 14 (vertices=101, convex hull=126, v0=78952)
After retessellation of defect 14 (v0=78952), euler #=-3 (122625,367792,245164) : difference with theory (-3) = 0
CORRECTING DEFECT 15 (vertices=42, convex hull=31, v0=81100)
After retessellation of defect 15 (v0=81100), euler #=-2 (122627,367812,245183) : difference with theory (-2) = 0
CORRECTING DEFECT 16 (vertices=23, convex hull=65, v0=89099)
After retessellation of defect 16 (v0=89099), euler #=-1 (122638,367868,245229) : difference with theory (-1) = 0
CORRECTING DEFECT 17 (vertices=197, convex hull=41, v0=90556)
After retessellation of defect 17 (v0=90556), euler #=0 (122650,367917,245267) : difference with theory (0) = 0
CORRECTING DEFECT 18 (vertices=6, convex hull=13, v0=90628)
After retessellation of defect 18 (v0=90628), euler #=1 (122650,367921,245272) : difference with theory (1) = 0
CORRECTING DEFECT 19 (vertices=34, convex hull=55, v0=115316)
After retessellation of defect 19 (v0=115316), euler #=2 (122665,367989,245326) : difference with theory (2) = 0
computing original vertex metric properties...
storing new metric properties...
computing tessellation statistics...
vertex spacing 0.89 +- 0.21 (0.08-->7.54) (max @ vno 2139 --> 2631)
face area -nan +- -nan (1000.00-->-1.00)
performing soap bubble on retessellated vertices for 0 iterations...
vertex spacing 0.89 +- 0.21 (0.08-->7.54) (max @ vno 2139 --> 2631)
face area -nan +- -nan (1000.00-->-1.00)
tessellation finished, orienting corrected surface...
56 mutations (35.9%), 100 crossovers (64.1%), 15 vertices were eliminated
building final representation...
921 vertices and 0 faces have been removed from triangulation
after topology correction, eno=2 (nv=122665, nf=245326, ne=367989, g=0)
writing corrected surface to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.premesh...
0.000 % of the vertices (0 vertices) exhibit an orientation change
removing intersecting faces
000: 156 intersecting
001: 4 intersecting
terminating search with 0 intersecting
topology fixing took 1.5 minutes
FSRUNTIME@ mris_fix_topology lh 0.0249 hours 1 threads
#VMPC# mris_fix_topology VmPeak 759004
#@# Fix Topology rh Thu Jun 12 14:13:35 UTC 2025
mris_fix_topology -mgz -sphere qsphere.nofix -inflated inflated.nofix -orig orig.nofix -out orig.premesh -ga -seed 1234 subjectname rh
reading spherical homeomorphism from 'qsphere.nofix'
reading inflated coordinates from 'inflated.nofix'
reading original coordinates from 'orig.nofix'
using genetic algorithm with optimized parameters
setting seed for random number genererator to 1234
*************************************************************
Topology Correction Parameters
retessellation mode: genetic search
number of patches/generation : 10
number of generations : 10
surface mri loglikelihood coefficient : 1.0
volume mri loglikelihood coefficient : 10.0
normal dot loglikelihood coefficient : 1.0
quadratic curvature loglikelihood coefficient : 1.0
volume resolution : 2
eliminate vertices during search : 1
initial patch selection : 1
select all defect vertices : 0
ordering dependant retessellation: 0
use precomputed edge table : 0
smooth retessellated patch : 2
match retessellated patch : 1
verbose mode : 0
*************************************************************
INFO: assuming .mgz format
writing corrected surface to 'orig.premesh'
7.3.2
7.3.2
before topology correction, eno=-24 (nv=123084, nf=246216, ne=369324, g=13)
using quasi-homeomorphic spherical map to tessellate cortical surface...
Correction of the Topology
Finding true center and radius of Spherical Surface...done
Surface centered at (0,0,0) with radius 100.0 in 9 iterations
marking ambiguous vertices...
1668 ambiguous faces found in tessellation
segmenting defects...
19 defects found, arbitrating ambiguous regions...
analyzing neighboring defects...
19 defects to be corrected
0 vertices coincident
reading input surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.qsphere.nofix...
reading brain volume from brain...
reading wm segmentation from wm...
Reading original properties of orig.nofix
Reading vertex positions of inflated.nofix
Computing Initial Surface Statistics
-face loglikelihood: -9.2790 (-4.6395)
-vertex loglikelihood: -6.3413 (-3.1706)
-normal dot loglikelihood: -3.4852 (-3.4852)
-quad curv loglikelihood: -6.0280 (-3.0140)
Total Loglikelihood : -25.1335
CORRECTING DEFECT 0 (vertices=84, convex hull=130, v0=48)
After retessellation of defect 0 (v0=48), euler #=-16 (122041,365633,243576) : difference with theory (-16) = 0
CORRECTING DEFECT 1 (vertices=127, convex hull=151, v0=243)
After retessellation of defect 1 (v0=243), euler #=-15 (122098,365879,243766) : difference with theory (-15) = 0
CORRECTING DEFECT 2 (vertices=118, convex hull=160, v0=6757)
After retessellation of defect 2 (v0=6757), euler #=-14 (122163,366141,243964) : difference with theory (-14) = 0
CORRECTING DEFECT 3 (vertices=254, convex hull=120, v0=11309)
After retessellation of defect 3 (v0=11309), euler #=-13 (122227,366392,244152) : difference with theory (-13) = 0
CORRECTING DEFECT 4 (vertices=26, convex hull=72, v0=13600)
After retessellation of defect 4 (v0=13600), euler #=-12 (122238,366450,244200) : difference with theory (-12) = 0
CORRECTING DEFECT 5 (vertices=5, convex hull=40, v0=16059)
After retessellation of defect 5 (v0=16059), euler #=-11 (122240,366466,244215) : difference with theory (-11) = 0
CORRECTING DEFECT 6 (vertices=45, convex hull=82, v0=27512)
After retessellation of defect 6 (v0=27512), euler #=-10 (122249,366530,244271) : difference with theory (-10) = 0
CORRECTING DEFECT 7 (vertices=6, convex hull=32, v0=32079)
After retessellation of defect 7 (v0=32079), euler #=-9 (122250,366544,244285) : difference with theory (-9) = 0
CORRECTING DEFECT 8 (vertices=72, convex hull=113, v0=37267)
After retessellation of defect 8 (v0=37267), euler #=-8 (122288,366703,244407) : difference with theory (-8) = 0
CORRECTING DEFECT 9 (vertices=7, convex hull=24, v0=52104)
After retessellation of defect 9 (v0=52104), euler #=-7 (122291,366719,244421) : difference with theory (-7) = 0
CORRECTING DEFECT 10 (vertices=36, convex hull=25, v0=52205)
After retessellation of defect 10 (v0=52205), euler #=-6 (122293,366733,244434) : difference with theory (-6) = 0
CORRECTING DEFECT 11 (vertices=22, convex hull=51, v0=64252)
After retessellation of defect 11 (v0=64252), euler #=-5 (122301,366776,244470) : difference with theory (-5) = 0
CORRECTING DEFECT 12 (vertices=87, convex hull=44, v0=67232)
After retessellation of defect 12 (v0=67232), euler #=-4 (122331,366881,244546) : difference with theory (-4) = 0
CORRECTING DEFECT 13 (vertices=75, convex hull=112, v0=79002)
After retessellation of defect 13 (v0=79002), euler #=-3 (122379,367068,244686) : difference with theory (-3) = 0
CORRECTING DEFECT 14 (vertices=11, convex hull=16, v0=80376)
After retessellation of defect 14 (v0=80376), euler #=-2 (122383,367084,244699) : difference with theory (-2) = 0
CORRECTING DEFECT 15 (vertices=44, convex hull=41, v0=83142)
After retessellation of defect 15 (v0=83142), euler #=-1 (122392,367126,244733) : difference with theory (-1) = 0
CORRECTING DEFECT 16 (vertices=6, convex hull=14, v0=86941)
After retessellation of defect 16 (v0=86941), euler #=0 (122393,367134,244741) : difference with theory (0) = 0
CORRECTING DEFECT 17 (vertices=23, convex hull=61, v0=88998)
After retessellation of defect 17 (v0=88998), euler #=1 (122402,367183,244782) : difference with theory (1) = 0
CORRECTING DEFECT 18 (vertices=33, convex hull=51, v0=94506)
After retessellation of defect 18 (v0=94506), euler #=2 (122417,367245,244830) : difference with theory (2) = 0
computing original vertex metric properties...
storing new metric properties...
computing tessellation statistics...
vertex spacing 0.89 +- 0.21 (0.12-->5.55) (max @ vno 26879 --> 31076)
face area -nan +- -nan (1000.00-->-1.00)
performing soap bubble on retessellated vertices for 0 iterations...
vertex spacing 0.89 +- 0.21 (0.12-->5.55) (max @ vno 26879 --> 31076)
face area -nan +- -nan (1000.00-->-1.00)
tessellation finished, orienting corrected surface...
70 mutations (37.6%), 116 crossovers (62.4%), 35 vertices were eliminated
building final representation...
667 vertices and 0 faces have been removed from triangulation
after topology correction, eno=2 (nv=122417, nf=244830, ne=367245, g=0)
writing corrected surface to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.premesh...
0.000 % of the vertices (0 vertices) exhibit an orientation change
removing intersecting faces
000: 104 intersecting
001: 4 intersecting
002: 3 intersecting
terminating search with 0 intersecting
topology fixing took 1.4 minutes
FSRUNTIME@ mris_fix_topology rh 0.0225 hours 1 threads
#VMPC# mris_fix_topology VmPeak 758412
mris_euler_number ../surf/lh.orig.premesh
euler # = v-e+f = 2g-2: 122665 - 367989 + 245326 = 2 --> 0 holes
F =2V-4: 245326 = 245330-4 (0)
2E=3F: 735978 = 735978 (0)
total defect index = 0
mris_euler_number ../surf/rh.orig.premesh
euler # = v-e+f = 2g-2: 122417 - 367245 + 244830 = 2 --> 0 holes
F =2V-4: 244830 = 244834-4 (0)
2E=3F: 734490 = 734490 (0)
total defect index = 0
Thu Jun 12 14:14:57 UTC 2025
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
/opt/freesurfer-7.3.2/bin/defect2seg --s subjectname --cortex
freesurfer-linux-centos8_x86_64-7.3.2-20220804-6354275
defect2seg 7.3.2
Linux jupyter-monidoerig 5.4.0-204-generic #224-Ubuntu SMP Thu Dec 5 13:38:28 UTC 2024 x86_64 x86_64 x86_64 GNU/Linux
pid 3540228
mri_label2label --label-cortex /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.nofix /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/aseg.presurf.mgz 0 /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/lh.nofix.cortex.label
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
13 non-cortical segments detected
only using segment with 7277 vertices
erasing segment 1 (vno[0] = 51162)
erasing segment 2 (vno[0] = 87031)
erasing segment 3 (vno[0] = 87917)
erasing segment 4 (vno[0] = 88800)
erasing segment 5 (vno[0] = 88816)
erasing segment 6 (vno[0] = 89716)
erasing segment 7 (vno[0] = 90465)
erasing segment 8 (vno[0] = 90506)
erasing segment 9 (vno[0] = 90553)
erasing segment 10 (vno[0] = 91254)
erasing segment 11 (vno[0] = 91336)
erasing segment 12 (vno[0] = 96325)
mri_label2vol --defects /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.nofix /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.defect_labels /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/orig.mgz 1000 0 /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/surface.defects.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/lh.nofix.cortex.label
mri_label2vol supposed to be reproducible but seed not set
Contraining to label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/lh.nofix.cortex.label
Changing input type 0 to MRI_INT
Converting defects to volume: offset=1000, merge=0
Writing to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/surface.defects.mgz
mris_defects_pointset -s /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.nofix -d /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.defect_labels -o /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.defects.pointset --label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/lh.nofix.cortex.label
Reading in surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.nofix
Reading in defect segmentation /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.defect_labels
Reading in label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/lh.nofix.cortex.label
#VMPC# mris_defects_pointset 183980
mris_defects_pointset done
mri_label2label --label-cortex /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.nofix /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/aseg.presurf.mgz 0 /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/rh.nofix.cortex.label
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
10 non-cortical segments detected
only using segment with 7387 vertices
erasing segment 1 (vno[0] = 77456)
erasing segment 2 (vno[0] = 80482)
erasing segment 3 (vno[0] = 81383)
erasing segment 4 (vno[0] = 83222)
erasing segment 5 (vno[0] = 90415)
erasing segment 6 (vno[0] = 92093)
erasing segment 7 (vno[0] = 92983)
erasing segment 8 (vno[0] = 103940)
erasing segment 9 (vno[0] = 107995)
mri_label2vol --defects /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.nofix /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.defect_labels /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/surface.defects.mgz 2000 1 /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/surface.defects.mgz /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/rh.nofix.cortex.label
mri_label2vol supposed to be reproducible but seed not set
Contraining to label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/rh.nofix.cortex.label
Converting defects to volume: offset=2000, merge=1
Writing to /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri/surface.defects.mgz
mris_defects_pointset -s /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.nofix -d /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.defect_labels -o /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.defects.pointset --label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/rh.nofix.cortex.label
Reading in surface /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.nofix
Reading in defect segmentation /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.defect_labels
Reading in label /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/label/rh.nofix.cortex.label
#VMPC# mris_defects_pointset 183400
mris_defects_pointset done
Started at Thu Jun 12 14:14:57 UTC 2025
Ended at Thu Jun 12 14:15:35 UTC 2025
Defect2seg-Run-Time-Sec 38
Defect2seg-Run-Time-Min 0.76
Defect2seg-Run-Time-Hours 0.01
tkmeditfv subjectname brain.finalsurfs.mgz -defect
defect2seg Done
mris_remesh --remesh --iters 3 --input /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig.premesh --output /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/lh.orig
iters = 3
standard remeshing without target
adjusted l: 0.714492
remeshing to edge length 0.714492 with 3 iterations
avg qual before : 0.893529 after: 0.971044
Removing intersections
Remeshed surface quality stats nv0 = 122665 nv = 128612 1.04848
Area 257220 0.30578 0.03410 0.073937 0.4764
Corner 771660 60.00000 8.82459 15.341482 145.9457
Edge 385830 0.84839 0.08303 0.458717 1.2680
Hinge 385830 8.78222 9.21839 0.000007 172.2284
mris_remesh done
mris_remesh --remesh --iters 3 --input /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig.premesh --output /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf/rh.orig
iters = 3
standard remeshing without target
adjusted l: 0.714266
remeshing to edge length 0.714266 with 3 iterations
avg qual before : 0.89368 after: 0.971137
Removing intersections
Remeshed surface quality stats nv0 = 122417 nv = 128513 1.0498
Area 257022 0.30539 0.03388 0.081315 0.4832
Corner 771066 60.00000 8.80726 16.307931 146.9377
Edge 385533 0.84783 0.08275 0.499427 1.2603
Hinge 385533 8.77716 9.20482 0.000002 126.6836
mris_remesh done
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_remove_intersection ../surf/lh.orig ../surf/lh.orig
intersection removal took 0.00 hours
writing corrected surface to ../surf/lh.orig
rm -f ../surf/lh.inflated
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_remove_intersection ../surf/rh.orig ../surf/rh.orig
intersection removal took 0.00 hours
writing corrected surface to ../surf/rh.orig
rm -f ../surf/rh.inflated
#--------------------------------------------
#@# AutoDetGWStats lh Thu Jun 12 14:17:03 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_autodet_gwstats --o ../surf/autodet.gw.stats.lh.dat --i brain.finalsurfs.mgz --wm wm.mgz --surf ../surf/lh.orig.premesh
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_autodet_gwstats --o ../surf/autodet.gw.stats.lh.dat --i brain.finalsurfs.mgz --wm wm.mgz --surf ../surf/lh.orig.premesh
border white: 237138 voxels (1.41%)
border gray 253932 voxels (1.51%)
Reading in intensity volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
Reading in surf ../surf/lh.orig.premesh
Auto detecting stats
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
Binarizing thresholding at 5
computing class statistics... low=30, hi=110.000000
CCS WM (100.0): 99.9 +- 7.7 [70.0 --> 110.0]
CCS GM (76.0) : 74.9 +- 10.5 [30.0 --> 110.0]
white_mean = 99.9153 +/- 7.71489, gray_mean = 74.9016 +/- 10.5005
using class modes intead of means, discounting robust sigmas....
MRIScomputeClassModes(): min=0 max=179 nbins=180
intensity peaks found at WM=104+-4.3, GM=71+-9.6
white_mode = 104, gray_mode = 71
std_scale = 1
Applying sanity checks, max_scale_down = 0.2
setting MIN_GRAY_AT_WHITE_BORDER to 60.5 (was 70.000000)
setting MAX_BORDER_WHITE to 111.7 (was 105.000000)
setting MIN_BORDER_WHITE to 71.0 (was 85.000000)
setting MAX_CSF to 50.0 (was 40.000000)
setting MAX_GRAY to 96.3 (was 95.000000)
setting MAX_GRAY_AT_CSF_BORDER to 60.5 (was 75.000000)
setting MIN_GRAY_AT_CSF_BORDER to 39.5 (was 40.000000)
When placing the white surface
white_border_hi = 111.715;
white_border_low = 71;
white_outside_low = 60.4995;
white_inside_hi = 120;
white_outside_hi = 111.715;
When placing the pial surface
pial_border_hi = 60.4995;
pial_border_low = 39.4984;
pial_outside_low = 10;
pial_inside_hi = 96.2851;
pial_outside_hi = 55.2492;
#VMPC# mris_autodet_gwstats VmPeak 255976
mris_autodet_gwstats done
#--------------------------------------------
#@# AutoDetGWStats rh Thu Jun 12 14:17:08 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_autodet_gwstats --o ../surf/autodet.gw.stats.rh.dat --i brain.finalsurfs.mgz --wm wm.mgz --surf ../surf/rh.orig.premesh
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_autodet_gwstats --o ../surf/autodet.gw.stats.rh.dat --i brain.finalsurfs.mgz --wm wm.mgz --surf ../surf/rh.orig.premesh
border white: 237138 voxels (1.41%)
border gray 253932 voxels (1.51%)
Reading in intensity volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
Reading in surf ../surf/rh.orig.premesh
Auto detecting stats
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
Binarizing thresholding at 5
computing class statistics... low=30, hi=110.000000
CCS WM (100.0): 99.9 +- 7.7 [70.0 --> 110.0]
CCS GM (76.0) : 74.9 +- 10.5 [30.0 --> 110.0]
white_mean = 99.9153 +/- 7.71489, gray_mean = 74.9016 +/- 10.5005
using class modes intead of means, discounting robust sigmas....
MRIScomputeClassModes(): min=0 max=179 nbins=180
intensity peaks found at WM=104+-4.3, GM=72+-9.6
white_mode = 104, gray_mode = 72
std_scale = 1
Applying sanity checks, max_scale_down = 0.2
setting MIN_GRAY_AT_WHITE_BORDER to 61.5 (was 70.000000)
setting MAX_BORDER_WHITE to 111.7 (was 105.000000)
setting MIN_BORDER_WHITE to 72.0 (was 85.000000)
setting MAX_CSF to 51.0 (was 40.000000)
setting MAX_GRAY to 96.3 (was 95.000000)
setting MAX_GRAY_AT_CSF_BORDER to 61.5 (was 75.000000)
setting MIN_GRAY_AT_CSF_BORDER to 40.5 (was 40.000000)
When placing the white surface
white_border_hi = 111.715;
white_border_low = 72;
white_outside_low = 61.4995;
white_inside_hi = 120;
white_outside_hi = 111.715;
When placing the pial surface
pial_border_hi = 61.4995;
pial_border_low = 40.4984;
pial_outside_low = 10;
pial_inside_hi = 96.2851;
pial_outside_hi = 56.2492;
#VMPC# mris_autodet_gwstats VmPeak 255628
mris_autodet_gwstats done
#--------------------------------------------
#@# WhitePreAparc lh Thu Jun 12 14:17:13 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --wm wm.mgz --threads 1 --invol brain.finalsurfs.mgz --lh --i ../surf/lh.orig --o ../surf/lh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --wm wm.mgz --threads 1 --invol brain.finalsurfs.mgz --lh --i ../surf/lh.orig --o ../surf/lh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
Reading in input surface ../surf/lh.orig
Smoothing surface before ripping with 5 iterations
Area 257220 0.27489 0.06201 0.001717 0.6114
Corner 771660 60.00000 9.08771 8.197482 148.5087
Edge 385830 0.80040 0.11111 0.048691 1.2764
Hinge 385830 5.94967 5.97972 0.000017 143.2159
Not reading in aparc
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6302, nmarked2=4, nripped=6302
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64306: xyz = (-30.9915,-10.7611,6.09681) oxyz = (-30.9915,-10.7611,6.09681) wxzy = (-30.9915,-10.7611,6.09681) pxyz = (0,0,0)
CBVO Creating mask 128612
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6302
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6302, nmarked2=4, nripped=6302
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 90 vertices, nripped=6302
mean border=81.8, 59 (59) missing vertices, mean dist 0.4 [0.6 (%33.7)->0.9 (%66.3))]
%61 local maxima, %34 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.2265 min
Finding expansion regions
mean absolute distance = 0.81 +- 0.98
2912 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 0
000: dt: 0.0000, sse=3281563.8, rms=11.329
001: dt: 0.5000, sse=1822606.0, rms=8.292 (26.805%)
002: dt: 0.5000, sse=1079192.5, rms=6.195 (25.295%)
003: dt: 0.5000, sse=686300.5, rms=4.720 (23.798%)
004: dt: 0.5000, sse=493150.2, rms=3.787 (19.770%)
005: dt: 0.5000, sse=405780.2, rms=3.270 (13.667%)
006: dt: 0.5000, sse=370697.5, rms=3.031 (7.291%)
007: dt: 0.5000, sse=358622.7, rms=2.925 (3.509%)
rms = 2.8792/2.9248, sse=354507.9/358622.7, time step reduction 1 of 3 to 0.250 0 0 1
008: dt: 0.5000, sse=354507.9, rms=2.879 (1.561%)
009: dt: 0.2500, sse=213733.7, rms=1.610 (44.096%)
010: dt: 0.2500, sse=197879.5, rms=1.351 (16.042%)
011: dt: 0.2500, sse=189937.9, rms=1.272 (5.896%)
rms = 1.2301/1.2717, sse=193458.7/189937.9, time step reduction 2 of 3 to 0.125 0 1 1
012: dt: 0.2500, sse=193458.7, rms=1.230 (3.269%)
013: dt: 0.1250, sse=186392.3, rms=1.171 (4.779%)
rms = 1.1603/1.1713, sse=183875.5/186392.3, time step reduction 3 of 3 to 0.062 0 0 1
014: dt: 0.1250, sse=183875.5, rms=1.160 (0.943%)
maximum number of reductions reached, breaking from loop
positioning took 1.5 minutes
Iteration 1 =========================================
n_averages=2, current_sigma=1
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6302
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6540, nmarked2=5, nripped=6540
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 42 vertices, nripped=6540
mean border=85.6, 63 (1) missing vertices, mean dist -0.3 [0.5 (%76.0)->0.2 (%24.0))]
%74 local maxima, %21 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1586 min
Finding expansion regions
mean absolute distance = 0.45 +- 0.71
4458 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=1032054.0, rms=5.861
015: dt: 0.5000, sse=627839.6, rms=4.200 (28.338%)
016: dt: 0.5000, sse=506827.0, rms=3.595 (14.409%)
017: dt: 0.5000, sse=445639.7, rms=3.210 (10.721%)
018: dt: 0.5000, sse=414914.5, rms=2.998 (6.590%)
019: dt: 0.5000, sse=384855.1, rms=2.794 (6.815%)
rms = 2.7768/2.7939, sse=379507.5/384855.1, time step reduction 1 of 3 to 0.250 0 0 1
020: dt: 0.5000, sse=379507.5, rms=2.777 (0.613%)
021: dt: 0.2500, sse=234942.8, rms=1.308 (52.890%)
022: dt: 0.2500, sse=220077.4, rms=1.037 (20.752%)
023: dt: 0.2500, sse=216465.2, rms=0.958 (7.563%)
rms = 0.9216/0.9583, sse=214156.9/216465.2, time step reduction 2 of 3 to 0.125 0 0 1
024: dt: 0.2500, sse=214156.9, rms=0.922 (3.824%)
rms = 0.8710/0.9216, sse=215440.1/214156.9, time step reduction 3 of 3 to 0.062 0 1 0
025: dt: 0.1250, sse=215440.1, rms=0.871 (5.494%)
maximum number of reductions reached, breaking from loop
positioning took 1.2 minutes
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6540
removing 4 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6590, nmarked2=6, nripped=6590
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 45 vertices, nripped=6590
mean border=87.9, 76 (0) missing vertices, mean dist -0.2 [0.3 (%73.6)->0.2 (%26.4))]
%83 local maxima, %12 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0883 min
Finding expansion regions
mean absolute distance = 0.27 +- 0.42
3083 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=465093.0, rms=3.356
026: dt: 0.5000, sse=334338.8, rms=2.414 (28.061%)
rms = 2.3865/2.4142, sse=330201.6/334338.8, time step reduction 1 of 3 to 0.250 0 0 1
027: dt: 0.5000, sse=330201.6, rms=2.387 (1.146%)
028: dt: 0.2500, sse=227477.6, rms=1.156 (51.541%)
029: dt: 0.2500, sse=217004.0, rms=0.963 (16.768%)
030: dt: 0.2500, sse=213663.0, rms=0.899 (6.571%)
rms = 0.8826/0.8993, sse=213604.9/213663.0, time step reduction 2 of 3 to 0.125 0 0 1
031: dt: 0.2500, sse=213604.9, rms=0.883 (1.862%)
032: dt: 0.1250, sse=207980.4, rms=0.797 (9.743%)
rms = 0.7903/0.7966, sse=208476.0/207980.4, time step reduction 3 of 3 to 0.062 0 1 1
033: dt: 0.1250, sse=208476.0, rms=0.790 (0.784%)
maximum number of reductions reached, breaking from loop
positioning took 0.8 minutes
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6590
removing 2 vertices from ripped group in thread:0
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6642, nmarked2=7, nripped=6642
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 34 vertices, nripped=6642
mean border=88.6, 86 (0) missing vertices, mean dist -0.0 [0.2 (%57.9)->0.2 (%42.1))]
%86 local maxima, % 9 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0567 min
Finding expansion regions
mean absolute distance = 0.20 +- 0.31
2923 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=235735.9, rms=1.369
034: dt: 0.5000, sse=226493.6, rms=1.230 (10.204%)
rms = 1.6233/1.2297, sse=251911.8/226493.6, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
035: dt: 0.2500, sse=203534.5, rms=0.784 (36.208%)
036: dt: 0.2500, sse=202779.4, rms=0.645 (17.778%)
rms = 0.6212/0.6450, sse=203488.2/202779.4, time step reduction 2 of 3 to 0.125 0 1 1
037: dt: 0.2500, sse=203488.2, rms=0.621 (3.691%)
rms = 0.5862/0.6212, sse=197685.3/203488.2, time step reduction 3 of 3 to 0.062 0 0 1
038: dt: 0.1250, sse=197685.3, rms=0.586 (5.636%)
maximum number of reductions reached, breaking from loop
positioning took 0.6 minutes
Writing output to ../surf/lh.white.preaparc
#ET# mris_place_surface 4.70 minutes
#VMPC# mris_place_surfaces VmPeak 2129624
mris_place_surface done
#--------------------------------------------
#@# WhitePreAparc rh Thu Jun 12 14:22:04 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --wm wm.mgz --threads 1 --invol brain.finalsurfs.mgz --rh --i ../surf/rh.orig --o ../surf/rh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --wm wm.mgz --threads 1 --invol brain.finalsurfs.mgz --rh --i ../surf/rh.orig --o ../surf/rh.white.preaparc --white --seg aseg.presurf.mgz --nsmooth 5
Reading in input surface ../surf/rh.orig
Smoothing surface before ripping with 5 iterations
Area 257022 0.27447 0.06184 0.011393 0.5992
Corner 771066 60.00000 9.04729 16.744599 137.0922
Edge 385533 0.79977 0.11083 0.138855 1.3532
Hinge 385533 5.95507 5.97774 0.000023 135.9850
Not reading in aparc
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6408, nmarked2=0, nripped=6408
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64257: xyz = (54.8197,-10.0048,35.8937) oxyz = (54.8197,-10.0048,35.8937) wxzy = (54.8197,-10.0048,35.8937) pxyz = (0,0,0)
CBVO Creating mask 128513
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6408
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6408, nmarked2=0, nripped=6408
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 25 vertices, nripped=6408
mean border=82.3, 27 (27) missing vertices, mean dist 0.3 [0.7 (%35.1)->0.9 (%64.9))]
%58 local maxima, %37 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.2226 min
Finding expansion regions
mean absolute distance = 0.80 +- 0.99
2807 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 0
000: dt: 0.0000, sse=3261747.0, rms=11.302
001: dt: 0.5000, sse=1781949.9, rms=8.197 (27.476%)
002: dt: 0.5000, sse=1042354.2, rms=6.076 (25.871%)
003: dt: 0.5000, sse=663757.8, rms=4.624 (23.896%)
004: dt: 0.5000, sse=477331.2, rms=3.704 (19.903%)
005: dt: 0.5000, sse=393044.5, rms=3.190 (13.861%)
006: dt: 0.5000, sse=357484.3, rms=2.951 (7.504%)
007: dt: 0.5000, sse=346692.7, rms=2.848 (3.494%)
rms = 2.8075/2.8479, sse=339116.0/346692.7, time step reduction 1 of 3 to 0.250 0 0 1
008: dt: 0.5000, sse=339116.0, rms=2.808 (1.419%)
009: dt: 0.2500, sse=201583.4, rms=1.461 (47.950%)
010: dt: 0.2500, sse=184584.9, rms=1.174 (19.694%)
011: dt: 0.2500, sse=179040.2, rms=1.091 (7.043%)
rms = 1.0461/1.0909, sse=176452.0/179040.2, time step reduction 2 of 3 to 0.125 0 0 1
012: dt: 0.2500, sse=176452.0, rms=1.046 (4.102%)
013: dt: 0.1250, sse=174330.8, rms=0.984 (5.912%)
rms = 0.9728/0.9843, sse=175666.7/174330.8, time step reduction 3 of 3 to 0.062 0 1 1
014: dt: 0.1250, sse=175666.7, rms=0.973 (1.163%)
maximum number of reductions reached, breaking from loop
positioning took 1.5 minutes
Iteration 1 =========================================
n_averages=2, current_sigma=1
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6408
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6652, nmarked2=0, nripped=6652
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 38 vertices, nripped=6652
mean border=86.0, 71 (6) missing vertices, mean dist -0.3 [0.5 (%75.2)->0.2 (%24.8))]
%72 local maxima, %23 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1567 min
Finding expansion regions
mean absolute distance = 0.44 +- 0.72
4302 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=982699.4, rms=5.688
015: dt: 0.5000, sse=599104.4, rms=4.094 (28.034%)
016: dt: 0.5000, sse=489103.0, rms=3.507 (14.328%)
017: dt: 0.5000, sse=433305.1, rms=3.160 (9.888%)
018: dt: 0.5000, sse=404448.4, rms=2.970 (6.006%)
019: dt: 0.5000, sse=379583.4, rms=2.784 (6.274%)
rms = 2.7653/2.7841, sse=376065.3/379583.4, time step reduction 1 of 3 to 0.250 0 0 1
020: dt: 0.5000, sse=376065.3, rms=2.765 (0.677%)
021: dt: 0.2500, sse=232462.6, rms=1.293 (53.238%)
022: dt: 0.2500, sse=216780.0, rms=1.015 (21.483%)
023: dt: 0.2500, sse=214919.4, rms=0.932 (8.188%)
rms = 0.8931/0.9322, sse=215157.1/214919.4, time step reduction 2 of 3 to 0.125 0 1 1
024: dt: 0.2500, sse=215157.1, rms=0.893 (4.189%)
rms = 0.8434/0.8931, sse=209640.9/215157.1, time step reduction 3 of 3 to 0.062 0 0 1
025: dt: 0.1250, sse=209640.9, rms=0.843 (5.563%)
maximum number of reductions reached, breaking from loop
positioning took 1.2 minutes
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6652
removing 3 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6676, nmarked2=0, nripped=6676
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 37 vertices, nripped=6676
mean border=88.2, 56 (3) missing vertices, mean dist -0.2 [0.3 (%73.2)->0.2 (%26.8))]
%81 local maxima, %14 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0862 min
Finding expansion regions
mean absolute distance = 0.26 +- 0.43
2817 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=441784.5, rms=3.240
026: dt: 0.5000, sse=318648.1, rms=2.327 (28.169%)
rms = 2.3291/2.3273, sse=318591.0/318648.1, time step reduction 1 of 3 to 0.250 0 0 1
RMS increased, rejecting step
027: dt: 0.2500, sse=250204.0, rms=1.618 (30.476%)
028: dt: 0.2500, sse=213275.4, rms=1.056 (34.730%)
029: dt: 0.2500, sse=204663.1, rms=0.871 (17.493%)
rms = 0.8228/0.8714, sse=202642.3/204663.1, time step reduction 2 of 3 to 0.125 0 0 1
030: dt: 0.2500, sse=202642.3, rms=0.823 (5.578%)
rms = 0.7806/0.8228, sse=200851.9/202642.3, time step reduction 3 of 3 to 0.062 0 0 1
031: dt: 0.1250, sse=200851.9, rms=0.781 (5.128%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Freezing midline and others
Ripping frozen voxels
INFO: rip surface needed but not specified, so using input surface
Freezing midline and others
Entering: MRISripMidline(): inhibiting deformation at non-cortical midline structures...
which=1, fix_mtl=0, using annot = 0
#FML0# MRISripMidline(): nripped=6676
removing 4 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 2 vertices from ripped group in thread:0
removing 3 vertices from ripped group in thread:0
#FML# MRISripMidline(): nmarked=6685, nmarked2=0, nripped=6685
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247 247 247 247 247
MRISripSegs(): -2 2 0.5 ripped 0
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 48 vertices, nripped=6685
mean border=88.9, 61 (2) missing vertices, mean dist -0.0 [0.2 (%57.8)->0.2 (%42.2))]
%84 local maxima, %11 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0552 min
Finding expansion regions
mean absolute distance = 0.20 +- 0.31
2721 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=227221.2, rms=1.343
032: dt: 0.5000, sse=221451.0, rms=1.229 (8.512%)
rms = 1.6243/1.2287, sse=247634.7/221451.0, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
033: dt: 0.2500, sse=199713.8, rms=0.798 (35.059%)
034: dt: 0.2500, sse=195598.2, rms=0.682 (14.562%)
rms = 0.6494/0.6817, sse=193634.6/195598.2, time step reduction 2 of 3 to 0.125 0 0 1
035: dt: 0.2500, sse=193634.6, rms=0.649 (4.741%)
rms = 0.6184/0.6494, sse=194650.0/193634.6, time step reduction 3 of 3 to 0.062 0 1 1
036: dt: 0.1250, sse=194650.0, rms=0.618 (4.771%)
maximum number of reductions reached, breaking from loop
positioning took 0.6 minutes
Writing output to ../surf/rh.white.preaparc
#ET# mris_place_surface 4.53 minutes
#VMPC# mris_place_surfaces VmPeak 2092460
mris_place_surface done
#--------------------------------------------
#@# CortexLabel lh Thu Jun 12 14:26:46 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_label2label --label-cortex ../surf/lh.white.preaparc aseg.presurf.mgz 0 ../label/lh.cortex.label
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
6 non-cortical segments detected
only using segment with 7999 vertices
erasing segment 0 (vno[0] = 27085)
erasing segment 1 (vno[0] = 28830)
erasing segment 3 (vno[0] = 46022)
erasing segment 4 (vno[0] = 77142)
erasing segment 5 (vno[0] = 79900)
#--------------------------------------------
#@# CortexLabel+HipAmyg lh Thu Jun 12 14:27:04 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_label2label --label-cortex ../surf/lh.white.preaparc aseg.presurf.mgz 1 ../label/lh.cortex+hipamyg.label
Generating cortex label... RemoveHipAmgy=1
NucAccIsMedialWall=0
mris->useRealRAS=0
9 non-cortical segments detected
only using segment with 6353 vertices
erasing segment 0 (vno[0] = 27085)
erasing segment 1 (vno[0] = 28830)
erasing segment 3 (vno[0] = 46022)
erasing segment 4 (vno[0] = 72454)
erasing segment 5 (vno[0] = 72475)
erasing segment 6 (vno[0] = 77142)
erasing segment 7 (vno[0] = 77149)
erasing segment 8 (vno[0] = 79900)
#--------------------------------------------
#@# CortexLabel rh Thu Jun 12 14:27:21 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_label2label --label-cortex ../surf/rh.white.preaparc aseg.presurf.mgz 0 ../label/rh.cortex.label
Generating cortex label... RemoveHipAmgy=0
NucAccIsMedialWall=0
mris->useRealRAS=0
2 non-cortical segments detected
only using segment with 8066 vertices
erasing segment 1 (vno[0] = 77348)
#--------------------------------------------
#@# CortexLabel+HipAmyg rh Thu Jun 12 14:27:39 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mri_label2label --label-cortex ../surf/rh.white.preaparc aseg.presurf.mgz 1 ../label/rh.cortex+hipamyg.label
Generating cortex label... RemoveHipAmgy=1
NucAccIsMedialWall=0
mris->useRealRAS=0
3 non-cortical segments detected
only using segment with 6327 vertices
erasing segment 1 (vno[0] = 77348)
erasing segment 2 (vno[0] = 117671)
#--------------------------------------------
#@# Smooth2 lh Thu Jun 12 14:27:56 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_smooth -n 3 -nw -seed 1234 ../surf/lh.white.preaparc ../surf/lh.smoothwm
smoothing for 3 iterations
setting seed for random number generator to 1234
smoothing surface tessellation for 3 iterations...
smoothing complete - recomputing first and second fundamental forms...
#--------------------------------------------
#@# Smooth2 rh Thu Jun 12 14:27:59 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_smooth -n 3 -nw -seed 1234 ../surf/rh.white.preaparc ../surf/rh.smoothwm
smoothing for 3 iterations
setting seed for random number generator to 1234
smoothing surface tessellation for 3 iterations...
smoothing complete - recomputing first and second fundamental forms...
#--------------------------------------------
#@# Inflation2 lh Thu Jun 12 14:28:03 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_inflate ../surf/lh.smoothwm ../surf/lh.inflated
Reading ../surf/lh.smoothwm
avg radius = 46.9 mm, total surface area = 79403 mm^2
step 060: RMS=0.019 (target=0.015) writing inflated surface to ../surf/lh.inflated
writing sulcal depths to ../surf/lh.sulc
inflation complete.
inflation took 0.5 minutes
mris_inflate utimesec 24.473599
mris_inflate stimesec 1.466720
mris_inflate ru_maxrss 197628
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 498486
mris_inflate ru_majflt 55
mris_inflate ru_nswap 0
mris_inflate ru_inblock 12936
mris_inflate ru_oublock 10056
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 325
mris_inflate ru_nivcsw 187
#--------------------------------------------
#@# Inflation2 rh Thu Jun 12 14:28:30 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_inflate ../surf/rh.smoothwm ../surf/rh.inflated
Reading ../surf/rh.smoothwm
avg radius = 46.8 mm, total surface area = 79169 mm^2
step 060: RMS=0.019 (target=0.015) writing inflated surface to ../surf/rh.inflated
writing sulcal depths to ../surf/rh.sulc
inflation complete.
inflation took 0.4 minutes
mris_inflate utimesec 24.661216
mris_inflate stimesec 0.103921
mris_inflate ru_maxrss 198184
mris_inflate ru_ixrss 0
mris_inflate ru_idrss 0
mris_inflate ru_isrss 0
mris_inflate ru_minflt 53139
mris_inflate ru_majflt 13
mris_inflate ru_nswap 0
mris_inflate ru_inblock 3424
mris_inflate ru_oublock 10048
mris_inflate ru_msgsnd 0
mris_inflate ru_msgrcv 0
mris_inflate ru_nsignals 0
mris_inflate ru_nvcsw 288
mris_inflate ru_nivcsw 1157
#--------------------------------------------
#@# Curv .H and .K lh Thu Jun 12 14:28:55 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf
mris_curvature -w -seed 1234 lh.white.preaparc
setting seed for random number generator to 1234
total integrated curvature = 23.376*4pi (293.755) --> -22 handles
ICI = 116.3, FI = 1118.8, variation=17585.305
writing Gaussian curvature to ./lh.white.preaparc.K...done.
writing mean curvature to ./lh.white.preaparc.H...done.
rm -f lh.white.H
ln -s lh.white.preaparc.H lh.white.H
rm -f lh.white.K
ln -s lh.white.preaparc.K lh.white.K
mris_curvature -seed 1234 -thresh .999 -n -a 5 -w -distances 10 10 lh.inflated
setting seed for random number generator to 1234
normalizing curvature values.
averaging curvature patterns 5 times.
sampling 10 neighbors out to a distance of 10 mm
209 vertices thresholded to be in k1 ~ [-0.21 0.33], k2 ~ [-0.10 0.07]
total integrated curvature = 0.511*4pi (6.418) --> 0 handles
ICI = 1.5, FI = 9.1, variation=160.850
125 vertices thresholded to be in [-0.01 0.02]
writing Gaussian curvature to ./lh.inflated.K...thresholding curvature at 99.90% level
curvature mean = 0.000, std = 0.001
154 vertices thresholded to be in [-0.13 0.17]
done.
writing mean curvature to ./lh.inflated.H...curvature mean = -0.016, std = 0.024
done.
#--------------------------------------------
#@# Curv .H and .K rh Thu Jun 12 14:29:44 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf
mris_curvature -w -seed 1234 rh.white.preaparc
setting seed for random number generator to 1234
total integrated curvature = 21.866*4pi (274.781) --> -21 handles
ICI = 114.1, FI = 1111.6, variation=17451.587
writing Gaussian curvature to ./rh.white.preaparc.K...done.
writing mean curvature to ./rh.white.preaparc.H...done.
rm -f rh.white.H
ln -s rh.white.preaparc.H rh.white.H
rm -f rh.white.K
ln -s rh.white.preaparc.K rh.white.K
mris_curvature -seed 1234 -thresh .999 -n -a 5 -w -distances 10 10 rh.inflated
setting seed for random number generator to 1234
normalizing curvature values.
averaging curvature patterns 5 times.
sampling 10 neighbors out to a distance of 10 mm
170 vertices thresholded to be in k1 ~ [-0.26 0.35], k2 ~ [-0.11 0.08]
total integrated curvature = 0.480*4pi (6.029) --> 1 handles
ICI = 1.5, FI = 9.2, variation=162.592
109 vertices thresholded to be in [-0.01 0.02]
writing Gaussian curvature to ./rh.inflated.K...thresholding curvature at 99.90% level
curvature mean = 0.000, std = 0.001
139 vertices thresholded to be in [-0.14 0.17]
done.
writing mean curvature to ./rh.inflated.H...curvature mean = -0.016, std = 0.024
done.
#--------------------------------------------
#@# Sphere lh Thu Jun 12 14:30:32 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_sphere -seed 1234 ../surf/lh.inflated ../surf/lh.sphere
setting seed for random number genererator to 1234
version: 7.3.2
available threads: 1
reading original vertex positions...
projecting onto sphere...
surface projected - minimizing metric distortion...
scaling brain by 0.296...
MRISunfold() max_passes = 1 -------
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=2, l_area=1.000, l_dist=1.000
using quadratic fit line minimization
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 1.000000
desired_rms_height -1.000000
momentum 0.900000
nbhd_size 7
max_nbrs 8
niterations 25
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 1234
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
--------------------
mrisRemoveNegativeArea()
pass 1: epoch 1 of 3 starting distance error %20.91
pass 1: epoch 2 of 3 starting distance error %20.87
unfolding complete - removing small folds...
starting distance error %20.81
removing remaining folds...
final distance error %20.85
MRISunfold() return, current seed 1234
-01: dt=0.0000, 4 negative triangles VmPeak 537588
174: dt=0.9900, 4 negative triangles
175: dt=0.9900, 2 negative triangles
writing spherical brain to ../surf/lh.sphere
spherical transformation took 0.1812 hours
FSRUNTIME@ mris_sphere 0.1812 hours 1 threads
#VMPC# mris_sphere VmPeak 537588
mris_sphere done
#--------------------------------------------
#@# Sphere rh Thu Jun 12 14:41:25 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_sphere -seed 1234 ../surf/rh.inflated ../surf/rh.sphere
setting seed for random number genererator to 1234
version: 7.3.2
available threads: 1
reading original vertex positions...
projecting onto sphere...
surface projected - minimizing metric distortion...
scaling brain by 0.296...
MRISunfold() max_passes = 1 -------
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=2, l_area=1.000, l_dist=1.000
using quadratic fit line minimization
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 1.000000
desired_rms_height -1.000000
momentum 0.900000
nbhd_size 7
max_nbrs 8
niterations 25
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 1234
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
singular matrix in quadratic form
--------------------
mrisRemoveNegativeArea()
pass 1: epoch 1 of 3 starting distance error %20.91
pass 1: epoch 2 of 3 starting distance error %20.90
unfolding complete - removing small folds...
starting distance error %20.85
removing remaining folds...
final distance error %20.85
MRISunfold() return, current seed 1234
-01: dt=0.0000, 141 negative triangles VmPeak 537372
091: dt=0.9900, 141 negative triangles
092: dt=0.9900, 72 negative triangles
093: dt=0.9900, 53 negative triangles
094: dt=0.9900, 36 negative triangles
095: dt=0.9900, 41 negative triangles
096: dt=0.9900, 27 negative triangles
097: dt=0.9900, 18 negative triangles
098: dt=0.9900, 11 negative triangles
099: dt=0.9900, 12 negative triangles
100: dt=0.9900, 14 negative triangles
101: dt=0.9900, 12 negative triangles
102: dt=0.9900, 7 negative triangles
103: dt=0.9900, 7 negative triangles
104: dt=0.9900, 5 negative triangles
105: dt=0.9900, 14 negative triangles
106: dt=0.9900, 7 negative triangles
107: dt=0.9900, 6 negative triangles
108: dt=0.9900, 3 negative triangles
109: dt=0.9900, 2 negative triangles
110: dt=0.9900, 5 negative triangles
111: dt=0.9900, 2 negative triangles
112: dt=0.9900, 1 negative triangles
writing spherical brain to ../surf/rh.sphere
spherical transformation took 0.1222 hours
FSRUNTIME@ mris_sphere 0.1222 hours 1 threads
#VMPC# mris_sphere VmPeak 537372
mris_sphere done
#--------------------------------------------
#@# Surf Reg lh Thu Jun 12 14:48:45 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_register -curv ../surf/lh.sphere /opt/freesurfer-7.3.2/average/lh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif ../surf/lh.sphere.reg
using smoothwm curvature for final alignment
cwd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
cmdline mris_register -curv ../surf/lh.sphere /opt/freesurfer-7.3.2/average/lh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif ../surf/lh.sphere.reg
0 inflated.H
1 sulc
2 smoothwm (computed)
7.3.2
7.3.2
reading surface from ../surf/lh.sphere...
reading template parameterization from /opt/freesurfer-7.3.2/average/lh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif...
MRISregister() -------
max_passes = 4
min_degrees = 0.500000
max_degrees = 64.000000
nangles = 8
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=1.000, l_dist=5.000
using quadratic fit line minimization
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height -1.000000
momentum 0.950000
nbhd_size -10
max_nbrs 10
niterations 25
nsurfaces 0
SURFACES 3
flags 16 (10)
use curv 16
no sulc 0
no rigid align 0
mris->nsize 1
mris->hemisphere 0
randomSeed 0
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=1.000, l_dist=5.000
using quadratic fit line minimization
--------------------
1 Reading lh.sulc
tol=1.0e+00, sigma=0.5, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=0.050, l_spring=0.500, l_dist=5.000
using quadratic fit line minimization
curvature mean = 0.000, std = 5.599
curvature mean = 0.040, std = 0.819
curvature mean = 0.012, std = 0.868
Starting MRISrigidBodyAlignGlobal()
Starting new MRISrigidBodyAlignGlobal_findMinSSE()
new MRISrigidBodyAlignGlobal_findMinSSE min @ (4.50, -7.50, -1.50) sse = 149085.1, elapsed since starting=0.3775 min
MRISrigidBodyAlignGlobal() done 0.38 min
curvature mean = 0.014, std = 0.838
curvature mean = 0.003, std = 0.951
curvature mean = 0.009, std = 0.849
curvature mean = 0.002, std = 0.980
curvature mean = 0.008, std = 0.851
curvature mean = 0.000, std = 0.992
2 Reading smoothwm
curvature mean = -0.021, std = 0.245
curvature mean = 0.041, std = 0.250
curvature mean = 0.043, std = 0.401
curvature mean = 0.037, std = 0.308
curvature mean = 0.022, std = 0.605
curvature mean = 0.036, std = 0.336
curvature mean = 0.010, std = 0.742
curvature mean = 0.036, std = 0.348
curvature mean = 0.003, std = 0.838
MRISregister() return, current seed 0
writing registered surface to ../surf/lh.sphere.reg...
-01: dt=0.0000, 0 negative triangles VmPeak 523536
registration took 0.13 hours
#VMPC# mris_register VmPeak 523536
FSRUNTIME@ mris_register 0.1303 hours 1 threads
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf ~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
ln -sf lh.sphere.reg lh.fsaverage.sphere.reg
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
#--------------------------------------------
#@# Surf Reg rh Thu Jun 12 14:56:34 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_register -curv ../surf/rh.sphere /opt/freesurfer-7.3.2/average/rh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif ../surf/rh.sphere.reg
using smoothwm curvature for final alignment
cwd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
cmdline mris_register -curv ../surf/rh.sphere /opt/freesurfer-7.3.2/average/rh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif ../surf/rh.sphere.reg
0 inflated.H
1 sulc
2 smoothwm (computed)
7.3.2
7.3.2
reading surface from ../surf/rh.sphere...
reading template parameterization from /opt/freesurfer-7.3.2/average/rh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif...
MRISregister() -------
max_passes = 4
min_degrees = 0.500000
max_degrees = 64.000000
nangles = 8
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=1.000, l_dist=5.000
using quadratic fit line minimization
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height -1.000000
momentum 0.950000
nbhd_size -10
max_nbrs 10
niterations 25
nsurfaces 0
SURFACES 3
flags 16 (10)
use curv 16
no sulc 0
no rigid align 0
mris->nsize 1
mris->hemisphere 1
randomSeed 0
tol=5.0e-01, sigma=0.0, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=1.000, l_dist=5.000
using quadratic fit line minimization
--------------------
1 Reading rh.sulc
tol=1.0e+00, sigma=0.5, host=jupyt, nav=1024, nbrs=1, l_extern=10000.000, l_parea=0.200, l_nlarea=1.000, l_corr=0.050, l_spring=0.500, l_dist=5.000
using quadratic fit line minimization
curvature mean = -0.000, std = 5.588
curvature mean = 0.017, std = 0.806
curvature mean = 0.013, std = 0.869
Starting MRISrigidBodyAlignGlobal()
Starting new MRISrigidBodyAlignGlobal_findMinSSE()
new MRISrigidBodyAlignGlobal_findMinSSE min @ (7.00, -5.50, 3.00) sse = 129613.3, elapsed since starting=0.3876 min
MRISrigidBodyAlignGlobal() done 0.39 min
curvature mean = -0.000, std = 0.833
curvature mean = 0.005, std = 0.953
curvature mean = -0.003, std = 0.843
curvature mean = 0.002, std = 0.982
curvature mean = -0.005, std = 0.844
curvature mean = 0.000, std = 0.993
2 Reading smoothwm
curvature mean = -0.022, std = 0.245
curvature mean = 0.033, std = 0.245
curvature mean = 0.049, std = 0.400
curvature mean = 0.031, std = 0.303
curvature mean = 0.023, std = 0.605
curvature mean = 0.029, std = 0.331
curvature mean = 0.011, std = 0.741
curvature mean = 0.029, std = 0.344
curvature mean = 0.003, std = 0.837
MRISregister() return, current seed 0
-01: dt=0.0000, 1 negative triangles VmPeak 522848
096: dt=0.9900, 1 negative triangles
writing registered surface to ../surf/rh.sphere.reg...
registration took 0.12 hours
#VMPC# mris_register VmPeak 522848
FSRUNTIME@ mris_register 0.1205 hours 1 threads
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/surf ~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
ln -sf rh.sphere.reg rh.fsaverage.sphere.reg
~/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
#--------------------------------------------
#@# Jacobian white lh Thu Jun 12 15:03:48 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_jacobian ../surf/lh.white.preaparc ../surf/lh.sphere.reg ../surf/lh.jacobian_white
reading surface from ../surf/lh.white.preaparc...
writing curvature file ../surf/lh.jacobian_white
#--------------------------------------------
#@# Jacobian white rh Thu Jun 12 15:03:50 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_jacobian ../surf/rh.white.preaparc ../surf/rh.sphere.reg ../surf/rh.jacobian_white
reading surface from ../surf/rh.white.preaparc...
writing curvature file ../surf/rh.jacobian_white
#--------------------------------------------
#@# AvgCurv lh Thu Jun 12 15:03:51 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mrisp_paint -a 5 /opt/freesurfer-7.3.2/average/lh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif#6 ../surf/lh.sphere.reg ../surf/lh.avg_curv
averaging curvature patterns 5 times...
reading surface from ../surf/lh.sphere.reg...
reading template parameterization from /opt/freesurfer-7.3.2/average/lh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif...
writing curvature file to ../surf/lh.avg_curv...
#--------------------------------------------
#@# AvgCurv rh Thu Jun 12 15:03:52 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mrisp_paint -a 5 /opt/freesurfer-7.3.2/average/rh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif#6 ../surf/rh.sphere.reg ../surf/rh.avg_curv
averaging curvature patterns 5 times...
reading surface from ../surf/rh.sphere.reg...
reading template parameterization from /opt/freesurfer-7.3.2/average/rh.folding.atlas.acfb40.noaparc.i12.2016-08-02.tif...
writing curvature file to ../surf/rh.avg_curv...
#-----------------------------------------
#@# Cortical Parc lh Thu Jun 12 15:03:54 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_ca_label -l ../label/lh.cortex.label -aseg ../mri/aseg.presurf.mgz -seed 1234 subjectname lh ../surf/lh.sphere.reg /opt/freesurfer-7.3.2/average/lh.DKaparc.atlas.acfb40.noaparc.i12.2016-08-02.gcs ../label/lh.aparc.annot
setting seed for random number generator to 1234
using ../mri/aseg.presurf.mgz aseg volume to correct midline
7.3.2
7.3.2
reading atlas from /opt/freesurfer-7.3.2/average/lh.DKaparc.atlas.acfb40.noaparc.i12.2016-08-02.gcs...
reading color table from GCSA file....
average std = 0.8 using min determinant for regularization = 0.006
0 singular and 342 ill-conditioned covariance matrices regularized
input 0: MEAN CURVATURE, flags 0, avgs 5, name mean_curvature
labeling surface...
1413 labels changed using aseg
relabeling using gibbs priors...
000: 2853 changed, 128612 examined...
001: 655 changed, 11979 examined...
002: 158 changed, 3717 examined...
003: 51 changed, 963 examined...
004: 22 changed, 306 examined...
005: 16 changed, 134 examined...
006: 10 changed, 82 examined...
007: 7 changed, 50 examined...
008: 1 changed, 37 examined...
009: 0 changed, 7 examined...
293 labels changed using aseg
000: 101 total segments, 62 labels (146 vertices) changed
001: 40 total segments, 2 labels (5 vertices) changed
002: 38 total segments, 0 labels (0 vertices) changed
10 filter iterations complete (10 requested, 2 changed)
rationalizing unknown annotations with cortex label
relabeling unknown label...
relabeling corpuscallosum label...
1741 vertices marked for relabeling...
1741 labels changed in reclassification.
writing output to ../label/lh.aparc.annot...
classification took 0 minutes and 18 seconds.
#-----------------------------------------
#@# Cortical Parc rh Thu Jun 12 15:04:13 UTC 2025
/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/scripts
mris_ca_label -l ../label/rh.cortex.label -aseg ../mri/aseg.presurf.mgz -seed 1234 subjectname rh ../surf/rh.sphere.reg /opt/freesurfer-7.3.2/average/rh.DKaparc.atlas.acfb40.noaparc.i12.2016-08-02.gcs ../label/rh.aparc.annot
setting seed for random number generator to 1234
using ../mri/aseg.presurf.mgz aseg volume to correct midline
7.3.2
7.3.2
reading atlas from /opt/freesurfer-7.3.2/average/rh.DKaparc.atlas.acfb40.noaparc.i12.2016-08-02.gcs...
reading color table from GCSA file....
average std = 0.7 using min determinant for regularization = 0.004
0 singular and 309 ill-conditioned covariance matrices regularized
input 0: MEAN CURVATURE, flags 0, avgs 5, name mean_curvature
labeling surface...
1235 labels changed using aseg
relabeling using gibbs priors...
000: 2490 changed, 128513 examined...
001: 504 changed, 10872 examined...
002: 98 changed, 3056 examined...
003: 33 changed, 612 examined...
004: 11 changed, 211 examined...
005: 4 changed, 69 examined...
006: 0 changed, 28 examined...
158 labels changed using aseg
000: 95 total segments, 57 labels (192 vertices) changed
001: 42 total segments, 4 labels (4 vertices) changed
002: 37 total segments, 0 labels (0 vertices) changed
10 filter iterations complete (10 requested, 2 changed)
rationalizing unknown annotations with cortex label
relabeling unknown label...
relabeling corpuscallosum label...
1541 vertices marked for relabeling...
1541 labels changed in reclassification.
writing output to ../label/rh.aparc.annot...
classification took 0 minutes and 13 seconds.
#--------------------------------------------
#@# WhiteSurfs lh Thu Jun 12 15:04:26 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --lh --i ../surf/lh.white.preaparc --o ../surf/lh.white --white --nsmooth 0 --rip-label ../label/lh.cortex.label --rip-bg --rip-surf ../surf/lh.white.preaparc --aparc ../label/lh.aparc.annot
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --lh --i ../surf/lh.white.preaparc --o ../surf/lh.white --white --nsmooth 0 --rip-label ../label/lh.cortex.label --rip-bg --rip-surf ../surf/lh.white.preaparc --aparc ../label/lh.aparc.annot
Reading in input surface ../surf/lh.white.preaparc
Not smoothing input surface
Area 257220 0.33392 0.10919 0.001133 1.8853
Corner 771660 60.00000 12.77267 0.197116 178.5246
Edge 385830 0.88662 0.18085 0.038419 3.8198
Hinge 385830 8.50431 9.59496 0.000003 179.8698
Reading in aparc ../label/lh.aparc.annot
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
Ripping vertices not in label ../label/lh.cortex.label
MRISripNotLabel() ripped 8249/128612 vertices (120363 unripped)
Reading in ripping surface ../surf/lh.white.preaparc
Reading in aparc ../label/lh.aparc.annot for ripsurf
Ripping BG
MRISripBasalGanglia(): 1 -2 2 0.5 ripped 342
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64306: xyz = (-31.0111,-10.7648,6.15307) oxyz = (-31.0111,-10.7648,6.15307) wxzy = (-31.0111,-10.7648,6.15307) pxyz = (0,0,0)
CBVO Creating mask 128612
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 95 vertices, nripped=8591
mean border=81.6, 111 (111) missing vertices, mean dist 0.4 [1.2 (%11.9)->0.7 (%88.1))]
%57 local maxima, %36 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.2106 min
Finding expansion regions
mean absolute distance = 0.74 +- 1.01
2383 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 0
000: dt: 0.0000, sse=2072407.4, rms=8.883
001: dt: 0.5000, sse=962413.2, rms=5.719 (35.622%)
002: dt: 0.5000, sse=568491.9, rms=4.039 (29.364%)
003: dt: 0.5000, sse=429833.1, rms=3.243 (19.719%)
004: dt: 0.5000, sse=364364.2, rms=2.775 (14.422%)
005: dt: 0.5000, sse=352972.6, rms=2.652 (4.448%)
006: dt: 0.5000, sse=345600.4, rms=2.505 (5.518%)
rms = 2.5449/2.5055, sse=335266.5/345600.4, time step reduction 1 of 3 to 0.250 0 0 1
RMS increased, rejecting step
007: dt: 0.2500, sse=249625.9, rms=1.640 (34.528%)
008: dt: 0.2500, sse=214982.7, rms=1.188 (27.594%)
009: dt: 0.2500, sse=207419.8, rms=1.045 (12.037%)
rms = 0.9977/1.0448, sse=205227.0/207419.8, time step reduction 2 of 3 to 0.125 0 0 1
010: dt: 0.2500, sse=205227.0, rms=0.998 (4.508%)
rms = 0.9545/0.9977, sse=203164.9/205227.0, time step reduction 3 of 3 to 0.062 0 0 1
011: dt: 0.1250, sse=203164.9, rms=0.954 (4.328%)
maximum number of reductions reached, breaking from loop
positioning took 1.3 minutes
Iteration 1 =========================================
n_averages=2, current_sigma=1
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 71 vertices, nripped=8591
mean border=85.4, 131 (55) missing vertices, mean dist -0.3 [0.5 (%74.6)->0.2 (%25.4))]
%73 local maxima, %20 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1574 min
Finding expansion regions
mean absolute distance = 0.46 +- 0.71
4157 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=996559.3, rms=5.781
012: dt: 0.5000, sse=603139.1, rms=4.122 (28.702%)
013: dt: 0.5000, sse=489101.3, rms=3.502 (15.036%)
014: dt: 0.5000, sse=425406.5, rms=3.115 (11.044%)
015: dt: 0.5000, sse=390606.6, rms=2.863 (8.106%)
016: dt: 0.5000, sse=367859.8, rms=2.680 (6.394%)
rms = 2.6585/2.6796, sse=366678.9/367859.8, time step reduction 1 of 3 to 0.250 0 0 1
017: dt: 0.5000, sse=366678.9, rms=2.659 (0.787%)
018: dt: 0.2500, sse=232516.0, rms=1.247 (53.091%)
019: dt: 0.2500, sse=219531.6, rms=1.011 (18.965%)
020: dt: 0.2500, sse=216204.0, rms=0.940 (6.995%)
rms = 0.9181/0.9399, sse=215278.9/216204.0, time step reduction 2 of 3 to 0.125 0 0 1
021: dt: 0.2500, sse=215278.9, rms=0.918 (2.321%)
022: dt: 0.1250, sse=212848.5, rms=0.858 (6.568%)
rms = 0.8562/0.8578, sse=212938.6/212848.5, time step reduction 3 of 3 to 0.062 0 1 1
023: dt: 0.1250, sse=212938.6, rms=0.856 (0.188%)
maximum number of reductions reached, breaking from loop
positioning took 1.2 minutes
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 47 vertices, nripped=8591
mean border=87.8, 97 (36) missing vertices, mean dist -0.2 [0.3 (%73.7)->0.2 (%26.3))]
%82 local maxima, %11 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0876 min
Finding expansion regions
mean absolute distance = 0.27 +- 0.43
3217 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=458879.7, rms=3.364
024: dt: 0.5000, sse=329321.0, rms=2.412 (28.306%)
025: dt: 0.5000, sse=317932.1, rms=2.325 (3.604%)
rms = 2.4057/2.3247, sse=327355.6/317932.1, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
026: dt: 0.2500, sse=240603.8, rms=1.480 (36.333%)
027: dt: 0.2500, sse=211509.7, rms=0.985 (33.444%)
028: dt: 0.2500, sse=206333.8, rms=0.875 (11.147%)
rms = 0.8449/0.8753, sse=205068.7/206333.8, time step reduction 2 of 3 to 0.125 0 0 1
029: dt: 0.2500, sse=205068.7, rms=0.845 (3.468%)
rms = 0.8101/0.8449, sse=203488.9/205068.7, time step reduction 3 of 3 to 0.062 0 0 1
030: dt: 0.1250, sse=203488.9, rms=0.810 (4.126%)
maximum number of reductions reached, breaking from loop
positioning took 0.8 minutes
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 71.0000000;
outside_low = 60.4994740;
outside_hi = 111.7148900;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 55 vertices, nripped=8591
mean border=88.5, 108 (29) missing vertices, mean dist -0.0 [0.2 (%57.8)->0.2 (%42.2))]
%85 local maxima, % 9 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0554 min
Finding expansion regions
mean absolute distance = 0.20 +- 0.32
2877 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=231820.7, rms=1.401
031: dt: 0.5000, sse=226625.7, rms=1.259 (10.187%)
rms = 1.6344/1.2585, sse=249895.6/226625.7, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
032: dt: 0.2500, sse=205711.2, rms=0.856 (31.970%)
033: dt: 0.2500, sse=200416.9, rms=0.725 (15.331%)
rms = 0.6942/0.7249, sse=198076.2/200416.9, time step reduction 2 of 3 to 0.125 0 0 1
034: dt: 0.2500, sse=198076.2, rms=0.694 (4.240%)
rms = 0.6706/0.6942, sse=197014.6/198076.2, time step reduction 3 of 3 to 0.062 0 0 1
035: dt: 0.1250, sse=197014.6, rms=0.671 (3.400%)
maximum number of reductions reached, breaking from loop
positioning took 0.6 minutes
Writing output to ../surf/lh.white
#ET# mris_place_surface 4.44 minutes
#VMPC# mris_place_surfaces VmPeak 2079212
mris_place_surface done
#--------------------------------------------
#@# WhiteSurfs rh Thu Jun 12 15:09:02 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --rh --i ../surf/rh.white.preaparc --o ../surf/rh.white --white --nsmooth 0 --rip-label ../label/rh.cortex.label --rip-bg --rip-surf ../surf/rh.white.preaparc --aparc ../label/rh.aparc.annot
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --rh --i ../surf/rh.white.preaparc --o ../surf/rh.white --white --nsmooth 0 --rip-label ../label/rh.cortex.label --rip-bg --rip-surf ../surf/rh.white.preaparc --aparc ../label/rh.aparc.annot
Reading in input surface ../surf/rh.white.preaparc
Not smoothing input surface
Area 257022 0.33315 0.11032 0.001483 4.1400
Corner 771066 60.00000 12.73578 0.378459 178.6430
Edge 385533 0.88555 0.18084 0.035539 3.9623
Hinge 385533 8.52032 9.62556 0.000028 179.9167
Reading in aparc ../label/rh.aparc.annot
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for white surface
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
Ripping vertices not in label ../label/rh.cortex.label
MRISripNotLabel() ripped 8261/128513 vertices (120252 unripped)
Reading in ripping surface ../surf/rh.white.preaparc
Reading in aparc ../label/rh.aparc.annot for ripsurf
Ripping BG
MRISripBasalGanglia(): 1 -2 2 0.5 ripped 420
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64257: xyz = (54.7528,-10.1277,35.7877) oxyz = (54.7528,-10.1277,35.7877) wxzy = (54.7528,-10.1277,35.7877) pxyz = (0,0,0)
CBVO Creating mask 128513
n_averages 4
Iteration 0 =========================================
n_averages=4, current_sigma=2
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 99 vertices, nripped=8681
mean border=82.1, 50 (50) missing vertices, mean dist 0.4 [1.3 (%12.3)->0.7 (%87.7))]
%53 local maxima, %40 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.2101 min
Finding expansion regions
mean absolute distance = 0.73 +- 1.02
2505 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=4, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 0
000: dt: 0.0000, sse=1948584.0, rms=8.597
001: dt: 0.5000, sse=911653.1, rms=5.536 (35.601%)
002: dt: 0.5000, sse=547065.5, rms=3.931 (28.999%)
003: dt: 0.5000, sse=420639.8, rms=3.194 (18.747%)
004: dt: 0.5000, sse=363216.7, rms=2.781 (12.919%)
005: dt: 0.5000, sse=350937.6, rms=2.675 (3.823%)
006: dt: 0.5000, sse=334442.5, rms=2.545 (4.862%)
rms = 2.5774/2.5449, sse=339233.1/334442.5, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
007: dt: 0.2500, sse=247943.9, rms=1.680 (33.985%)
008: dt: 0.2500, sse=217677.9, rms=1.235 (26.485%)
009: dt: 0.2500, sse=209963.8, rms=1.090 (11.706%)
rms = 1.0497/1.0905, sse=208558.8/209963.7, time step reduction 2 of 3 to 0.125 0 0 1
010: dt: 0.2500, sse=208558.8, rms=1.050 (3.737%)
rms = 1.0134/1.0497, sse=207137.4/208558.8, time step reduction 3 of 3 to 0.062 0 0 1
011: dt: 0.1250, sse=207137.4, rms=1.013 (3.460%)
maximum number of reductions reached, breaking from loop
positioning took 1.3 minutes
Iteration 1 =========================================
n_averages=2, current_sigma=1
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 66 vertices, nripped=8681
mean border=85.8, 83 (10) missing vertices, mean dist -0.3 [0.5 (%73.6)->0.2 (%26.4))]
%70 local maxima, %23 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1549 min
Finding expansion regions
mean absolute distance = 0.45 +- 0.73
3994 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=2, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=947789.4, rms=5.602
012: dt: 0.5000, sse=582160.6, rms=4.021 (28.223%)
013: dt: 0.5000, sse=472545.2, rms=3.421 (14.915%)
014: dt: 0.5000, sse=416706.6, rms=3.072 (10.217%)
015: dt: 0.5000, sse=384077.9, rms=2.832 (7.800%)
016: dt: 0.5000, sse=362347.1, rms=2.667 (5.815%)
rms = 2.6380/2.6674, sse=360691.7/362347.1, time step reduction 1 of 3 to 0.250 0 0 1
017: dt: 0.5000, sse=360691.7, rms=2.638 (1.101%)
018: dt: 0.2500, sse=229834.1, rms=1.214 (53.984%)
019: dt: 0.2500, sse=216701.5, rms=0.961 (20.853%)
020: dt: 0.2500, sse=212969.7, rms=0.873 (9.130%)
rms = 0.8411/0.8731, sse=211507.6/212969.7, time step reduction 2 of 3 to 0.125 0 0 1
021: dt: 0.2500, sse=211507.6, rms=0.841 (3.666%)
022: dt: 0.1250, sse=209313.7, rms=0.782 (6.979%)
rms = 0.7754/0.7824, sse=208894.7/209313.7, time step reduction 3 of 3 to 0.062 0 0 1
023: dt: 0.1250, sse=208894.7, rms=0.775 (0.896%)
maximum number of reductions reached, breaking from loop
positioning took 1.2 minutes
Iteration 2 =========================================
n_averages=1, current_sigma=0.5
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 42 vertices, nripped=8681
mean border=88.1, 94 (8) missing vertices, mean dist -0.2 [0.3 (%73.3)->0.2 (%26.7))]
%80 local maxima, %13 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0854 min
Finding expansion regions
mean absolute distance = 0.27 +- 0.43
2911 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=1, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=435150.5, rms=3.219
024: dt: 0.5000, sse=313274.4, rms=2.281 (29.145%)
rms = 2.2356/2.2811, sse=306748.5/313274.4, time step reduction 1 of 3 to 0.250 0 0 1
025: dt: 0.5000, sse=306748.5, rms=2.236 (1.994%)
026: dt: 0.2500, sse=212972.1, rms=1.030 (53.912%)
027: dt: 0.2500, sse=204169.2, rms=0.838 (18.689%)
rms = 0.7917/0.8378, sse=202369.2/204169.2, time step reduction 2 of 3 to 0.125 0 0 1
028: dt: 0.2500, sse=202369.2, rms=0.792 (5.504%)
029: dt: 0.1250, sse=200103.3, rms=0.733 (7.473%)
rms = 0.7284/0.7325, sse=199924.0/200103.3, time step reduction 3 of 3 to 0.062 0 0 1
030: dt: 0.1250, sse=199924.0, rms=0.728 (0.567%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
Iteration 3 =========================================
n_averages=0, current_sigma=0.25
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 120.0000000;
border_hi = 111.7148900;
border_low = 72.0000000;
outside_low = 61.4994740;
outside_hi = 111.7148900;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 1
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 45 vertices, nripped=8681
mean border=88.8, 114 (6) missing vertices, mean dist -0.0 [0.2 (%57.8)->0.2 (%42.2))]
%83 local maxima, %10 large gradients and % 0 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0547 min
Finding expansion regions
mean absolute distance = 0.20 +- 0.32
2702 vertices more than 2 sigmas from mean.
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 5, l_surf_repulse = 0, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=0, nbrs=2, l_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=227661.6, rms=1.346
031: dt: 0.5000, sse=222462.8, rms=1.192 (11.475%)
rms = 1.5519/1.1917, sse=241500.2/222462.8, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
032: dt: 0.2500, sse=200720.6, rms=0.772 (35.190%)
033: dt: 0.2500, sse=194840.4, rms=0.635 (17.821%)
rms = 0.6080/0.6347, sse=194135.0/194840.4, time step reduction 2 of 3 to 0.125 0 0 1
034: dt: 0.2500, sse=194135.0, rms=0.608 (4.205%)
rms = 0.5944/0.6080, sse=192607.9/194135.0, time step reduction 3 of 3 to 0.062 0 0 1
035: dt: 0.1250, sse=192607.9, rms=0.594 (2.238%)
maximum number of reductions reached, breaking from loop
positioning took 0.6 minutes
Writing output to ../surf/rh.white
#ET# mris_place_surface 4.34 minutes
#VMPC# mris_place_surfaces VmPeak 2094112
mris_place_surface done
#--------------------------------------------
#@# T1PialSurf lh Thu Jun 12 15:13:32 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --lh --i ../surf/lh.white --o ../surf/lh.pial.T1 --pial --nsmooth 0 --rip-label ../label/lh.cortex+hipamyg.label --pin-medial-wall ../label/lh.cortex.label --aparc ../label/lh.aparc.annot --repulse-surf ../surf/lh.white --white-surf ../surf/lh.white
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.lh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --lh --i ../surf/lh.white --o ../surf/lh.pial.T1 --pial --nsmooth 0 --rip-label ../label/lh.cortex+hipamyg.label --pin-medial-wall ../label/lh.cortex.label --aparc ../label/lh.aparc.annot --repulse-surf ../surf/lh.white --white-surf ../surf/lh.white
Reading in input surface ../surf/lh.white
Not smoothing input surface
Area 257220 0.33538 0.12074 0.000208 2.4830
Corner 771660 60.00000 13.93859 0.041284 178.5246
Edge 385830 0.88960 0.19820 0.015512 3.9128
Hinge 385830 8.54357 9.72700 0.000098 179.9957
Reading white surface coordinates from ../surf/lh.white
Reading repulsion surface coordinates from ../surf/lh.white
Reading in aparc ../label/lh.aparc.annot
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for pial surface mid_gray = 67.8918
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
Ripping vertices not in label ../label/lh.cortex+hipamyg.label
MRISripNotLabel() ripped 6669/128612 vertices (121943 unripped)
INFO: rip surface needed but not specified, so using input surface
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64306: xyz = (-31.0111,-10.7648,6.15307) oxyz = (-31.0111,-10.7648,6.15307) wxzy = (-31.0111,-10.7648,6.15307) pxyz = (-31.0111,-10.7648,6.15307)
CBVO Creating mask 128612
n_averages 16
Iteration 0 =========================================
n_averages=16, current_sigma=2
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 60.4994740;
border_low = 39.4984210;
outside_low = 10.0000000;
outside_hi = 55.2492100;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 136 vertices, nripped=6669
mean border=57.7, 117 (117) missing vertices, mean dist 2.2 [1.2 (%0.0)->2.7 (%100.0))]
%19 local maxima, %58 large gradients and %18 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1730 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=16, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 0
randomSeed 0
000: dt: 0.0000, sse=24388168.0, rms=31.520
001: dt: 0.5000, sse=17481378.0, rms=26.647 (15.461%)
002: dt: 0.5000, sse=12427076.0, rms=22.418 (15.869%)
003: dt: 0.5000, sse=9038692.0, rms=19.064 (14.961%)
004: dt: 0.5000, sse=6818608.0, rms=16.500 (13.452%)
005: dt: 0.5000, sse=5279241.5, rms=14.456 (12.389%)
006: dt: 0.5000, sse=4122297.5, rms=12.703 (12.123%)
007: dt: 0.5000, sse=3215822.0, rms=11.138 (12.319%)
008: dt: 0.5000, sse=2487525.5, rms=9.699 (12.923%)
009: dt: 0.5000, sse=1907502.6, rms=8.377 (13.631%)
010: dt: 0.5000, sse=1441324.4, rms=7.138 (14.786%)
011: dt: 0.5000, sse=1090044.5, rms=6.039 (15.401%)
012: dt: 0.5000, sse=863133.3, rms=5.206 (13.799%)
013: dt: 0.5000, sse=749018.7, rms=4.730 (9.140%)
014: dt: 0.5000, sse=681625.5, rms=4.423 (6.485%)
015: dt: 0.5000, sse=657704.9, rms=4.307 (2.626%)
016: dt: 0.5000, sse=634997.2, rms=4.193 (2.642%)
rms = 4.1746/4.1931, sse=631841.7/634997.2, time step reduction 1 of 3 to 0.250 0 0 1
017: dt: 0.5000, sse=631841.7, rms=4.175 (0.442%)
018: dt: 0.2500, sse=485639.2, rms=3.360 (19.511%)
019: dt: 0.2500, sse=459525.1, rms=3.193 (4.961%)
rms = 3.1717/3.1934, sse=456560.4/459525.1, time step reduction 2 of 3 to 0.125 0 0 1
020: dt: 0.2500, sse=456560.3, rms=3.172 (0.678%)
021: dt: 0.1250, sse=426729.7, rms=2.969 (6.400%)
rms = 2.9311/2.9687, sse=421501.3/426729.7, time step reduction 3 of 3 to 0.062 0 0 1
022: dt: 0.1250, sse=421501.2, rms=2.931 (1.267%)
maximum number of reductions reached, breaking from loop
positioning took 2.0 minutes
Iteration 1 =========================================
n_averages=8, current_sigma=1
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 60.4994740;
border_low = 39.4984210;
outside_low = 10.0000000;
outside_hi = 55.2492100;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 3821 vertices, nripped=6669
mean border=55.3, 1057 (26) missing vertices, mean dist 0.2 [0.2 (%43.2)->0.6 (%56.8))]
%38 local maxima, %42 large gradients and %15 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0613 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=8, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=840234.6, rms=4.909
023: dt: 0.5000, sse=701732.8, rms=4.285 (12.720%)
024: dt: 0.5000, sse=662317.2, rms=4.101 (4.293%)
rms = 4.1060/4.1007, sse=664376.3/662317.3, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
025: dt: 0.2500, sse=532715.1, rms=3.380 (17.567%)
026: dt: 0.2500, sse=485775.9, rms=3.075 (9.021%)
027: dt: 0.2500, sse=474838.8, rms=2.999 (2.469%)
028: dt: 0.2500, sse=466296.0, rms=2.939 (2.027%)
rms = 2.9223/2.9386, sse=464156.7/466296.0, time step reduction 2 of 3 to 0.125 0 0 1
029: dt: 0.2500, sse=464156.7, rms=2.922 (0.554%)
030: dt: 0.1250, sse=435439.0, rms=2.709 (7.288%)
rms = 2.6711/2.7093, sse=430556.1/435439.0, time step reduction 3 of 3 to 0.062 0 0 1
031: dt: 0.1250, sse=430556.2, rms=2.671 (1.413%)
maximum number of reductions reached, breaking from loop
positioning took 0.9 minutes
Iteration 2 =========================================
n_averages=4, current_sigma=0.5
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 60.4994740;
border_low = 39.4984210;
outside_low = 10.0000000;
outside_hi = 55.2492100;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 4227 vertices, nripped=6669
mean border=53.3, 1455 (21) missing vertices, mean dist 0.2 [0.2 (%33.4)->0.4 (%66.6))]
%55 local maxima, %24 large gradients and %14 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0356 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=4, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=640133.9, rms=3.941
rms = 3.8993/3.9414, sse=633741.3/640133.9, time step reduction 1 of 3 to 0.250 0 0 1
032: dt: 0.5000, sse=633741.3, rms=3.899 (1.069%)
033: dt: 0.2500, sse=485563.1, rms=3.023 (22.472%)
034: dt: 0.2500, sse=463115.8, rms=2.866 (5.192%)
rms = 2.8392/2.8661, sse=459482.9/463115.8, time step reduction 2 of 3 to 0.125 0 0 1
035: dt: 0.2500, sse=459482.9, rms=2.839 (0.939%)
036: dt: 0.1250, sse=423298.7, rms=2.561 (9.808%)
rms = 2.5161/2.5607, sse=417829.9/423298.7, time step reduction 3 of 3 to 0.062 0 0 1
037: dt: 0.1250, sse=417829.9, rms=2.516 (1.742%)
maximum number of reductions reached, breaking from loop
positioning took 0.5 minutes
Iteration 3 =========================================
n_averages=2, current_sigma=0.25
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 60.4994740;
border_low = 39.4984210;
outside_low = 10.0000000;
outside_hi = 55.2492100;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128612
Gdiag_no=-1
vno start=0, stop=128612
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 3731 vertices, nripped=6669
mean border=52.3, 2284 (20) missing vertices, mean dist 0.1 [0.2 (%37.3)->0.3 (%62.7))]
%63 local maxima, %16 large gradients and %14 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0235 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=2, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=468110.6, rms=2.899
rms = 3.2588/2.8995, sse=522830.0/468110.6, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
038: dt: 0.2500, sse=434245.0, rms=2.645 (8.773%)
039: dt: 0.2500, sse=418323.5, rms=2.516 (4.875%)
rms = 2.4909/2.5161, sse=415082.3/418323.5, time step reduction 2 of 3 to 0.125 0 0 1
040: dt: 0.2500, sse=415082.2, rms=2.491 (1.004%)
041: dt: 0.1250, sse=405114.0, rms=2.406 (3.424%)
rms = 2.3796/2.4056, sse=402067.0/405114.0, time step reduction 3 of 3 to 0.062 0 0 1
042: dt: 0.1250, sse=402067.0, rms=2.380 (1.080%)
maximum number of reductions reached, breaking from loop
positioning took 0.5 minutes
Pinning medial wall to white surface
removing intersecting faces
000: 13 intersecting
terminating search with 0 intersecting
Writing output to ../surf/lh.pial.T1
#ET# mris_place_surface 4.26 minutes
#VMPC# mris_place_surfaces VmPeak 1344124
mris_place_surface done
#--------------------------------------------
#@# T1PialSurf rh Thu Jun 12 15:17:57 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --rh --i ../surf/rh.white --o ../surf/rh.pial.T1 --pial --nsmooth 0 --rip-label ../label/rh.cortex+hipamyg.label --pin-medial-wall ../label/rh.cortex.label --aparc ../label/rh.aparc.annot --repulse-surf ../surf/rh.white --white-surf ../surf/rh.white
7.3.2
7.3.2
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
setenv SUBJECTS_DIR /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output
mris_place_surface --adgws-in ../surf/autodet.gw.stats.rh.dat --seg aseg.presurf.mgz --threads 1 --wm wm.mgz --invol brain.finalsurfs.mgz --rh --i ../surf/rh.white --o ../surf/rh.pial.T1 --pial --nsmooth 0 --rip-label ../label/rh.cortex+hipamyg.label --pin-medial-wall ../label/rh.cortex.label --aparc ../label/rh.aparc.annot --repulse-surf ../surf/rh.white --white-surf ../surf/rh.white
Reading in input surface ../surf/rh.white
Not smoothing input surface
Area 257022 0.33470 0.12289 0.001255 4.1479
Corner 771066 60.00000 13.89162 0.206599 179.4338
Edge 385533 0.88859 0.19836 0.025653 4.0625
Hinge 385533 8.56625 9.79708 0.000042 179.9721
Reading white surface coordinates from ../surf/rh.white
Reading repulsion surface coordinates from ../surf/rh.white
Reading in aparc ../label/rh.aparc.annot
Reading in input volume brain.finalsurfs.mgz
Reading in wm volume wm.mgz
MRIclipBrightWM(): nthresh=15301, wmmin=5, clip=110
MRIfindBrightNonWM(): 1607 bright non-wm voxels segmented.
Masking bright non-wm for pial surface mid_gray = 68.3918
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
MRImask(): AllowDiffGeom = 1
Reading in seg volume aseg.presurf.mgz
Ripping frozen voxels
Ripping vertices not in label ../label/rh.cortex+hipamyg.label
MRISripNotLabel() ripped 6811/128513 vertices (121702 unripped)
INFO: rip surface needed but not specified, so using input surface
Ripping segs (eg, WMSA, BG, frozen)
Starting MRISripSegs() d = (-2 2 0.5) segnos: 247
MRISripSegs(): -2 2 0.5 ripped 0
vertex 64257: xyz = (54.7828,-10.2038,35.7793) oxyz = (54.7828,-10.2038,35.7793) wxzy = (54.7828,-10.2038,35.7793) pxyz = (54.7828,-10.2038,35.7793)
CBVO Creating mask 128513
n_averages 16
Iteration 0 =========================================
n_averages=16, current_sigma=2
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 61.4994740;
border_low = 40.4984210;
outside_low = 10.0000000;
outside_hi = 56.2492100;
sigma = 2;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=2 had to be increased for 138 vertices, nripped=6811
mean border=58.5, 101 (101) missing vertices, mean dist 2.2 [0.6 (%0.0)->2.6 (%100.0))]
%19 local maxima, %59 large gradients and %16 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.1746 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=2.0, host=jupyt, nav=16, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
complete_dist_mat 0
rms 0
smooth_averages 0
remove_neg 0
ico_order 0
which_surface 0
target_radius 0.000000
nfields 0
scale 0.000000
desired_rms_height 0.000000
momentum 0.000000
nbhd_size 0
max_nbrs 0
niterations 100
nsurfaces 0
SURFACES 3
flags 0 (0)
use curv 0
no sulc 0
no rigid align 0
mris->nsize 2
mris->hemisphere 1
randomSeed 0
000: dt: 0.0000, sse=23554578.0, rms=31.002
001: dt: 0.5000, sse=16784692.0, rms=26.130 (15.716%)
002: dt: 0.5000, sse=11834183.0, rms=21.890 (16.226%)
003: dt: 0.5000, sse=8528209.0, rms=18.525 (15.373%)
004: dt: 0.5000, sse=6393952.0, rms=15.979 (13.743%)
005: dt: 0.5000, sse=4940414.5, rms=13.981 (12.505%)
006: dt: 0.5000, sse=3858533.5, rms=12.283 (12.144%)
007: dt: 0.5000, sse=3019278.0, rms=10.782 (12.217%)
008: dt: 0.5000, sse=2353217.5, rms=9.422 (12.619%)
009: dt: 0.5000, sse=1819856.8, rms=8.170 (13.287%)
010: dt: 0.5000, sse=1379682.6, rms=6.969 (14.700%)
011: dt: 0.5000, sse=1049517.1, rms=5.909 (15.202%)
012: dt: 0.5000, sse=830212.5, rms=5.084 (13.973%)
013: dt: 0.5000, sse=719418.8, rms=4.609 (9.337%)
014: dt: 0.5000, sse=660474.6, rms=4.333 (5.981%)
015: dt: 0.5000, sse=640033.5, rms=4.231 (2.363%)
016: dt: 0.5000, sse=620504.2, rms=4.131 (2.369%)
rms = 4.1130/4.1308, sse=617656.1/620504.2, time step reduction 1 of 3 to 0.250 0 0 1
017: dt: 0.5000, sse=617656.1, rms=4.113 (0.432%)
018: dt: 0.2500, sse=473323.8, rms=3.294 (19.917%)
019: dt: 0.2500, sse=446952.4, rms=3.122 (5.230%)
rms = 3.0912/3.1215, sse=442742.5/446952.4, time step reduction 2 of 3 to 0.125 0 0 1
020: dt: 0.2500, sse=442742.5, rms=3.091 (0.971%)
021: dt: 0.1250, sse=415856.7, rms=2.903 (6.076%)
rms = 2.8679/2.9034, sse=411033.0/415856.7, time step reduction 3 of 3 to 0.062 0 0 1
022: dt: 0.1250, sse=411033.0, rms=2.868 (1.222%)
maximum number of reductions reached, breaking from loop
positioning took 2.0 minutes
Iteration 1 =========================================
n_averages=8, current_sigma=1
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 61.4994740;
border_low = 40.4984210;
outside_low = 10.0000000;
outside_hi = 56.2492100;
sigma = 1;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=1 had to be increased for 3219 vertices, nripped=6811
mean border=56.0, 919 (26) missing vertices, mean dist 0.2 [0.2 (%42.6)->0.6 (%57.4))]
%39 local maxima, %43 large gradients and %13 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0645 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=1.0, host=jupyt, nav=8, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=867179.2, rms=5.028
023: dt: 0.5000, sse=699609.2, rms=4.281 (14.848%)
024: dt: 0.5000, sse=654912.9, rms=4.070 (4.926%)
rms = 4.0471/4.0704, sse=651240.2/654912.9, time step reduction 1 of 3 to 0.250 0 0 1
025: dt: 0.5000, sse=651240.2, rms=4.047 (0.573%)
026: dt: 0.2500, sse=485345.0, rms=3.079 (23.914%)
027: dt: 0.2500, sse=454453.8, rms=2.863 (7.028%)
rms = 2.8447/2.8628, sse=452181.8/454453.7, time step reduction 2 of 3 to 0.125 0 0 1
028: dt: 0.2500, sse=452181.8, rms=2.845 (0.634%)
029: dt: 0.1250, sse=422005.4, rms=2.614 (8.124%)
rms = 2.5723/2.6136, sse=416911.5/422005.4, time step reduction 3 of 3 to 0.062 0 0 1
030: dt: 0.1250, sse=416911.5, rms=2.572 (1.579%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
Iteration 2 =========================================
n_averages=4, current_sigma=0.5
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 61.4994740;
border_low = 40.4984210;
outside_low = 10.0000000;
outside_hi = 56.2492100;
sigma = 0.5;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.5 had to be increased for 3654 vertices, nripped=6811
mean border=53.9, 1203 (23) missing vertices, mean dist 0.2 [0.2 (%33.3)->0.4 (%66.7))]
%56 local maxima, %25 large gradients and %13 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0372 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.5, host=jupyt, nav=4, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=644639.2, rms=3.971
031: dt: 0.5000, sse=625780.6, rms=3.862 (2.733%)
rms = 3.9240/3.8623, sse=634568.0/625780.6, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
032: dt: 0.2500, sse=494187.5, rms=3.082 (20.195%)
033: dt: 0.2500, sse=450274.9, rms=2.777 (9.903%)
034: dt: 0.2500, sse=443713.7, rms=2.727 (1.812%)
rms = 2.6917/2.7268, sse=438959.9/443713.7, time step reduction 2 of 3 to 0.125 0 0 1
035: dt: 0.2500, sse=438959.9, rms=2.692 (1.286%)
036: dt: 0.1250, sse=412416.7, rms=2.477 (7.984%)
rms = 2.4375/2.4768, sse=407744.0/412416.7, time step reduction 3 of 3 to 0.062 0 0 1
037: dt: 0.1250, sse=407744.0, rms=2.437 (1.589%)
maximum number of reductions reached, breaking from loop
positioning took 0.7 minutes
Iteration 3 =========================================
n_averages=2, current_sigma=0.25
Computing target border values
Entering MRIScomputeBorderValues_new():
inside_hi = 96.2851100;
border_hi = 61.4994740;
border_low = 40.4984210;
outside_low = 10.0000000;
outside_hi = 56.2492100;
sigma = 0.25;
max_thickness = 10;
step_size=0.5;
STEP_SIZE=0.1;
which = 2
thresh = 0
flags = 0
CBVfindFirstPeakD1=0
CBVfindFirstPeakD2=0
nvertices=128513
Gdiag_no=-1
vno start=0, stop=128513
Replacing 255s with 0s
#SI# sigma=0.25 had to be increased for 3388 vertices, nripped=6811
mean border=52.8, 1833 (20) missing vertices, mean dist 0.1 [0.2 (%36.5)->0.3 (%63.5))]
%65 local maxima, %16 large gradients and %13 min vals, 0 gradients ignored
nFirstPeakD1 0
MRIScomputeBorderValues_new() finished in 0.0246 min
Averaging target values for 5 iterations...
Positioning Surface: tspring = 0.3, nspring = 0.3, spring = 0, niters = 100 l_repulse = 0, l_surf_repulse = 5, checktol = 0
Positioning pial surface
Entering MRISpositionSurface()
max_mm = 0.3
MAX_REDUCTIONS = 2, REDUCTION_PCT = 0.5
parms->check_tol = 0, niterations = 100
tol=1.0e-04, sigma=0.2, host=jupyt, nav=2, nbrs=2, l_surf_repulse=5.000, l_tspring=0.300, l_nspring=0.300, l_intensity=0.200, l_curv=1.000
mom=0.00, dt=0.50
000: dt: 0.0000, sse=461868.7, rms=2.861
rms = 3.1951/2.8609, sse=512028.5/461868.7, time step reduction 1 of 3 to 0.250 0 1 1
RMS increased, rejecting step
038: dt: 0.2500, sse=425401.6, rms=2.581 (9.771%)
039: dt: 0.2500, sse=407793.3, rms=2.434 (5.710%)
rms = 2.4111/2.4340, sse=404976.3/407793.3, time step reduction 2 of 3 to 0.125 0 0 1
040: dt: 0.2500, sse=404976.3, rms=2.411 (0.939%)
041: dt: 0.1250, sse=395850.5, rms=2.330 (3.359%)
rms = 2.3102/2.3301, sse=393576.2/395850.5, time step reduction 3 of 3 to 0.062 0 0 1
042: dt: 0.1250, sse=393576.2, rms=2.310 (0.856%)
maximum number of reductions reached, breaking from loop
positioning took 0.5 minutes
Pinning medial wall to white surface
removing intersecting faces
000: 6 intersecting
001: 4 intersecting
terminating search with 0 intersecting
Writing output to ../surf/rh.pial.T1
#ET# mris_place_surface 4.35 minutes
#VMPC# mris_place_surfaces VmPeak 1342884
mris_place_surface done
#@# white curv lh Thu Jun 12 15:22:27 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --curv-map ../surf/lh.white 2 10 ../surf/lh.curv
insurf ../surf/lh.white, nbrs 2, curvature_avgs 10
writing curvature file ../surf/lh.curv
#@# white area lh Thu Jun 12 15:22:29 UTC 2025
cd /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/freesurfer_output/subjectname/mri
mris_place_surface --area-map ../surf/lh.white ../surf/lh.area
!ls ./freesurfer_output/subjectname/mri
T1.mgz mri_nu_correct.mni.log.bak
antsdn.brain.mgz norm.mgz
aparc+aseg.mgz nu.mgz
aparc.DKTatlas+aseg.mgz orig
aparc.a2009s+aseg.mgz orig.mgz
aseg.auto.mgz orig_nu.mgz
aseg.auto_noCCseg.label_intensities.txt rawavg.mgz
aseg.auto_noCCseg.mgz rh.ribbon.mgz
aseg.mgz ribbon.mgz
aseg.presurf.hypos.mgz segment.dat
aseg.presurf.mgz surface.defects.mgz
brain.finalsurfs.mgz talairach.label_intensities.txt
brain.mgz talairach.log
brainmask.auto.mgz talairach_with_skull.log
brainmask.mgz transforms
ctrl_pts.mgz wm.asegedit.mgz
filled.auto.mgz wm.mgz
filled.mgz wm.seg.mgz
lh.ribbon.mgz wmparc.mgz
mri_nu_correct.mni.log
!ls ./freesurfer_output/subjectname/surf
autodet.gw.stats.lh.dat lh.smoothwm.K1.crv rh.inflated.nofix
autodet.gw.stats.rh.dat lh.smoothwm.K2.crv rh.jacobian_white
lh.area lh.smoothwm.S.crv rh.orig
lh.area.mid lh.smoothwm.nofix rh.orig.nofix
lh.area.pial lh.sphere rh.orig.premesh
lh.avg_curv lh.sphere.reg rh.pial
lh.curv lh.sulc rh.pial.T1
lh.curv.pial lh.thickness rh.qsphere.nofix
lh.defect_borders lh.volume rh.smoothwm
lh.defect_chull lh.w-g.pct.mgh rh.smoothwm.BE.crv
lh.defect_labels lh.white rh.smoothwm.C.crv
lh.defects.pointset lh.white.H rh.smoothwm.FI.crv
lh.fsaverage.sphere.reg lh.white.K rh.smoothwm.H.crv
lh.inflated lh.white.preaparc rh.smoothwm.K.crv
lh.inflated.H lh.white.preaparc.H rh.smoothwm.K1.crv
lh.inflated.K lh.white.preaparc.K rh.smoothwm.K2.crv
lh.inflated.nofix rh.area rh.smoothwm.S.crv
lh.jacobian_white rh.area.mid rh.smoothwm.nofix
lh.orig rh.area.pial rh.sphere
lh.orig.nofix rh.avg_curv rh.sphere.reg
lh.orig.premesh rh.curv rh.sulc
lh.pial rh.curv.pial rh.thickness
lh.pial.T1 rh.defect_borders rh.volume
lh.qsphere.nofix rh.defect_chull rh.w-g.pct.mgh
lh.smoothwm rh.defect_labels rh.white
lh.smoothwm.BE.crv rh.defects.pointset rh.white.H
lh.smoothwm.C.crv rh.fsaverage.sphere.reg rh.white.K
lh.smoothwm.FI.crv rh.inflated rh.white.preaparc
lh.smoothwm.H.crv rh.inflated.H rh.white.preaparc.H
lh.smoothwm.K.crv rh.inflated.K rh.white.preaparc.K
Results#
from ipyniivue import NiiVue
nv = NiiVue(crosshair_color=[0,1,0,1])
nv.load_volumes([{"path": "./freesurfer_output/subjectname/mri/orig.mgz"},
{"path": "./freesurfer_output/subjectname/mri/aseg.mgz"}])
nv
from IPython.display import Image
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/freesurfer_orig_aseg.png')
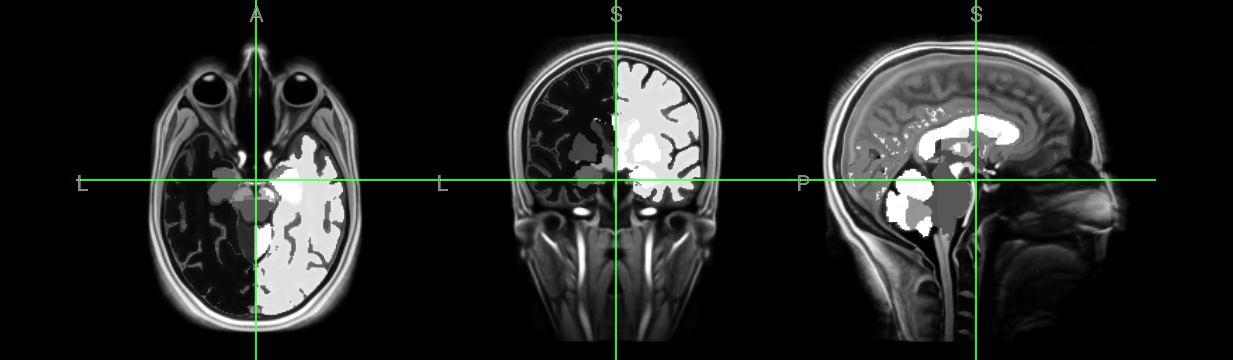
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
The watermark extension is already loaded. To reload it, use:
%reload_ext watermark
Last updated: 2025-11-02T23:13:08.528046+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
ipyniivue: 2.3.2
IPython : 8.16.1
