AFNI Preprocessing & Group Analysis#
Author: Monika Doerig
Citation:#
Tools included in this workflow#
AFNI
Cox RW (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29(3):162-173. doi:10.1006/cbmr.1996.0014
RW Cox, JS Hyde (1997). Software tools for analysis and visualization of FMRI Data. NMR in Biomedicine, 10: 171-178., https://pubmed.ncbi.nlm.nih.gov/9430344/
Workflows this work is based on#
Educational resources#
Andy’s Brain Book:
This AFNI example is based on the AFNI Tutorial: Statistics and Modeling from Andy’s Brain Book (Jahn, 2022. doi:10.5281/zenodo.5879293)
Dataset#
Flanker Dataset from OpenNeuro:
Kelly AMC and Uddin LQ and Biswal BB and Castellanos FX and Milham MP (2018). Flanker task (event-related). OpenNeuro Dataset ds000102. [Dataset] doi: null
Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Competition between functional brain networks mediates behavioral variability. Neuroimage. 2008 Jan 1;39(1):527-37. doi: 10.1016/j.neuroimage.2007.08.008. Epub 2007 Aug 23. PMID: 17919929.
Mennes, M., Kelly, C., Zuo, X.N., Di Martino, A., Biswal, B.B., Castellanos, F.X., Milham, M.P. (2010). Inter-individual differences in resting-state functional connectivity predict task-induced BOLD activity. Neuroimage, 50(4):1690-701. doi: 10.1016/j.neuroimage.2010.01.002. Epub 2010 Jan 15. Erratum in: Neuroimage. 2011 Mar 1;55(1):434
Mennes, M., Zuo, X.N., Kelly, C., Di Martino, A., Zang, Y.F., Biswal, B., Castellanos, F.X., Milham, M.P. (2011). Linking inter-individual differences in neural activation and behavior to intrinsic brain dynamics. Neuroimage, 54(4):2950-9. doi: 10.1016/j.neuroimage.2010.10.046ed in your example
Load packages#
import module
await module.load('afni/21.2.00')
await module.list()
['afni/21.2.00']
Import Python Modules#
%%capture
! pip install nibabel numpy nilearn scipy
import os
from glob import glob
import subprocess
import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
from nilearn import image, plotting
from ipyniivue import NiiVue
from IPython.display import display, Markdown, Image
from scipy.stats import ttest_1samp
from concurrent.futures import ProcessPoolExecutor, as_completed
from tqdm.notebook import tqdm
1. Download Data#
# 9 subjects from the Flanker Dataset
PATTERN = "sub-0*"
!datalad install https://github.com/OpenNeuroDatasets/ds000102.git
!cd ds000102 && datalad get $PATTERN
Cloning: 0%| | 0.00/2.00 [00:00<?, ? candidates/s]
Enumerating: 0.00 Objects [00:00, ? Objects/s]
Counting: 0%| | 0.00/27.0 [00:00<?, ? Objects/s]
Compressing: 0%| | 0.00/23.0 [00:00<?, ? Objects/s]
Receiving: 0%| | 0.00/2.15k [00:00<?, ? Objects/s]
Resolving: 0%| | 0.00/537 [00:00<?, ? Deltas/s]
[INFO ] scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
[INFO ] access to 1 dataset sibling s3-PRIVATE not auto-enabled, enable with:
| datalad siblings -d "/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102" enable -s s3-PRIVATE
install(ok): /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102 (dataset)
Total: 0%| | 0.00/618M [00:00<?, ? Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 0%| | 33.3k/10.6M [00:00<00:34, 304k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 1%| | 85.6k/10.6M [00:00<00:25, 409k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 2%| | 190k/10.6M [00:00<00:15, 665k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 4%|▏ | 382k/10.6M [00:00<00:09, 1.11M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 8%|▎ | 799k/10.6M [00:00<00:04, 2.11M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 15%|▍ | 1.60M/10.6M [00:00<00:02, 3.92M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 27%|▊ | 2.82M/10.6M [00:00<00:01, 6.35M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 42%|█▏ | 4.39M/10.6M [00:00<00:00, 8.73M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 64%|█▉ | 6.75M/10.6M [00:00<00:00, 13.0M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 79%|██▍| 8.39M/10.6M [00:01<00:00, 13.8M Bytes/s]
Total: 2%|▍ | 10.6M/618M [00:01<01:47, 5.64M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 0%| | 0.00/28.1M [00:00<?, ? Bytes/s]
Get sub-01/f .. _bold.nii.gz: 10%|▎ | 2.77M/28.1M [00:00<00:01, 12.9M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 19%|▌ | 5.31M/28.1M [00:00<00:01, 17.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 32%|▉ | 9.11M/28.1M [00:00<00:01, 17.5M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 46%|█▍ | 13.0M/28.1M [00:00<00:00, 17.9M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 53%|█▌ | 14.9M/28.1M [00:00<00:00, 17.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 61%|█▊ | 17.1M/28.1M [00:00<00:00, 18.3M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 75%|██▏| 20.9M/28.1M [00:01<00:00, 18.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 82%|██▍| 22.9M/28.1M [00:01<00:00, 18.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 88%|██▋| 24.8M/28.1M [00:01<00:00, 18.0M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 96%|██▉| 27.0M/28.1M [00:01<00:00, 18.7M Bytes/s]
Total: 6%|█▋ | 38.6M/618M [00:03<00:55, 10.5M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 0%| | 0.00/28.1M [00:00<?, ? Bytes/s]
Get sub-01/f .. _bold.nii.gz: 5%|▏ | 1.48M/28.1M [00:00<00:02, 12.6M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 10%|▎ | 2.75M/28.1M [00:00<00:02, 10.8M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 21%|▌ | 5.81M/28.1M [00:00<00:01, 14.6M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 27%|▊ | 7.48M/28.1M [00:00<00:01, 14.9M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 33%|▉ | 9.18M/28.1M [00:00<00:01, 15.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 39%|█▏ | 10.9M/28.1M [00:00<00:01, 15.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 45%|█▎ | 12.6M/28.1M [00:00<00:00, 15.6M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 51%|█▌ | 14.4M/28.1M [00:00<00:00, 15.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 57%|█▋ | 16.1M/28.1M [00:01<00:00, 16.0M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 63%|█▉ | 17.8M/28.1M [00:01<00:00, 16.1M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 70%|██ | 19.6M/28.1M [00:01<00:00, 16.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 76%|██▎| 21.4M/28.1M [00:01<00:00, 16.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 82%|██▍| 23.2M/28.1M [00:01<00:00, 16.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 89%|██▋| 25.0M/28.1M [00:01<00:00, 16.3M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 95%|██▊| 26.8M/28.1M [00:01<00:00, 16.6M Bytes/s]
Total: 11%|██▉ | 66.8M/618M [00:05<00:47, 11.5M Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 0%| | 0.00/10.7M [00:00<?, ? Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 17%|▌ | 1.82M/10.7M [00:00<00:00, 16.9M Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 34%|█ | 3.63M/10.7M [00:00<00:00, 16.8M Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 51%|█▌ | 5.46M/10.7M [00:00<00:00, 16.9M Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 68%|██ | 7.32M/10.7M [00:00<00:00, 17.0M Bytes/s]
Get sub-03/a .. 3_T1w.nii.gz: 86%|██▌| 9.17M/10.7M [00:00<00:00, 17.0M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 0%| | 0.00/28.8M [00:00<?, ? Bytes/s]
Get sub-03/f .. _bold.nii.gz: 6%|▏ | 1.87M/28.8M [00:00<00:01, 17.4M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 13%|▍ | 3.73M/28.8M [00:00<00:01, 17.3M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 19%|▌ | 5.60M/28.8M [00:00<00:01, 17.3M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 26%|▊ | 7.48M/28.8M [00:00<00:01, 17.4M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 33%|▉ | 9.37M/28.8M [00:00<00:01, 17.5M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 39%|█▏ | 11.3M/28.8M [00:00<00:00, 17.5M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 46%|█▎ | 13.2M/28.8M [00:00<00:00, 17.6M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 52%|█▌ | 15.1M/28.8M [00:00<00:00, 17.5M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 59%|█▊ | 17.0M/28.8M [00:00<00:00, 17.8M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 66%|█▉ | 18.8M/28.8M [00:01<00:00, 17.6M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 72%|██▏| 20.8M/28.8M [00:01<00:00, 17.7M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 79%|██▎| 22.7M/28.8M [00:01<00:00, 17.8M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 86%|██▌| 24.6M/28.8M [00:01<00:00, 17.8M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 92%|██▊| 26.6M/28.8M [00:01<00:00, 17.8M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 99%|██▉| 28.5M/28.8M [00:01<00:00, 17.9M Bytes/s]
Total: 17%|████▊ | 106M/618M [00:08<00:41, 12.4M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 0%| | 0.00/28.8M [00:00<?, ? Bytes/s]
Get sub-03/f .. _bold.nii.gz: 7%|▏ | 1.95M/28.8M [00:00<00:01, 18.2M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 14%|▍ | 3.89M/28.8M [00:00<00:01, 18.2M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 20%|▌ | 5.86M/28.8M [00:00<00:01, 18.2M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 27%|▊ | 7.83M/28.8M [00:00<00:01, 18.2M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 34%|█ | 9.79M/28.8M [00:00<00:01, 18.1M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 41%|█▏ | 11.8M/28.8M [00:00<00:00, 18.1M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 48%|█▍ | 13.7M/28.8M [00:00<00:00, 18.4M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 55%|█▋ | 15.7M/28.8M [00:00<00:00, 18.2M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 68%|██ | 19.7M/28.8M [00:01<00:00, 18.3M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 75%|██▎| 21.6M/28.8M [00:01<00:00, 18.3M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 82%|██▍| 23.6M/28.8M [00:01<00:00, 18.4M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 89%|██▋| 25.6M/28.8M [00:01<00:00, 18.3M Bytes/s]
Get sub-03/f .. _bold.nii.gz: 96%|██▉| 27.6M/28.8M [00:01<00:00, 18.3M Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 0%| | 0.00/10.7M [00:00<?, ? Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 18%|▌ | 1.94M/10.7M [00:00<00:00, 18.2M Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 36%|█ | 3.87M/10.7M [00:00<00:00, 18.0M Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 55%|█▋ | 5.87M/10.7M [00:00<00:00, 18.2M Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 73%|██▏| 7.87M/10.7M [00:00<00:00, 18.3M Bytes/s]
Get sub-02/a .. 2_T1w.nii.gz: 92%|██▊| 9.87M/10.7M [00:00<00:00, 18.5M Bytes/s]
Total: 24%|██████▌ | 146M/618M [00:11<00:36, 12.9M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 0%| | 0.00/29.2M [00:00<?, ? Bytes/s]
Get sub-02/f .. _bold.nii.gz: 7%|▏ | 1.98M/29.2M [00:00<00:01, 18.5M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 14%|▍ | 3.95M/29.2M [00:00<00:01, 18.4M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 20%|▌ | 5.96M/29.2M [00:00<00:01, 18.5M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 27%|▊ | 7.98M/29.2M [00:00<00:01, 18.5M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 34%|█ | 9.99M/29.2M [00:00<00:01, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 41%|█▏ | 12.0M/29.2M [00:00<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 48%|█▍ | 14.0M/29.2M [00:00<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 55%|█▋ | 16.0M/29.2M [00:00<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 62%|█▊ | 18.0M/29.2M [00:00<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 69%|██ | 20.1M/29.2M [00:01<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 76%|██▎| 22.1M/29.2M [00:01<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 83%|██▍| 24.1M/29.2M [00:01<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 89%|██▋| 26.1M/29.2M [00:01<00:00, 18.8M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 96%|██▉| 28.1M/29.2M [00:01<00:00, 18.9M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 0%| | 0.00/29.2M [00:00<?, ? Bytes/s]
Get sub-02/f .. _bold.nii.gz: 7%|▏ | 1.97M/29.2M [00:00<00:01, 18.4M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 13%|▍ | 3.89M/29.2M [00:00<00:01, 18.1M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 20%|▌ | 5.92M/29.2M [00:00<00:01, 18.5M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 27%|▊ | 7.89M/29.2M [00:00<00:01, 18.4M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 34%|█ | 9.90M/29.2M [00:00<00:01, 18.5M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 41%|█▏ | 11.9M/29.2M [00:00<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 48%|█▍ | 13.9M/29.2M [00:00<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 55%|█▋ | 15.9M/29.2M [00:00<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 61%|█▊ | 17.9M/29.2M [00:00<00:00, 18.6M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 68%|██ | 20.0M/29.2M [00:01<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 75%|██▎| 22.0M/29.2M [00:01<00:00, 18.8M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 82%|██▍| 24.0M/29.2M [00:01<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 89%|██▋| 26.0M/29.2M [00:01<00:00, 18.7M Bytes/s]
Get sub-02/f .. _bold.nii.gz: 96%|██▉| 28.0M/29.2M [00:01<00:00, 18.8M Bytes/s]
Total: 33%|█████████▏ | 204M/618M [00:15<00:30, 13.5M Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 19%|▌ | 2.02M/10.6M [00:00<00:00, 18.9M Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 37%|█ | 3.95M/10.6M [00:00<00:00, 18.3M Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 56%|█▋ | 5.92M/10.6M [00:00<00:00, 18.3M Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 74%|██▏| 7.85M/10.6M [00:00<00:00, 18.2M Bytes/s]
Get sub-06/a .. 6_T1w.nii.gz: 93%|██▊| 9.88M/10.6M [00:00<00:00, 18.0M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 0%| | 0.00/29.4M [00:00<?, ? Bytes/s]
Get sub-06/f .. _bold.nii.gz: 7%|▏ | 1.98M/29.4M [00:00<00:01, 18.5M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 13%|▍ | 3.93M/29.4M [00:00<00:01, 18.3M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 20%|▌ | 5.93M/29.4M [00:00<00:01, 18.4M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 27%|▊ | 7.86M/29.4M [00:00<00:01, 18.2M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 34%|█ | 9.89M/29.4M [00:00<00:01, 18.4M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 47%|█▍ | 13.7M/29.4M [00:00<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 54%|█▌ | 15.7M/29.4M [00:00<00:00, 18.1M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 60%|█▊ | 17.8M/29.4M [00:00<00:00, 18.3M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 67%|██ | 19.8M/29.4M [00:01<00:00, 18.4M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 74%|██▏| 21.8M/29.4M [00:01<00:00, 18.5M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 81%|██▍| 23.8M/29.4M [00:01<00:00, 18.6M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 88%|██▋| 25.9M/29.4M [00:01<00:00, 18.6M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 95%|██▊| 27.9M/29.4M [00:01<00:00, 18.7M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 0%| | 0.00/29.4M [00:00<?, ? Bytes/s]
Get sub-06/f .. _bold.nii.gz: 7%|▏ | 1.96M/29.4M [00:00<00:01, 18.3M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 13%|▍ | 3.89M/29.4M [00:00<00:01, 18.1M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 20%|▌ | 5.84M/29.4M [00:00<00:01, 18.1M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 27%|▊ | 7.87M/29.4M [00:00<00:01, 18.4M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 34%|█ | 9.88M/29.4M [00:00<00:01, 18.5M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 40%|█▏ | 11.9M/29.4M [00:00<00:00, 18.6M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 47%|█▍ | 13.9M/29.4M [00:00<00:00, 18.7M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 54%|█▋ | 16.0M/29.4M [00:00<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 61%|█▊ | 18.0M/29.4M [00:00<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 68%|██ | 20.0M/29.4M [00:01<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 75%|██▎| 22.0M/29.4M [00:01<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 82%|██▍| 24.1M/29.4M [00:01<00:00, 18.8M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 89%|██▋| 26.1M/29.4M [00:01<00:00, 18.9M Bytes/s]
Get sub-06/f .. _bold.nii.gz: 96%|██▊| 28.2M/29.4M [00:01<00:00, 18.8M Bytes/s]
Total: 44%|████████████▍ | 274M/618M [00:19<00:24, 13.8M Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 0%| | 0.00/10.7M [00:00<?, ? Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 18%|▌ | 1.91M/10.7M [00:00<00:00, 19.1M Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 36%|█ | 3.91M/10.7M [00:00<00:00, 18.1M Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 55%|█▋ | 5.93M/10.7M [00:00<00:00, 18.4M Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 73%|██▏| 7.85M/10.7M [00:00<00:00, 18.2M Bytes/s]
Get sub-07/a .. 7_T1w.nii.gz: 92%|██▊| 9.86M/10.7M [00:00<00:00, 18.4M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 0%| | 0.00/28.9M [00:00<?, ? Bytes/s]
Get sub-07/f .. _bold.nii.gz: 7%|▏ | 2.04M/28.9M [00:00<00:01, 19.1M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 14%|▍ | 3.97M/28.9M [00:00<00:01, 18.4M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 20%|▌ | 5.92M/28.9M [00:00<00:01, 18.3M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 27%|▊ | 7.86M/28.9M [00:00<00:01, 18.2M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 34%|█ | 9.79M/28.9M [00:00<00:01, 18.1M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 41%|█▏ | 11.8M/28.9M [00:00<00:00, 18.2M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 47%|█▍ | 13.7M/28.9M [00:00<00:00, 18.2M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 55%|█▋ | 15.8M/28.9M [00:00<00:00, 18.5M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 61%|█▊ | 17.7M/28.9M [00:00<00:00, 18.2M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 68%|██ | 19.7M/28.9M [00:01<00:00, 18.4M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 75%|██▎| 21.8M/28.9M [00:01<00:00, 18.7M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 82%|██▍| 23.8M/28.9M [00:01<00:00, 18.8M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 89%|██▋| 25.8M/28.9M [00:01<00:00, 18.7M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 96%|██▉| 27.9M/28.9M [00:01<00:00, 18.8M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 0%| | 0.00/29.0M [00:00<?, ? Bytes/s]
Get sub-07/f .. _bold.nii.gz: 7%|▏ | 1.97M/29.0M [00:00<00:01, 16.7M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 16%|▍ | 4.72M/29.0M [00:00<00:01, 14.6M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 22%|▋ | 6.49M/29.0M [00:00<00:01, 15.0M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 33%|▉ | 9.53M/29.0M [00:00<00:01, 14.6M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 38%|█▏ | 11.1M/29.0M [00:00<00:01, 14.6M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 44%|█▎ | 12.6M/29.0M [00:00<00:01, 14.5M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 49%|█▍ | 14.2M/29.0M [00:00<00:01, 14.6M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 55%|█▋ | 15.8M/29.0M [00:01<00:00, 14.6M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 60%|█▊ | 17.4M/29.0M [00:01<00:00, 14.8M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 66%|█▉ | 19.0M/29.0M [00:01<00:00, 14.9M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 71%|██▏| 20.7M/29.0M [00:01<00:00, 15.0M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 77%|██▎| 22.3M/29.0M [00:01<00:00, 15.1M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 83%|██▍| 24.0M/29.0M [00:01<00:00, 15.2M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 89%|██▋| 25.7M/29.0M [00:01<00:00, 15.4M Bytes/s]
Get sub-07/f .. _bold.nii.gz: 95%|██▊| 27.4M/29.0M [00:01<00:00, 15.4M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 16%|▍ | 1.71M/10.6M [00:00<00:00, 17.0M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 33%|▉ | 3.46M/10.6M [00:00<00:00, 16.5M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 49%|█▍ | 5.22M/10.6M [00:00<00:00, 16.4M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 66%|█▉ | 6.98M/10.6M [00:00<00:00, 16.5M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 83%|██▍| 8.77M/10.6M [00:00<00:00, 16.5M Bytes/s]
Total: 57%|███████████████▉ | 353M/618M [00:25<00:19, 13.6M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 0%| | 0.00/28.6M [00:00<?, ? Bytes/s]
Get sub-08/f .. _bold.nii.gz: 6%|▏ | 1.79M/28.6M [00:00<00:01, 16.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 13%|▍ | 3.62M/28.6M [00:00<00:01, 16.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 19%|▌ | 5.43M/28.6M [00:00<00:01, 16.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 25%|▊ | 7.28M/28.6M [00:00<00:01, 17.0M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 32%|▉ | 9.14M/28.6M [00:00<00:01, 17.0M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 38%|█▏ | 11.0M/28.6M [00:00<00:01, 12.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 48%|█▍ | 13.7M/28.6M [00:00<00:00, 16.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 58%|█▊ | 16.8M/28.6M [00:01<00:00, 15.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 68%|██ | 19.4M/28.6M [00:01<00:00, 14.7M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 77%|██▎| 22.2M/28.6M [00:01<00:00, 13.4M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 87%|██▌| 24.9M/28.6M [00:01<00:00, 13.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 97%|██▉| 27.7M/28.6M [00:01<00:00, 13.1M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 0%| | 0.00/28.6M [00:00<?, ? Bytes/s]
Get sub-08/f .. _bold.nii.gz: 5%|▏ | 1.41M/28.6M [00:00<00:02, 13.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 10%|▎ | 2.84M/28.6M [00:00<00:01, 13.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 15%|▍ | 4.27M/28.6M [00:00<00:01, 13.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 20%|▌ | 5.71M/28.6M [00:00<00:01, 13.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 25%|▋ | 7.15M/28.6M [00:00<00:01, 13.4M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 30%|▉ | 8.61M/28.6M [00:00<00:01, 13.4M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 35%|█ | 10.1M/28.6M [00:00<00:01, 13.4M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 40%|█▏ | 11.5M/28.6M [00:00<00:01, 13.6M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 45%|█▎ | 13.0M/28.6M [00:00<00:01, 13.6M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 51%|█▌ | 14.5M/28.6M [00:01<00:01, 13.5M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 56%|█▋ | 16.0M/28.6M [00:01<00:00, 13.7M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 61%|█▊ | 17.5M/28.6M [00:01<00:00, 13.7M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 66%|█▉ | 18.9M/28.6M [00:01<00:00, 13.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 71%|██▏| 20.5M/28.6M [00:01<00:00, 13.7M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 77%|██▎| 22.0M/28.6M [00:01<00:00, 13.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 82%|██▍| 23.5M/28.6M [00:01<00:00, 13.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 87%|██▌| 25.0M/28.6M [00:01<00:00, 13.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 93%|██▊| 26.5M/28.6M [00:01<00:00, 14.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 98%|██▉| 28.0M/28.6M [00:02<00:00, 14.2M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 0%| | 0.00/10.7M [00:00<?, ? Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 14%|▍ | 1.53M/10.7M [00:00<00:00, 14.3M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 28%|▊ | 3.06M/10.7M [00:00<00:00, 14.2M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 43%|█▎ | 4.60M/10.7M [00:00<00:00, 14.3M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 57%|█▋ | 6.14M/10.7M [00:00<00:00, 14.2M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 72%|██▏| 7.68M/10.7M [00:00<00:00, 14.2M Bytes/s]
Get sub-04/a .. 4_T1w.nii.gz: 86%|██▌| 9.22M/10.7M [00:00<00:00, 14.3M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 0%| | 0.00/29.1M [00:00<?, ? Bytes/s]
Get sub-04/f .. _bold.nii.gz: 5%|▏ | 1.55M/29.1M [00:00<00:01, 14.5M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 11%|▎ | 3.10M/29.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 16%|▍ | 4.66M/29.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 21%|▋ | 6.22M/29.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 27%|▊ | 7.77M/29.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 32%|▉ | 9.33M/29.1M [00:00<00:01, 14.5M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 38%|█▏ | 10.9M/29.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 43%|█▎ | 12.5M/29.1M [00:00<00:01, 14.5M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 48%|█▍ | 14.0M/29.1M [00:00<00:01, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 54%|█▌ | 15.6M/29.1M [00:01<00:00, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 59%|█▊ | 17.2M/29.1M [00:01<00:00, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 65%|█▉ | 18.8M/29.1M [00:01<00:00, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 70%|██ | 20.3M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 75%|██▎| 21.9M/29.1M [00:01<00:00, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 81%|██▍| 23.5M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 86%|██▌| 25.1M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 92%|██▊| 26.6M/29.1M [00:01<00:00, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 97%|██▉| 28.2M/29.1M [00:01<00:00, 14.7M Bytes/s]
Total: 73%|████████████████████▎ | 450M/618M [00:33<00:12, 13.3M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 0%| | 0.00/29.1M [00:00<?, ? Bytes/s]
Get sub-04/f .. _bold.nii.gz: 5%|▏ | 1.55M/29.1M [00:00<00:01, 14.6M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 11%|▎ | 3.14M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 16%|▍ | 4.72M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 22%|▋ | 6.31M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 27%|▊ | 7.90M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 33%|▉ | 9.49M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 38%|█▏ | 11.1M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 44%|█▎ | 12.7M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 49%|█▍ | 14.2M/29.1M [00:00<00:01, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 54%|█▋ | 15.8M/29.1M [00:01<00:00, 14.8M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 60%|█▊ | 17.4M/29.1M [00:01<00:00, 14.8M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 65%|█▉ | 19.0M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 71%|██▏| 20.6M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 76%|██▎| 22.2M/29.1M [00:01<00:00, 14.8M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 82%|██▍| 23.8M/29.1M [00:01<00:00, 14.8M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 87%|██▌| 25.4M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 93%|██▊| 27.0M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-04/f .. _bold.nii.gz: 98%|██▉| 28.5M/29.1M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 0%| | 0.00/10.8M [00:00<?, ? Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 15%|▍ | 1.57M/10.8M [00:00<00:00, 14.6M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 29%|▉ | 3.15M/10.8M [00:00<00:00, 14.7M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 44%|█▎ | 4.75M/10.8M [00:00<00:00, 14.7M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 59%|█▊ | 6.33M/10.8M [00:00<00:00, 14.7M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 74%|██▏| 7.92M/10.8M [00:00<00:00, 14.7M Bytes/s]
Get sub-05/a .. 5_T1w.nii.gz: 88%|██▋| 9.51M/10.8M [00:00<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 0%| | 0.00/29.7M [00:00<?, ? Bytes/s]
Get sub-05/f .. _bold.nii.gz: 5%|▏ | 1.58M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 11%|▎ | 3.17M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 16%|▍ | 4.77M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 21%|▋ | 6.36M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 27%|▊ | 7.95M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 32%|▉ | 9.53M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 37%|█ | 11.1M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 43%|█▎ | 12.7M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 48%|█▍ | 14.3M/29.7M [00:00<00:01, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 54%|█▌ | 15.9M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 59%|█▊ | 17.5M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 64%|█▉ | 19.1M/29.7M [00:01<00:00, 14.4M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 70%|██ | 20.7M/29.7M [00:01<00:00, 14.5M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 75%|██▏| 22.2M/29.7M [00:01<00:00, 14.6M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 80%|██▍| 23.8M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 86%|██▌| 25.4M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 91%|██▋| 27.0M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 96%|██▉| 28.6M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 0%| | 0.00/29.7M [00:00<?, ? Bytes/s]
Get sub-05/f .. _bold.nii.gz: 5%|▏ | 1.60M/29.7M [00:00<00:01, 15.0M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 11%|▎ | 3.19M/29.7M [00:00<00:01, 14.9M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 16%|▍ | 4.78M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 21%|▋ | 6.37M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 27%|▊ | 7.98M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 32%|▉ | 9.59M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 38%|█▏ | 11.2M/29.7M [00:00<00:01, 14.9M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 43%|█▎ | 12.8M/29.7M [00:00<00:01, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 49%|█▍ | 14.4M/29.7M [00:00<00:01, 14.9M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 54%|█▌ | 16.0M/29.7M [00:01<00:00, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 59%|█▊ | 17.6M/29.7M [00:01<00:00, 14.9M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 65%|█▉ | 19.2M/29.7M [00:01<00:00, 15.0M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 70%|██ | 20.8M/29.7M [00:01<00:00, 14.7M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 76%|██▎| 22.4M/29.7M [00:01<00:00, 14.8M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 81%|██▍| 24.1M/29.7M [00:01<00:00, 14.9M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 87%|██▌| 25.7M/29.7M [00:01<00:00, 15.2M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 92%|██▊| 27.3M/29.7M [00:01<00:00, 15.2M Bytes/s]
Get sub-05/f .. _bold.nii.gz: 97%|██▉| 28.9M/29.7M [00:01<00:00, 15.1M Bytes/s]
Total: 89%|████████████████████████▊ | 549M/618M [00:41<00:05, 13.1M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 0%| | 0.00/10.8M [00:00<?, ? Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 15%|▍ | 1.61M/10.8M [00:00<00:00, 15.0M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 30%|▉ | 3.24M/10.8M [00:00<00:00, 15.1M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 45%|█▎ | 4.87M/10.8M [00:00<00:00, 15.1M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 60%|█▊ | 6.50M/10.8M [00:00<00:00, 15.1M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 76%|██▎| 8.14M/10.8M [00:00<00:00, 15.2M Bytes/s]
Get sub-09/a .. 9_T1w.nii.gz: 91%|██▋| 9.78M/10.8M [00:00<00:00, 15.2M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 0%| | 0.00/29.2M [00:00<?, ? Bytes/s]
Get sub-09/f .. _bold.nii.gz: 6%|▏ | 1.64M/29.2M [00:00<00:01, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 11%|▎ | 3.28M/29.2M [00:00<00:01, 15.4M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 17%|▌ | 4.95M/29.2M [00:00<00:01, 15.4M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 23%|▋ | 6.61M/29.2M [00:00<00:01, 15.4M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 28%|▊ | 8.28M/29.2M [00:00<00:01, 15.4M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 34%|█ | 9.95M/29.2M [00:00<00:01, 15.5M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 40%|█▏ | 11.6M/29.2M [00:00<00:01, 15.5M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 46%|█▎ | 13.3M/29.2M [00:00<00:01, 15.4M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 51%|█▌ | 15.0M/29.2M [00:00<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 57%|█▋ | 16.7M/29.2M [00:01<00:00, 15.6M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 63%|█▉ | 18.4M/29.2M [00:01<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 69%|██ | 20.1M/29.2M [00:01<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 75%|██▏| 21.8M/29.2M [00:01<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 80%|██▍| 23.5M/29.2M [00:01<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 86%|██▌| 25.2M/29.2M [00:01<00:00, 15.9M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 92%|██▊| 26.9M/29.2M [00:01<00:00, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 98%|██▉| 28.6M/29.2M [00:01<00:00, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 0%| | 0.00/29.2M [00:00<?, ? Bytes/s]
Get sub-09/f .. _bold.nii.gz: 6%|▏ | 1.70M/29.2M [00:00<00:01, 15.9M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 12%|▎ | 3.39M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 17%|▌ | 5.11M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 23%|▋ | 6.81M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 29%|▊ | 8.51M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 35%|█ | 10.2M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 41%|█▏ | 11.9M/29.2M [00:00<00:01, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 47%|█▍ | 13.6M/29.2M [00:00<00:00, 15.9M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 53%|█▌ | 15.3M/29.2M [00:00<00:00, 15.7M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 58%|█▊ | 17.0M/29.2M [00:01<00:00, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 64%|█▉ | 18.8M/29.2M [00:01<00:00, 16.0M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 70%|██ | 20.5M/29.2M [00:01<00:00, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 76%|██▎| 22.2M/29.2M [00:01<00:00, 16.0M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 82%|██▍| 23.9M/29.2M [00:01<00:00, 15.8M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 88%|██▋| 25.6M/29.2M [00:01<00:00, 15.9M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 93%|██▊| 27.3M/29.2M [00:01<00:00, 15.9M Bytes/s]
Get sub-09/f .. _bold.nii.gz: 99%|██▉| 29.0M/29.2M [00:01<00:00, 16.0M Bytes/s]
get(ok): sub-01/anat/sub-01_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-01/func/sub-01_task-flanker_run-2_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-03/anat/sub-03_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-03/func/sub-03_task-flanker_run-1_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-03/func/sub-03_task-flanker_run-2_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-02/anat/sub-02_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-02/func/sub-02_task-flanker_run-1_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-02/func/sub-02_task-flanker_run-2_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-06/anat/sub-06_T1w.nii.gz (file) [from s3-PUBLIC...]
[17 similar messages have been suppressed; disable with datalad.ui.suppress-similar-results=off]
get(ok): sub-01 (directory)
get(ok): sub-03 (directory)
get(ok): sub-02 (directory)
get(ok): sub-06 (directory)
get(ok): sub-07 (directory)
get(ok): sub-08 (directory)
get(ok): sub-04 (directory)
get(ok): sub-05 (directory)
get(ok): sub-09 (directory)
action summary:
get (ok: 36)
The data is structured in BIDS format:
!tree -L 4 ds000102
ds000102
├── CHANGES
├── README
├── T1w.json
├── dataset_description.json
├── derivatives
│ └── mriqc
│ ├── aMRIQC.csv -> ../../.git/annex/objects/Q4/jv/MD5E-s14180--3addf0456b803b7c5ec5147481ecdd62.csv/MD5E-s14180--3addf0456b803b7c5ec5147481ecdd62.csv
│ ├── anatomical_group.pdf -> ../../.git/annex/objects/6m/q9/MD5E-s98927--d11151f65ae061833e7fd4373adfec3f.pdf/MD5E-s98927--d11151f65ae061833e7fd4373adfec3f.pdf
│ ├── anatomical_sub-01.pdf -> ../../.git/annex/objects/K3/7x/MD5E-s2747349--5d40f2a54fb4194ac4a79f0295ff51c0.pdf/MD5E-s2747349--5d40f2a54fb4194ac4a79f0295ff51c0.pdf
│ ├── anatomical_sub-02.pdf -> ../../.git/annex/objects/Kx/Kv/MD5E-s2803965--56f6b768362bd9b7f0ef501b8cb6dde6.pdf/MD5E-s2803965--56f6b768362bd9b7f0ef501b8cb6dde6.pdf
│ ├── anatomical_sub-03.pdf -> ../../.git/annex/objects/kx/g5/MD5E-s2809843--e90d7a4859ed4be986b55e23f93ca89d.pdf/MD5E-s2809843--e90d7a4859ed4be986b55e23f93ca89d.pdf
│ ├── anatomical_sub-04.pdf -> ../../.git/annex/objects/JK/Zm/MD5E-s2846770--4869146771178dbb01ac79b95b35a8a1.pdf/MD5E-s2846770--4869146771178dbb01ac79b95b35a8a1.pdf
│ ├── anatomical_sub-05.pdf -> ../../.git/annex/objects/zm/FG/MD5E-s2824086--fda634d34556c83005a5eb2ca8c498dd.pdf/MD5E-s2824086--fda634d34556c83005a5eb2ca8c498dd.pdf
│ ├── anatomical_sub-06.pdf -> ../../.git/annex/objects/92/q0/MD5E-s2798058--fdddf0aff1eca8f61ed7c8b04ada9735.pdf/MD5E-s2798058--fdddf0aff1eca8f61ed7c8b04ada9735.pdf
│ ├── anatomical_sub-07.pdf -> ../../.git/annex/objects/39/3K/MD5E-s2795270--29ce2e2352596df940e5f3fae45b5a38.pdf/MD5E-s2795270--29ce2e2352596df940e5f3fae45b5a38.pdf
│ ├── anatomical_sub-08.pdf -> ../../.git/annex/objects/Fx/F4/MD5E-s2727492--b55dad8ffe22fc035110ecf4119d2960.pdf/MD5E-s2727492--b55dad8ffe22fc035110ecf4119d2960.pdf
│ ├── anatomical_sub-09.pdf -> ../../.git/annex/objects/4M/pz/MD5E-s2887144--28ea830af2a4d741147d18ea9c7fda84.pdf/MD5E-s2887144--28ea830af2a4d741147d18ea9c7fda84.pdf
│ ├── anatomical_sub-10.pdf -> ../../.git/annex/objects/0z/Vw/MD5E-s2874045--6542a57a9fc58f97f2a03c2384663c62.pdf/MD5E-s2874045--6542a57a9fc58f97f2a03c2384663c62.pdf
│ ├── anatomical_sub-11.pdf -> ../../.git/annex/objects/wm/76/MD5E-s2781221--1071b83e3c1b4532879521c37c3329da.pdf/MD5E-s2781221--1071b83e3c1b4532879521c37c3329da.pdf
│ ├── anatomical_sub-12.pdf -> ../../.git/annex/objects/GF/19/MD5E-s2817233--bfd24ca3274fa5efd654e2afd927f9ef.pdf/MD5E-s2817233--bfd24ca3274fa5efd654e2afd927f9ef.pdf
│ ├── anatomical_sub-13.pdf -> ../../.git/annex/objects/9Q/X2/MD5E-s2796088--ed299ab7e1662cb03aa01299eed2602b.pdf/MD5E-s2796088--ed299ab7e1662cb03aa01299eed2602b.pdf
│ ├── anatomical_sub-14.pdf -> ../../.git/annex/objects/Wg/55/MD5E-s2558074--cadc9bd81856dcd02677de84e7e6ca90.pdf/MD5E-s2558074--cadc9bd81856dcd02677de84e7e6ca90.pdf
│ ├── anatomical_sub-15.pdf -> ../../.git/annex/objects/43/Q3/MD5E-s2847293--0c678a4b309d055ad9ba4ba25b77351b.pdf/MD5E-s2847293--0c678a4b309d055ad9ba4ba25b77351b.pdf
│ ├── anatomical_sub-16.pdf -> ../../.git/annex/objects/xq/qj/MD5E-s2890454--87c62253c1711f30d53c41b3ac38dc66.pdf/MD5E-s2890454--87c62253c1711f30d53c41b3ac38dc66.pdf
│ ├── anatomical_sub-17.pdf -> ../../.git/annex/objects/zK/M1/MD5E-s2825765--0a91015e22836a3076641b963e1ccfc6.pdf/MD5E-s2825765--0a91015e22836a3076641b963e1ccfc6.pdf
│ ├── anatomical_sub-18.pdf -> ../../.git/annex/objects/w2/Vk/MD5E-s2821624--1d9a3f0b21ce1f9a3b490d44d36f1f11.pdf/MD5E-s2821624--1d9a3f0b21ce1f9a3b490d44d36f1f11.pdf
│ ├── anatomical_sub-19.pdf -> ../../.git/annex/objects/J2/Jq/MD5E-s2453814--533411f3353cb3fa0264485e81f3fcf6.pdf/MD5E-s2453814--533411f3353cb3fa0264485e81f3fcf6.pdf
│ ├── anatomical_sub-20.pdf -> ../../.git/annex/objects/MF/9X/MD5E-s2881144--176c560778c55db87e8468b3246d373c.pdf/MD5E-s2881144--176c560778c55db87e8468b3246d373c.pdf
│ ├── anatomical_sub-21.pdf -> ../../.git/annex/objects/XQ/p1/MD5E-s2330589--b7546dfe5fb43a974cd23111b860c493.pdf/MD5E-s2330589--b7546dfe5fb43a974cd23111b860c493.pdf
│ ├── anatomical_sub-22.pdf -> ../../.git/annex/objects/Fx/k8/MD5E-s2505165--55f0661ad209b742c517cc5b5469436a.pdf/MD5E-s2505165--55f0661ad209b742c517cc5b5469436a.pdf
│ ├── anatomical_sub-23.pdf -> ../../.git/annex/objects/qj/8K/MD5E-s2784018--7e8697a7d4601547a899a27af132166d.pdf/MD5E-s2784018--7e8697a7d4601547a899a27af132166d.pdf
│ ├── anatomical_sub-24.pdf -> ../../.git/annex/objects/G8/Kw/MD5E-s2828817--e86be931adef2a7b0297d557d827d629.pdf/MD5E-s2828817--e86be931adef2a7b0297d557d827d629.pdf
│ ├── anatomical_sub-25.pdf -> ../../.git/annex/objects/XG/kg/MD5E-s2447908--3d392b9d27929dc4146d2b47be16e8dc.pdf/MD5E-s2447908--3d392b9d27929dc4146d2b47be16e8dc.pdf
│ ├── anatomical_sub-26.pdf -> ../../.git/annex/objects/8P/42/MD5E-s2850007--6d2f87a305b30d5704aaf4be9b8ff1e6.pdf/MD5E-s2850007--6d2f87a305b30d5704aaf4be9b8ff1e6.pdf
│ ├── fMRIQC.csv -> ../../.git/annex/objects/2Z/Ff/MD5E-s21038--cbe73db3db1beb0a1977583cff2a724b.csv/MD5E-s21038--cbe73db3db1beb0a1977583cff2a724b.csv
│ ├── functional_group.pdf -> ../../.git/annex/objects/Kq/xg/MD5E-s90712--7058c3db328fecb86303bc27a9ef0110.pdf/MD5E-s90712--7058c3db328fecb86303bc27a9ef0110.pdf
│ ├── functional_sub-01.pdf -> ../../.git/annex/objects/k2/vQ/MD5E-s1157925--e055f942b72b9aabad7a5e3d7b25b201.pdf/MD5E-s1157925--e055f942b72b9aabad7a5e3d7b25b201.pdf
│ ├── functional_sub-02.pdf -> ../../.git/annex/objects/X3/X6/MD5E-s1235840--cb32b7f8f1274af250b4f0fc15dacecb.pdf/MD5E-s1235840--cb32b7f8f1274af250b4f0fc15dacecb.pdf
│ ├── functional_sub-03.pdf -> ../../.git/annex/objects/Vp/0x/MD5E-s1228507--73ab1cc4cb27712892fcb10a0853ba7c.pdf/MD5E-s1228507--73ab1cc4cb27712892fcb10a0853ba7c.pdf
│ ├── functional_sub-04.pdf -> ../../.git/annex/objects/xk/jz/MD5E-s1252659--8ae6d1b02767c1ddb72dd7e6afefe696.pdf/MD5E-s1252659--8ae6d1b02767c1ddb72dd7e6afefe696.pdf
│ ├── functional_sub-05.pdf -> ../../.git/annex/objects/Zm/VJ/MD5E-s1258815--dd043691d548a501dd63d1aaf420e43c.pdf/MD5E-s1258815--dd043691d548a501dd63d1aaf420e43c.pdf
│ ├── functional_sub-06.pdf -> ../../.git/annex/objects/M5/gq/MD5E-s1247345--9c06bc69792b812ab8deffb01c6656c2.pdf/MD5E-s1247345--9c06bc69792b812ab8deffb01c6656c2.pdf
│ ├── functional_sub-07.pdf -> ../../.git/annex/objects/25/35/MD5E-s1229731--99cc64e99df0025ccb0341cd0dcf688b.pdf/MD5E-s1229731--99cc64e99df0025ccb0341cd0dcf688b.pdf
│ ├── functional_sub-08.pdf -> ../../.git/annex/objects/MX/vQ/MD5E-s1222308--e13c56f17109d3f142c9c4db60fea674.pdf/MD5E-s1222308--e13c56f17109d3f142c9c4db60fea674.pdf
│ ├── functional_sub-09.pdf -> ../../.git/annex/objects/90/0F/MD5E-s1265097--41a69211a0569413917ce3825eac95d6.pdf/MD5E-s1265097--41a69211a0569413917ce3825eac95d6.pdf
│ ├── functional_sub-10.pdf -> ../../.git/annex/objects/FZ/gq/MD5E-s1299358--12ccfc4a5f52b077b99481fe25aa8ef1.pdf/MD5E-s1299358--12ccfc4a5f52b077b99481fe25aa8ef1.pdf
│ ├── functional_sub-11.pdf -> ../../.git/annex/objects/MJ/mQ/MD5E-s1166014--3465ef6b18514d3cd361c0bffe2b73fc.pdf/MD5E-s1166014--3465ef6b18514d3cd361c0bffe2b73fc.pdf
│ ├── functional_sub-12.pdf -> ../../.git/annex/objects/xp/1f/MD5E-s1177325--6fe4937d5aa567fb5b3c3977362fc9af.pdf/MD5E-s1177325--6fe4937d5aa567fb5b3c3977362fc9af.pdf
│ ├── functional_sub-13.pdf -> ../../.git/annex/objects/4g/vW/MD5E-s1178873--96f341322d21e2bdeb709edc5b047df0.pdf/MD5E-s1178873--96f341322d21e2bdeb709edc5b047df0.pdf
│ ├── functional_sub-14.pdf -> ../../.git/annex/objects/5p/6X/MD5E-s1206987--729f64cf514c9103556c53ccb5430bc4.pdf/MD5E-s1206987--729f64cf514c9103556c53ccb5430bc4.pdf
│ ├── functional_sub-15.pdf -> ../../.git/annex/objects/m1/k9/MD5E-s1223617--9239a1c2d968ed18093b69d28fd9e654.pdf/MD5E-s1223617--9239a1c2d968ed18093b69d28fd9e654.pdf
│ ├── functional_sub-16.pdf -> ../../.git/annex/objects/jq/wP/MD5E-s1294856--5eb7ec97924a22c7e68fd95373694e7e.pdf/MD5E-s1294856--5eb7ec97924a22c7e68fd95373694e7e.pdf
│ ├── functional_sub-17.pdf -> ../../.git/annex/objects/0m/3Q/MD5E-s1238563--129db424a50b7889278024828c08c736.pdf/MD5E-s1238563--129db424a50b7889278024828c08c736.pdf
│ ├── functional_sub-18.pdf -> ../../.git/annex/objects/VF/Fm/MD5E-s1197868--3b23e8d53b11d98d49b1adf62ff559df.pdf/MD5E-s1197868--3b23e8d53b11d98d49b1adf62ff559df.pdf
│ ├── functional_sub-19.pdf -> ../../.git/annex/objects/Jj/m8/MD5E-s1164028--aea7dfa78e9be2e83a9b313f2ebdc4bd.pdf/MD5E-s1164028--aea7dfa78e9be2e83a9b313f2ebdc4bd.pdf
│ ├── functional_sub-20.pdf -> ../../.git/annex/objects/x1/ZQ/MD5E-s1292308--8869b1b640797a2be2aa03be69b89840.pdf/MD5E-s1292308--8869b1b640797a2be2aa03be69b89840.pdf
│ ├── functional_sub-21.pdf -> ../../.git/annex/objects/MG/zW/MD5E-s1216271--3d5c5ca0f8f4ba06b3289e197a40defd.pdf/MD5E-s1216271--3d5c5ca0f8f4ba06b3289e197a40defd.pdf
│ ├── functional_sub-22.pdf -> ../../.git/annex/objects/5m/pj/MD5E-s1142289--4f9e57d8bfe3d39881c43b959189d69f.pdf/MD5E-s1142289--4f9e57d8bfe3d39881c43b959189d69f.pdf
│ ├── functional_sub-23.pdf -> ../../.git/annex/objects/m7/Z2/MD5E-s1233046--7758914aecbf2b5d01cd0825952609be.pdf/MD5E-s1233046--7758914aecbf2b5d01cd0825952609be.pdf
│ ├── functional_sub-24.pdf -> ../../.git/annex/objects/mz/5m/MD5E-s1265224--c188bd88fc1c99308389f528ea4df71e.pdf/MD5E-s1265224--c188bd88fc1c99308389f528ea4df71e.pdf
│ ├── functional_sub-25.pdf -> ../../.git/annex/objects/Mk/G6/MD5E-s1260984--1b16abcbbf55ccc7763f1d704d76628f.pdf/MD5E-s1260984--1b16abcbbf55ccc7763f1d704d76628f.pdf
│ └── functional_sub-26.pdf -> ../../.git/annex/objects/1m/zq/MD5E-s1285726--6838f727d5c4b5593a7b5e0e6b20483a.pdf/MD5E-s1285726--6838f727d5c4b5593a7b5e0e6b20483a.pdf
├── participants.tsv
├── sub-01
│ ├── anat
│ │ └── sub-01_T1w.nii.gz -> ../../.git/annex/objects/Pf/6k/MD5E-s10581116--757e697a01eeea5c97a7d6fbc7153373.nii.gz/MD5E-s10581116--757e697a01eeea5c97a7d6fbc7153373.nii.gz
│ └── func
│ ├── sub-01_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/5m/w9/MD5E-s28061534--8e8c44ff53f9b5d46f2caae5916fa4ef.nii.gz/MD5E-s28061534--8e8c44ff53f9b5d46f2caae5916fa4ef.nii.gz
│ ├── sub-01_task-flanker_run-1_events.tsv
│ ├── sub-01_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/2F/58/MD5E-s28143286--f0bcf782c3688e2cf7149b4665949484.nii.gz/MD5E-s28143286--f0bcf782c3688e2cf7149b4665949484.nii.gz
│ └── sub-01_task-flanker_run-2_events.tsv
├── sub-02
│ ├── anat
│ │ └── sub-02_T1w.nii.gz -> ../../.git/annex/objects/3m/FF/MD5E-s10737123--cbd4181ee26559e8ec0a441fa2f834a7.nii.gz/MD5E-s10737123--cbd4181ee26559e8ec0a441fa2f834a7.nii.gz
│ └── func
│ ├── sub-02_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/8v/2j/MD5E-s29188378--80050f0deb13562c24f2fc23f8d095bd.nii.gz/MD5E-s29188378--80050f0deb13562c24f2fc23f8d095bd.nii.gz
│ ├── sub-02_task-flanker_run-1_events.tsv
│ ├── sub-02_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/fM/Kw/MD5E-s29193540--cc013f2d7d148b448edca8aada349d02.nii.gz/MD5E-s29193540--cc013f2d7d148b448edca8aada349d02.nii.gz
│ └── sub-02_task-flanker_run-2_events.tsv
├── sub-03
│ ├── anat
│ │ └── sub-03_T1w.nii.gz -> ../../.git/annex/objects/7W/9z/MD5E-s10707026--8f1858934cc7c7457e3a4a71cc2131fc.nii.gz/MD5E-s10707026--8f1858934cc7c7457e3a4a71cc2131fc.nii.gz
│ └── func
│ ├── sub-03_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/q6/kF/MD5E-s28755729--b19466702eee6b9385bd6e19e362f94c.nii.gz/MD5E-s28755729--b19466702eee6b9385bd6e19e362f94c.nii.gz
│ ├── sub-03_task-flanker_run-1_events.tsv
│ ├── sub-03_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/zV/K1/MD5E-s28782544--8d9700a435d08c90f0c1d534efdc8b69.nii.gz/MD5E-s28782544--8d9700a435d08c90f0c1d534efdc8b69.nii.gz
│ └── sub-03_task-flanker_run-2_events.tsv
├── sub-04
│ ├── anat
│ │ └── sub-04_T1w.nii.gz -> ../../.git/annex/objects/FW/14/MD5E-s10738444--2a9a2ba4ea7d2324c84bf5a2882f196c.nii.gz/MD5E-s10738444--2a9a2ba4ea7d2324c84bf5a2882f196c.nii.gz
│ └── func
│ ├── sub-04_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/9Z/0Q/MD5E-s29062799--27171406951ea275cb5857ea0dc32345.nii.gz/MD5E-s29062799--27171406951ea275cb5857ea0dc32345.nii.gz
│ ├── sub-04_task-flanker_run-1_events.tsv
│ ├── sub-04_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/FW/FZ/MD5E-s29071279--f89b61fe3ebab26df1374f2564bd95c2.nii.gz/MD5E-s29071279--f89b61fe3ebab26df1374f2564bd95c2.nii.gz
│ └── sub-04_task-flanker_run-2_events.tsv
├── sub-05
│ ├── anat
│ │ └── sub-05_T1w.nii.gz -> ../../.git/annex/objects/k2/Kj/MD5E-s10753867--c4b5788da5f4c627f0f5862da5f46c35.nii.gz/MD5E-s10753867--c4b5788da5f4c627f0f5862da5f46c35.nii.gz
│ └── func
│ ├── sub-05_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/VZ/z5/MD5E-s29667270--0ce9ac78b6aa9a77fc94c655a6ff5a06.nii.gz/MD5E-s29667270--0ce9ac78b6aa9a77fc94c655a6ff5a06.nii.gz
│ ├── sub-05_task-flanker_run-1_events.tsv
│ ├── sub-05_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/z7/MP/MD5E-s29660544--752750dabb21e2cf28e87d1d550a71b9.nii.gz/MD5E-s29660544--752750dabb21e2cf28e87d1d550a71b9.nii.gz
│ └── sub-05_task-flanker_run-2_events.tsv
├── sub-06
│ ├── anat
│ │ └── sub-06_T1w.nii.gz -> ../../.git/annex/objects/5w/G0/MD5E-s10620585--1132eab3830fe59b8a10b6582bb49004.nii.gz/MD5E-s10620585--1132eab3830fe59b8a10b6582bb49004.nii.gz
│ └── func
│ ├── sub-06_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/3x/qj/MD5E-s29386982--e671c0c647ce7d0d4596e35b702ee970.nii.gz/MD5E-s29386982--e671c0c647ce7d0d4596e35b702ee970.nii.gz
│ ├── sub-06_task-flanker_run-1_events.tsv
│ ├── sub-06_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/9j/6P/MD5E-s29379265--e513a2746d2b5c603f96044cf48c557c.nii.gz/MD5E-s29379265--e513a2746d2b5c603f96044cf48c557c.nii.gz
│ └── sub-06_task-flanker_run-2_events.tsv
├── sub-07
│ ├── anat
│ │ └── sub-07_T1w.nii.gz -> ../../.git/annex/objects/08/fF/MD5E-s10718092--38481fbc489dfb1ec4b174b57591a074.nii.gz/MD5E-s10718092--38481fbc489dfb1ec4b174b57591a074.nii.gz
│ └── func
│ ├── sub-07_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/z1/7W/MD5E-s28946009--5baf7a314874b280543fc0f91f2731af.nii.gz/MD5E-s28946009--5baf7a314874b280543fc0f91f2731af.nii.gz
│ ├── sub-07_task-flanker_run-1_events.tsv
│ ├── sub-07_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Jf/W7/MD5E-s28960603--682e13963bfc49cc6ae05e9ba5c62619.nii.gz/MD5E-s28960603--682e13963bfc49cc6ae05e9ba5c62619.nii.gz
│ └── sub-07_task-flanker_run-2_events.tsv
├── sub-08
│ ├── anat
│ │ └── sub-08_T1w.nii.gz -> ../../.git/annex/objects/mw/MM/MD5E-s10561256--b94dddd8dc1c146aa8cd97f8d9994146.nii.gz/MD5E-s10561256--b94dddd8dc1c146aa8cd97f8d9994146.nii.gz
│ └── func
│ ├── sub-08_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/zX/v9/MD5E-s28641609--47314e6d1a14b8545686110b5b67f8b8.nii.gz/MD5E-s28641609--47314e6d1a14b8545686110b5b67f8b8.nii.gz
│ ├── sub-08_task-flanker_run-1_events.tsv
│ ├── sub-08_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/WZ/F0/MD5E-s28636310--4535bf26281e1c5556ad0d3468e7fe4e.nii.gz/MD5E-s28636310--4535bf26281e1c5556ad0d3468e7fe4e.nii.gz
│ └── sub-08_task-flanker_run-2_events.tsv
├── sub-09
│ ├── anat
│ │ └── sub-09_T1w.nii.gz -> ../../.git/annex/objects/QJ/ZZ/MD5E-s10775967--e6a18e64bc0a6b17254a9564cf9b8f82.nii.gz/MD5E-s10775967--e6a18e64bc0a6b17254a9564cf9b8f82.nii.gz
│ └── func
│ ├── sub-09_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/k9/1X/MD5E-s29200533--59e86a903e0ab3d1d320c794ba1f0777.nii.gz/MD5E-s29200533--59e86a903e0ab3d1d320c794ba1f0777.nii.gz
│ ├── sub-09_task-flanker_run-1_events.tsv
│ ├── sub-09_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/W3/94/MD5E-s29223017--7f3fb9e260d3bd28e29b0b586ce4c344.nii.gz/MD5E-s29223017--7f3fb9e260d3bd28e29b0b586ce4c344.nii.gz
│ └── sub-09_task-flanker_run-2_events.tsv
├── sub-10
│ ├── anat
│ │ └── sub-10_T1w.nii.gz -> ../../.git/annex/objects/5F/3f/MD5E-s10750712--bde2309077bffe22cb65e42ebdce5bfa.nii.gz/MD5E-s10750712--bde2309077bffe22cb65e42ebdce5bfa.nii.gz
│ └── func
│ ├── sub-10_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/3p/qp/MD5E-s29732696--339715d5cec387f4d44dfe94f304a429.nii.gz/MD5E-s29732696--339715d5cec387f4d44dfe94f304a429.nii.gz
│ ├── sub-10_task-flanker_run-1_events.tsv
│ ├── sub-10_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/11/Zx/MD5E-s29724034--16f2bf452524a315182f188becc1866d.nii.gz/MD5E-s29724034--16f2bf452524a315182f188becc1866d.nii.gz
│ └── sub-10_task-flanker_run-2_events.tsv
├── sub-11
│ ├── anat
│ │ └── sub-11_T1w.nii.gz -> ../../.git/annex/objects/kj/xX/MD5E-s10534963--9e5bff7ec0b5df2850e1d05b1af281ba.nii.gz/MD5E-s10534963--9e5bff7ec0b5df2850e1d05b1af281ba.nii.gz
│ └── func
│ ├── sub-11_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/35/fk/MD5E-s28226875--d5012074c2c7a0a394861b010bcf9a8f.nii.gz/MD5E-s28226875--d5012074c2c7a0a394861b010bcf9a8f.nii.gz
│ ├── sub-11_task-flanker_run-1_events.tsv
│ ├── sub-11_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/j7/ff/MD5E-s28198976--c0a64e3b549568c44bb40b1588027c9a.nii.gz/MD5E-s28198976--c0a64e3b549568c44bb40b1588027c9a.nii.gz
│ └── sub-11_task-flanker_run-2_events.tsv
├── sub-12
│ ├── anat
│ │ └── sub-12_T1w.nii.gz -> ../../.git/annex/objects/kx/2F/MD5E-s10550168--a7f651adc817b6678148b575654532a4.nii.gz/MD5E-s10550168--a7f651adc817b6678148b575654532a4.nii.gz
│ └── func
│ ├── sub-12_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/M0/fX/MD5E-s28403807--f1c3eb2e519020f4315a696ea845fc01.nii.gz/MD5E-s28403807--f1c3eb2e519020f4315a696ea845fc01.nii.gz
│ ├── sub-12_task-flanker_run-1_events.tsv
│ ├── sub-12_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/vW/V0/MD5E-s28424992--8740628349be3c056a0411bf4a852b25.nii.gz/MD5E-s28424992--8740628349be3c056a0411bf4a852b25.nii.gz
│ └── sub-12_task-flanker_run-2_events.tsv
├── sub-13
│ ├── anat
│ │ └── sub-13_T1w.nii.gz -> ../../.git/annex/objects/wM/Xw/MD5E-s10609761--440413c3251d182086105649164222c6.nii.gz/MD5E-s10609761--440413c3251d182086105649164222c6.nii.gz
│ └── func
│ ├── sub-13_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/mf/M4/MD5E-s28180916--aa35f4ad0cf630d6396a8a2dd1f3dda6.nii.gz/MD5E-s28180916--aa35f4ad0cf630d6396a8a2dd1f3dda6.nii.gz
│ ├── sub-13_task-flanker_run-1_events.tsv
│ ├── sub-13_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/XP/76/MD5E-s28202786--8caf1ac548c87b2b35f85e8ae2bf72c1.nii.gz/MD5E-s28202786--8caf1ac548c87b2b35f85e8ae2bf72c1.nii.gz
│ └── sub-13_task-flanker_run-2_events.tsv
├── sub-14
│ ├── anat
│ │ └── sub-14_T1w.nii.gz -> ../../.git/annex/objects/Zw/0z/MD5E-s9223596--33abfb5da565f3487e3a7aebc15f940c.nii.gz/MD5E-s9223596--33abfb5da565f3487e3a7aebc15f940c.nii.gz
│ └── func
│ ├── sub-14_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Jp/29/MD5E-s29001492--250f1e4daa9be1d95e06af0d56629cc9.nii.gz/MD5E-s29001492--250f1e4daa9be1d95e06af0d56629cc9.nii.gz
│ ├── sub-14_task-flanker_run-1_events.tsv
│ ├── sub-14_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/PK/V2/MD5E-s29068193--5621a3b0af8132c509420b4ad9aaf8fb.nii.gz/MD5E-s29068193--5621a3b0af8132c509420b4ad9aaf8fb.nii.gz
│ └── sub-14_task-flanker_run-2_events.tsv
├── sub-15
│ ├── anat
│ │ └── sub-15_T1w.nii.gz -> ../../.git/annex/objects/Mz/qq/MD5E-s10752891--ddd2622f115ec0d29a0c7ab2366f6f95.nii.gz/MD5E-s10752891--ddd2622f115ec0d29a0c7ab2366f6f95.nii.gz
│ └── func
│ ├── sub-15_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/08/JJ/MD5E-s28285239--feda22c4526af1910fcee58d4c42f07e.nii.gz/MD5E-s28285239--feda22c4526af1910fcee58d4c42f07e.nii.gz
│ ├── sub-15_task-flanker_run-1_events.tsv
│ ├── sub-15_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/9f/0W/MD5E-s28289760--433000a1def662e72d8433dba151c61b.nii.gz/MD5E-s28289760--433000a1def662e72d8433dba151c61b.nii.gz
│ └── sub-15_task-flanker_run-2_events.tsv
├── sub-16
│ ├── anat
│ │ └── sub-16_T1w.nii.gz -> ../../.git/annex/objects/4g/8k/MD5E-s10927450--a196f7075c793328dd6ff3cebf36ea6b.nii.gz/MD5E-s10927450--a196f7075c793328dd6ff3cebf36ea6b.nii.gz
│ └── func
│ ├── sub-16_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/9z/g2/MD5E-s29757991--1a1648b2fa6cc74e31c94f109d8137ba.nii.gz/MD5E-s29757991--1a1648b2fa6cc74e31c94f109d8137ba.nii.gz
│ ├── sub-16_task-flanker_run-1_events.tsv
│ ├── sub-16_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/k8/4F/MD5E-s29773832--fe08739ea816254395b985ee704aaa99.nii.gz/MD5E-s29773832--fe08739ea816254395b985ee704aaa99.nii.gz
│ └── sub-16_task-flanker_run-2_events.tsv
├── sub-17
│ ├── anat
│ │ └── sub-17_T1w.nii.gz -> ../../.git/annex/objects/jQ/MQ/MD5E-s10826014--8e2a6b062df4d1c4327802f2b905ef36.nii.gz/MD5E-s10826014--8e2a6b062df4d1c4327802f2b905ef36.nii.gz
│ └── func
│ ├── sub-17_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Wz/2P/MD5E-s28991563--9845f461a017a39d1f6e18baaa0c9c41.nii.gz/MD5E-s28991563--9845f461a017a39d1f6e18baaa0c9c41.nii.gz
│ ├── sub-17_task-flanker_run-1_events.tsv
│ ├── sub-17_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/jF/3m/MD5E-s29057821--84ccc041163bcc5b3a9443951e2a5a78.nii.gz/MD5E-s29057821--84ccc041163bcc5b3a9443951e2a5a78.nii.gz
│ └── sub-17_task-flanker_run-2_events.tsv
├── sub-18
│ ├── anat
│ │ └── sub-18_T1w.nii.gz -> ../../.git/annex/objects/3v/pK/MD5E-s10571510--6fc4b5792bc50ea4d14eb5247676fafe.nii.gz/MD5E-s10571510--6fc4b5792bc50ea4d14eb5247676fafe.nii.gz
│ └── func
│ ├── sub-18_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/94/P2/MD5E-s28185776--5b3879ec6fc4bbe1e48efc64984f88cf.nii.gz/MD5E-s28185776--5b3879ec6fc4bbe1e48efc64984f88cf.nii.gz
│ ├── sub-18_task-flanker_run-1_events.tsv
│ ├── sub-18_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/qp/6K/MD5E-s28234699--58019d798a133e5d7806569374dd8160.nii.gz/MD5E-s28234699--58019d798a133e5d7806569374dd8160.nii.gz
│ └── sub-18_task-flanker_run-2_events.tsv
├── sub-19
│ ├── anat
│ │ └── sub-19_T1w.nii.gz -> ../../.git/annex/objects/Zw/p8/MD5E-s8861893--d338005753d8af3f3d7bd8dc293e2a97.nii.gz/MD5E-s8861893--d338005753d8af3f3d7bd8dc293e2a97.nii.gz
│ └── func
│ ├── sub-19_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/04/k6/MD5E-s28178448--3874e748258cf19aa69a05a7c37ad137.nii.gz/MD5E-s28178448--3874e748258cf19aa69a05a7c37ad137.nii.gz
│ ├── sub-19_task-flanker_run-1_events.tsv
│ ├── sub-19_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/mz/P4/MD5E-s28190932--91e6b3e4318ca28f01de8cb967cf8421.nii.gz/MD5E-s28190932--91e6b3e4318ca28f01de8cb967cf8421.nii.gz
│ └── sub-19_task-flanker_run-2_events.tsv
├── sub-20
│ ├── anat
│ │ └── sub-20_T1w.nii.gz -> ../../.git/annex/objects/g1/FF/MD5E-s11025608--5929806a7aa5720fc755687e1450b06c.nii.gz/MD5E-s11025608--5929806a7aa5720fc755687e1450b06c.nii.gz
│ └── func
│ ├── sub-20_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/v5/ZJ/MD5E-s29931631--bf9abb057367ce66961f0b7913e8e707.nii.gz/MD5E-s29931631--bf9abb057367ce66961f0b7913e8e707.nii.gz
│ ├── sub-20_task-flanker_run-1_events.tsv
│ ├── sub-20_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/J3/KW/MD5E-s29945590--96cfd5b77cd096f6c6a3530015fea32d.nii.gz/MD5E-s29945590--96cfd5b77cd096f6c6a3530015fea32d.nii.gz
│ └── sub-20_task-flanker_run-2_events.tsv
├── sub-21
│ ├── anat
│ │ └── sub-21_T1w.nii.gz -> ../../.git/annex/objects/K6/6K/MD5E-s8662805--77b262ddd929fa08d78591bfbe558ac6.nii.gz/MD5E-s8662805--77b262ddd929fa08d78591bfbe558ac6.nii.gz
│ └── func
│ ├── sub-21_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Wz/p9/MD5E-s28756041--9ae556d4e3042532d25af5dc4ab31840.nii.gz/MD5E-s28756041--9ae556d4e3042532d25af5dc4ab31840.nii.gz
│ ├── sub-21_task-flanker_run-1_events.tsv
│ ├── sub-21_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/xF/M3/MD5E-s28758438--81866411fc6b6333ec382a20ff0be718.nii.gz/MD5E-s28758438--81866411fc6b6333ec382a20ff0be718.nii.gz
│ └── sub-21_task-flanker_run-2_events.tsv
├── sub-22
│ ├── anat
│ │ └── sub-22_T1w.nii.gz -> ../../.git/annex/objects/JG/ZV/MD5E-s9282392--9e7296a6a5b68df46b77836182b6681a.nii.gz/MD5E-s9282392--9e7296a6a5b68df46b77836182b6681a.nii.gz
│ └── func
│ ├── sub-22_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/qW/Gw/MD5E-s28002098--c6bea10177a38667ceea3261a642b3c6.nii.gz/MD5E-s28002098--c6bea10177a38667ceea3261a642b3c6.nii.gz
│ ├── sub-22_task-flanker_run-1_events.tsv
│ ├── sub-22_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/VX/Zj/MD5E-s28027568--b34d0df9ad62485aba25296939429885.nii.gz/MD5E-s28027568--b34d0df9ad62485aba25296939429885.nii.gz
│ └── sub-22_task-flanker_run-2_events.tsv
├── sub-23
│ ├── anat
│ │ └── sub-23_T1w.nii.gz -> ../../.git/annex/objects/4Z/4x/MD5E-s10626062--db5a6ba6730b319c6425f2e847ce9b14.nii.gz/MD5E-s10626062--db5a6ba6730b319c6425f2e847ce9b14.nii.gz
│ └── func
│ ├── sub-23_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/VK/8F/MD5E-s28965005--4a9a96d9322563510ca14439e7fd6cea.nii.gz/MD5E-s28965005--4a9a96d9322563510ca14439e7fd6cea.nii.gz
│ ├── sub-23_task-flanker_run-1_events.tsv
│ ├── sub-23_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/56/20/MD5E-s29050413--753b0d2c23c4af6592501219c2e2c6bd.nii.gz/MD5E-s29050413--753b0d2c23c4af6592501219c2e2c6bd.nii.gz
│ └── sub-23_task-flanker_run-2_events.tsv
├── sub-24
│ ├── anat
│ │ └── sub-24_T1w.nii.gz -> ../../.git/annex/objects/jQ/fV/MD5E-s10739691--458f0046eff18ee8c43456637766a819.nii.gz/MD5E-s10739691--458f0046eff18ee8c43456637766a819.nii.gz
│ └── func
│ ├── sub-24_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/km/fV/MD5E-s29354610--29ebfa60e52d49f7dac6814cb5fdc2bc.nii.gz/MD5E-s29354610--29ebfa60e52d49f7dac6814cb5fdc2bc.nii.gz
│ ├── sub-24_task-flanker_run-1_events.tsv
│ ├── sub-24_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Wj/KK/MD5E-s29423307--fedaa1d7c6e34420735bb3bbe5a2fe38.nii.gz/MD5E-s29423307--fedaa1d7c6e34420735bb3bbe5a2fe38.nii.gz
│ └── sub-24_task-flanker_run-2_events.tsv
├── sub-25
│ ├── anat
│ │ └── sub-25_T1w.nii.gz -> ../../.git/annex/objects/Gk/FQ/MD5E-s8998578--f560d832f13e757b485c16d570bf6ebc.nii.gz/MD5E-s8998578--f560d832f13e757b485c16d570bf6ebc.nii.gz
│ └── func
│ ├── sub-25_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/XW/1v/MD5E-s29473003--49b04e7e4b450ec5ef93ff02d4158775.nii.gz/MD5E-s29473003--49b04e7e4b450ec5ef93ff02d4158775.nii.gz
│ ├── sub-25_task-flanker_run-1_events.tsv
│ ├── sub-25_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Qm/M7/MD5E-s29460132--b0e9039e9f33510631f229c8c2193285.nii.gz/MD5E-s29460132--b0e9039e9f33510631f229c8c2193285.nii.gz
│ └── sub-25_task-flanker_run-2_events.tsv
├── sub-26
│ ├── anat
│ │ └── sub-26_T1w.nii.gz -> ../../.git/annex/objects/kf/9F/MD5E-s10850250--5f103b2660f488e4afa193f9307c1291.nii.gz/MD5E-s10850250--5f103b2660f488e4afa193f9307c1291.nii.gz
│ └── func
│ ├── sub-26_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/QV/10/MD5E-s30127491--8e30aa4bbfcc461bac8598bf621283c5.nii.gz/MD5E-s30127491--8e30aa4bbfcc461bac8598bf621283c5.nii.gz
│ ├── sub-26_task-flanker_run-1_events.tsv
│ ├── sub-26_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/3G/Q6/MD5E-s30162480--80fd132e7cb1600ab248249e78f6f1aa.nii.gz/MD5E-s30162480--80fd132e7cb1600ab248249e78f6f1aa.nii.gz
│ └── sub-26_task-flanker_run-2_events.tsv
└── task-flanker_bold.json
80 directories, 192 files
Creating Timing Files#
Condition-specific timing files are generated using the same approach described in the Preprocessing and GLM notebook. In brief:
Onset times and durations are extracted from each subject’s
events.tsvfile.Trials are categorized into congruent and incongruent conditions.
The
make_Timings.shscript (from Andy’s AFNI_Scripts repository) is used to convert this event information into AFNI-compatible.1Dtiming files.For each subject, two timing files are created (one per condition), spanning both runs, and saved in the subject’s
func/directory.
These .1D timing files will then be used in the first-level GLM to model condition-specific brain responses.
!wget -O ds000102/make_Timings.sh https://raw.githubusercontent.com/andrewjahn/AFNI_Scripts/master/make_Timings.sh
--2025-11-04 04:55:45-- https://raw.githubusercontent.com/andrewjahn/AFNI_Scripts/master/make_Timings.sh
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.109.133, 185.199.108.133, 185.199.111.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.109.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 953 [text/plain]
Saving to: ‘ds000102/make_Timings.sh’
ds000102/make_Timin 100%[===================>] 953 --.-KB/s in 0s
2025-11-04 04:55:45 (63.2 MB/s) - ‘ds000102/make_Timings.sh’ saved [953/953]
Running the Timing File Script#
After placing the make_Timings.sh script into the ds000102/ directory, we can execute it from within the notebook:
!cd ds000102 && chmod +x make_Timings.sh && bash make_Timings.sh
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
You may see a SyntaxWarning from AFNI’s internal Python scripts when running this command - this can be safely ignored and does not affect the output.
Check the output:
!cat ds000102/sub-01/func/incongruent.1D
0 10 40 76 102 150 164 174 208 220 232 260
0 54 64 76 88 130 144 154 196 246 274
2. Running Preprocessing and First-Level Analysis for All Subjects#
All of the preprocessing and regression steps for subject sub-08 were introduced and explained in the example notebooks Preprocessing with AFNI and AFNI Preprocessing and GLM, both of which are highly inspired by Andy’s Brain Book’s tutorial.
These notebooks demonstrate how to use afni_proc.py to generate an automated pipeline and interpret each preprocessing and regression block.
➡️ setup ➡️ tcat ➡️ align ➡️ volreg ➡️ tlrc ➡️ blur ➡️ mask ➡️ scale ➡️ regress 🧠
✅ Outputs: fitted time series, beta weights, and statistical maps from the GLM.
To replicate the preprocessing and GLM steps used for sub-08 across multiple participants, we loop over all 9 subjects (sub-01 to sub-09) and generate an individualized AFNI pipeline for each one using afni_proc.py. This automated approach performs several key operations:
Loads each subject’s anatomical image and both functional runs.
Specifies the full processing pipeline: alignment, linear registration to MNI space, motion correction, blurring, masking, scaling, and regression modeling.
Models two stimulus conditions (congruent and incongruent) using a gamma basis function.
Specifies two general linear tests (GLTs): incongruent - congruent and congruent - incongruent.
Censors time points with excessive motion (> 0.3 mm), disables motion derivatives, and estimates spatial smoothness for later use in group analysis.
Runs
3dREMLfitfor improved model estimation and produces a Pythonic HTML review report for each subject.
To improve alignment robustness, the script is post-edited to insert the -init_xform AUTO_CENTER option into the @auto_tlrc command as this works better on this dataset.
Processing is executed in parallel using a process pool, with up to 3 subjects processed simultaneously. Each subject’s script is saved and executed in a controlled logging environment, which captures terminal output into a log file for transparency and troubleshooting.
This block ensures consistent preprocessing and first-level GLM estimation across all subjects in the dataset.
# Create directory for output
os.makedirs("afni_pro_glm", exist_ok=True)
# Main subject processing function
def process_subject(subj_id):
subj = f"sub-{subj_id:02d}"
anat = f"./ds000102/{subj}/anat/{subj}_T1w.nii.gz"
func1 = f"./ds000102/{subj}/func/{subj}_task-flanker_run-1_bold.nii.gz"
func2 = f"./ds000102/{subj}/func/{subj}_task-flanker_run-2_bold.nii.gz"
stim1D_c = f"./ds000102/{subj}/func/congruent.1D"
stim1D_i = f"./ds000102/{subj}/func/incongruent.1D"
out_dir = f"./afni_pro_glm/{subj}.results"
script_name = f"./afni_pro_glm/proc.{subj}.tcsh"
log_file = f"./afni_pro_glm/output.{subj}.log"
# Skip subject if already processed
if os.path.exists(out_dir):
return f"{subj} skipped (already processed)"
cmd = [
"afni_proc.py",
"-subj_id", subj,
"-script", script_name,
"-scr_overwrite",
"-out_dir", out_dir,
"-blocks", "align", "tlrc", "volreg", "blur", "mask", "scale", "regress",
"-copy_anat", anat,
"-dsets", func1, func2,
"-tcat_remove_first_trs", "0",
"-align_opts_aea", "-giant_move",
"-tlrc_base", "MNI_avg152T1+tlrc",
"-volreg_align_to", "MIN_OUTLIER",
"-volreg_align_e2a",
"-volreg_tlrc_warp",
"-blur_size", "4.0",
"-regress_stim_times", stim1D_c, stim1D_i,
"-regress_stim_labels", "congruent", "incongruent",
"-regress_stim_types", "times", "times",
"-regress_basis", "GAM",
"-regress_opts_3dD",
"-gltsym", "SYM: +incongruent -congruent", "-glt_label", "1", "incongruent-congruent",
"-gltsym", "SYM: +congruent -incongruent", "-glt_label", "2", "congruent-incongruent",
"-jobs", "8", # speedup with multithreading
"-GOFORIT", "0", # safe default
"-regress_no_motion_deriv", # disables motion derivatives (simpler model)
"-regress_censor_motion", "0.3", # standard threshold
"-regress_motion_per_run", # run-wise motion parameters
"-regress_est_blur_epits",
"-regress_est_blur_errts",
"-regress_make_ideal_sum", "sum_ideal.1D",
"-regress_reml_exec",
"-regress_run_clustsim", "no", # skip, since you'll do group-level stats separately
"-html_review_style", "pythonic"
]
try:
# Step 1: Create the AFNI processing script
subprocess.run(cmd, check=True)
# Step 2: Insert -init_xform AUTO_CENTER into the @auto_tlrc command
with open(script_name, 'r') as f:
lines = f.readlines()
for i, line in enumerate(lines):
if line.strip().startswith("@auto_tlrc") and "AUTO_CENTER" not in line:
lines[i] = line.strip() + " -init_xform AUTO_CENTER\n"
with open(script_name, 'w') as f:
f.writelines(lines)
# Step 3: Run the AFNI tcsh script, logging output
with open(log_file, "w") as logfile:
proc = subprocess.Popen(
["tcsh", "-xef", script_name],
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
text=True
)
for line in proc.stdout:
print(line, end="") # Stream to console
logfile.write(line) # Log to file
proc.wait()
return f"{subj} {'done' if proc.returncode == 0 else 'failed'}"
except subprocess.CalledProcessError as e:
return f"{subj} failed: afni_proc.py error"
except Exception as e:
return f"{subj} failed: unexpected error\n{e}"
# ──────────────────────────────────────────────────────────────────────────────
# Main loop to process multiple subjects in parallel
if __name__ == "__main__":
subject_ids = list(range(1, 10)) # sub-01 to sub-09
results = []
with ProcessPoolExecutor(max_workers=3) as executor:
futures = {executor.submit(process_subject, sid): sid for sid in subject_ids}
for future in tqdm(as_completed(futures), total=len(futures), desc="Subjects processed"):
results.append(future.result())
print("\nSummary of results:")
for r in results:
print(r)
echo auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
echo auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
echo auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
echo (version 7.16, May 19, 2021)
auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
(version 7.16, May 19, 2021)
auto-generated by afni_proc.py, Tue Nov 4 04:56:01 2025
echo execution started: `date`
echo (version 7.16, May 19, 2021)
echo (version 7.16, May 19, 2021)
date
(version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
echo execution started: `date`
execution started: Tue Nov 4 04:56:01 UTC 2025
date
date
execution started: Tue Nov 4 04:56:01 UTC 2025
afni -ver
execution started: Tue Nov 4 04:56:01 UTC 2025
afni -ver
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-03_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-03.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-03.tcsh |& tee ./afni_pro_glm/output.proc.sub-03.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-03.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-03.tcsh
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-02_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-02.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-02.tcsh |& tee ./afni_pro_glm/output.proc.sub-02.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-02.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-02.tcsh
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-01_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-01.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-01.tcsh |& tee ./afni_pro_glm/output.proc.sub-01.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-01.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-01.tcsh
-- is current: afni_history as new as: 27 Jun 2019
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
most recent entry is: 30 Jun 2021
if ( 0 ) then
-- is current: afni_history as new as: 27 Jun 2019
if ( 0 > 0 ) then
most recent entry is: 30 Jun 2021
if ( 0 ) then
set subj = sub-03
if ( 0 > 0 ) then
endif
set subj = sub-01
if ( 0 ) then
set output_dir = ./afni_pro_glm/sub-03.results
endif
if ( 0 > 0 ) then
if ( -d ./afni_pro_glm/sub-03.results ) then
set output_dir = ./afni_pro_glm/sub-01.results
set subj = sub-02
set runs = ( `count -digits 2 1 2` )
endif
if ( -d ./afni_pro_glm/sub-01.results ) then
count -digits 2 1 2
set runs = ( `count -digits 2 1 2` )
set output_dir = ./afni_pro_glm/sub-02.results
count -digits 2 1 2
if ( -d ./afni_pro_glm/sub-02.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_pro_glm/sub-01.results
mkdir -p ./afni_pro_glm/sub-03.results
mkdir ./afni_pro_glm/sub-01.results/stimuli
mkdir ./afni_pro_glm/sub-03.results/stimuli
cp ./ds000102/sub-01/func/congruent.1D ./ds000102/sub-01/func/incongruent.1D ./afni_pro_glm/sub-01.results/stimuli
cp ./ds000102/sub-03/func/congruent.1D ./ds000102/sub-03/func/incongruent.1D ./afni_pro_glm/sub-03.results/stimuli
3dcopy ds000102/sub-01/anat/sub-01_T1w.nii.gz ./afni_pro_glm/sub-01.results/sub-01_T1w
3dcopy ds000102/sub-03/anat/sub-03_T1w.nii.gz ./afni_pro_glm/sub-03.results/sub-03_T1w
mkdir -p ./afni_pro_glm/sub-02.results
mkdir ./afni_pro_glm/sub-02.results/stimuli
cp ./ds000102/sub-02/func/congruent.1D ./ds000102/sub-02/func/incongruent.1D ./afni_pro_glm/sub-02.results/stimuli
3dcopy ds000102/sub-02/anat/sub-02_T1w.nii.gz ./afni_pro_glm/sub-02.results/sub-02_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_pro_glm/sub-02.results/pb00.sub-02.r01.tcat ds000102/sub-02/func/sub-02_task-flanker_run-1_bold.nii.gz[0..$]
3dTcat -prefix ./afni_pro_glm/sub-03.results/pb00.sub-03.r01.tcat ds000102/sub-03/func/sub-03_task-flanker_run-1_bold.nii.gz[0..$]
3dTcat -prefix ./afni_pro_glm/sub-01.results/pb00.sub-01.r01.tcat ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 1.0 s
++ elapsed time = 1.0 s
++ elapsed time = 1.0 s
3dTcat -prefix ./afni_pro_glm/sub-03.results/pb00.sub-03.r02.tcat ds000102/sub-03/func/sub-03_task-flanker_run-2_bold.nii.gz[0..$]
3dTcat -prefix ./afni_pro_glm/sub-01.results/pb00.sub-01.r02.tcat ds000102/sub-01/func/sub-01_task-flanker_run-2_bold.nii.gz[0..$]
3dTcat -prefix ./afni_pro_glm/sub-02.results/pb00.sub-02.r02.tcat ds000102/sub-02/func/sub-02_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.9 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-01.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-01.r01.tcat+orig
++ elapsed time = 0.9 s
++ elapsed time = 0.9 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-02.results
set tr_counts = ( 146 146 )
touch out.pre_ss_warn.txt
cd ./afni_pro_glm/sub-03.results
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-02.r01.tcat+orig
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-03.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 36604 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
++ 36324 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
++ 37617 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-02.r02.tcat+orig
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-03.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-01.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 36573 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
++ 36278 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
++ 37649 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 82
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 184
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 39
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 01
set minouttr = 82
echo min outlier: run 01, TR 82
tee out.min_outlier.txt
set minoutrun = 02
min outlier: run 01, TR 82
set minouttr = 38
3dbucket -prefix vr_base_min_outlier pb00.sub-02.r01.tcat+orig[82]
echo min outlier: run 02, TR 38
tee out.min_outlier.txt
min outlier: run 02, TR 38
3dbucket -prefix vr_base_min_outlier pb00.sub-03.r02.tcat+orig[38]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 01
set minouttr = 39
echo min outlier: run 01, TR 39
tee out.min_outlier.txt
min outlier: run 01, TR 39
3dbucket -prefix vr_base_min_outlier pb00.sub-01.r01.tcat+orig[39]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
align_epi_anat.py -anat2epi -anat sub-02_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
align_epi_anat.py -anat2epi -anat sub-03_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
align_epi_anat.py -anat2epi -anat sub-01_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-02_T1w+orig
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
#Script is running (command trimmed):
3dAttribute DELTA ./sub-03_T1w+orig
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-01_T1w+orig
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-02_T1w*
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-03_T1w*
#Script is running (command trimmed):
3dcopy ./sub-02_T1w+orig ./__tt_sub-02_T1w+orig
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-01_T1w*
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#Script is running (command trimmed):
3dcopy ./sub-03_T1w+orig ./__tt_sub-03_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#Script is running (command trimmed):
3dcopy ./sub-01_T1w+orig ./__tt_sub-01_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-02_T1w+orig -prefix ./__tt_sub-02_T1w_ns
#++ Removing skull from anat data
#++ Removing skull from anat data
#Script is running (command trimmed):
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-01_T1w+orig -prefix ./__tt_sub-01_T1w_ns
3dSkullStrip -orig_vol -input ./__tt_sub-03_T1w+orig -prefix ./__tt_sub-03_T1w_ns
#Script is running (command trimmed):
3dinfo ./__tt_sub-03_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/__tt_sub-03_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 358.490295
+ Used gradual clip level = 338.484009 .. 380.472412
+ Number voxels above clip level = 37209
+ Clustering voxels ...
+ Largest cluster has 36737 voxels
+ Clustering voxels ...
+ Largest cluster has 35853 voxels
+ Filled 391 voxels in small holes; now have 36244 voxels
+ Filled 4 voxels in large holes; now have 36248 voxels
+ Clustering voxels ...
+ Largest cluster has 36248 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 127592 voxels
+ Mask now has 36248 voxels
++ 36248 voxels in the mask [out of 163840: 22.12%]
++ first 8 x-planes are zero [from L]
++ last 10 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 6 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 862.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/862.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-03_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-03_T1w_ns+orig -prefix ./sub-03_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-03_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-03_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 1471023 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=2 ytop=1
++ Zero-pad: zbot=8 ztop=2
++ 36247 voxels [16.9%] in weight mask
++ Output dataset ./__tt_sub-03_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 36247
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.083 30.723 26.578 (index)
+ source center of mass = 86.969 122.492 105.519 (index)
+ source-target CM = 85.719 -4.768 -1.665 (xyz)
+ estimated center of mass shifts = 85.719 -4.768 -1.665
++ shift param auto-range: 25.1..146.4 -68.3..58.8 -64.6..61.3
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 75.719246 .. 95.719246 center = 85.719246
+ Range param#2 [y-shift] = -14.767860 .. 5.232140 center = -4.767860
+ Range param#3 [z-shift] = -11.664719 .. 8.335281 center = -1.664719
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 45.719246 .. 125.719246 center = 85.719246
+ Range param#2 [y-shift] = -44.767860 .. 35.232140 center = -4.767860
+ Range param#3 [z-shift] = -41.664719 .. 38.335281 center = -1.664719
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000092
++ Final parameter search ranges:
+ x-shift = 45.719 .. 125.719
+ y-shift = -44.768 .. 35.232
+ z-shift = -41.665 .. 38.335
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1471023 [out of 11534336] voxels
+ base mask has 48787 [out of 214400] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=65 usiz=160
+ - copying weight image
+ - using 36247 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 30537 total points stored in 61 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
#Script is running (command trimmed):
3dinfo ./__tt_sub-02_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/__tt_sub-02_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 367.570007
+ Used gradual clip level = 355.493591 .. 385.553711
+ Number voxels above clip level = 37698
+ Clustering voxels ...
+ Largest cluster has 36997 voxels
+ Clustering voxels ...
+ Largest cluster has 36269 voxels
+ Filled 316 voxels in small holes; now have 36585 voxels
+ Clustering voxels ...
+ Largest cluster has 36585 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 127255 voxels
+ Mask now has 36585 voxels
++ 36585 voxels in the mask [out of 163840: 22.33%]
++ first 9 x-planes are zero [from L]
++ last 12 x-planes are zero [from R]
++ first 4 y-planes are zero [from P]
++ last 6 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 4 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 873.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/873.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-02_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-02_T1w_ns+orig -prefix ./sub-02_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-02_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-02_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
+ - Test (64+191)*64 params [top5=*o+-.]:#ooo.-++-o-o.-o++--o-..o+-.ooo-....-..----+++$--.+-o + - best 88 costs found:
o= 0 v=-0.133650: 85.72 -4.77 -1.66 0.00 0.00 0.00 [grid]
o= 1 v=-0.025201: 68.95 16.01 -30.95 12.96 3.05 -9.17 [rand]
o= 2 v=-0.023611: 92.39 -11.43 -28.33 7.50 7.50 7.50 [grid]
o= 3 v=-0.023216: 99.72 10.77 -37.35 -14.61 -39.00 28.71 [rand]
o= 4 v=-0.022957: 74.23 11.54 -35.42 -19.17 13.96 2.56 [rand]
o= 5 v=-0.022315: 68.95 16.01 -30.95 12.96 -3.05 -9.17 [rand]
o= 6 v=-0.021787: 92.39 21.90 -28.33 -7.50 -30.00 30.00 [grid]
o= 7 v=-0.021469: 97.21 11.54 -35.42 19.17 13.96 -2.56 [rand]
o= 8 v=-0.021166: 94.38 19.39 -38.24 -27.41 -42.10 24.35 [rand]
o= 9 v=-0.021068: 94.38 19.39 -38.24 -27.41 -42.10 -24.35 [rand]
o=10 v=-0.020770: 74.23 11.54 -35.42 -19.17 13.96 -2.56 [rand]
o=11 v=-0.020424: 97.21 11.54 -35.42 19.17 13.96 2.56 [rand]
o=12 v=-0.019175: 102.49 16.01 -30.95 -12.96 3.05 9.17 [rand]
o=13 v=-0.018614: 77.06 19.39 -38.24 27.41 -42.10 24.35 [rand]
o=14 v=-0.018597: 92.39 21.90 -28.33 -30.00 -7.50 30.00 [grid]
o=15 v=-0.018396: 68.95 16.01 -30.95 12.96 3.05 9.17 [rand]
o=16 v=-0.018330: 74.23 11.54 -35.42 -19.17 -13.96 -2.56 [rand]
o=17 v=-0.018023: 104.83 18.65 -34.96 -37.88 -13.88 18.42 [rand]
o=18 v=-0.017901: 101.36 3.99 -35.50 23.16 17.17 -3.59 [rand]
o=19 v=-0.017890: 94.32 16.86 -30.37 -14.86 -11.51 26.00 [rand]
o=20 v=-0.017788: 99.06 19.48 -32.46 -6.84 -32.76 20.90 [rand]
o=21 v=-0.017676: 92.39 21.90 -28.33 -7.50 -7.50 30.00 [grid]
o=22 v=-0.017523: 92.39 1.90 -28.33 7.50 7.50 7.50 [grid]
o=23 v=-0.017188: 89.86 -28.90 -4.29 26.16 25.44 13.47 [rand]
o=24 v=-0.016811: 74.23 11.54 -35.42 -19.17 -13.96 2.56 [rand]
o=25 v=-0.016809: 68.95 16.01 -30.95 12.96 -3.05 9.17 [rand]
o=26 v=-0.016275: 83.42 2.41 -33.36 -10.97 18.37 -5.04 [rand]
o=27 v=-0.016087: 89.86 19.04 -33.56 -31.52 -11.26 33.63 [rand]
o=28 v=-0.016065: 97.21 11.54 -35.42 19.17 -13.96 -2.56 [rand]
o=29 v=-0.016060: 79.05 1.90 -28.33 -30.00 7.50 -7.50 [grid]
o=30 v=-0.015989: 79.90 9.42 -31.30 38.21 15.41 13.51 [rand]
o=31 v=-0.015862: 101.01 16.12 -32.05 -14.56 -29.40 15.92 [rand]
o=32 v=-0.015760: 97.21 11.54 -35.42 -19.17 -13.96 2.56 [rand]
o=33 v=-0.015729: 101.94 9.34 -36.80 -15.74 -34.09 7.78 [rand]
o=34 v=-0.015704: 92.39 -11.43 -28.33 30.00 7.50 7.50 [grid]
o=35 v=-0.015617: 92.39 21.90 -28.33 -30.00 -30.00 30.00 [grid]
o=36 v=-0.015556: 92.39 1.90 -28.33 30.00 7.50 7.50 [grid]
o=37 v=-0.015466: 79.05 -31.43 -8.33 30.00 30.00 30.00 [grid]
o=38 v=-0.015299: 102.49 16.01 -30.95 -12.96 -3.05 9.17 [rand]
o=39 v=-0.015221: 105.97 22.51 -36.15 -37.90 5.03 -3.08 [rand]
o=40 v=-0.015136: 92.39 21.90 -28.33 7.50 -30.00 -7.50 [grid]
o=41 v=-0.014978: 92.39 21.90 -28.33 -7.50 -30.00 -7.50 [grid]
o=42 v=-0.014950: 52.69 0.25 -38.48 11.98 -4.28 19.20 [rand]
o=43 v=-0.014928: 74.23 11.54 -35.42 19.17 -13.96 2.56 [rand]
o=44 v=-0.014910: 97.21 11.54 -35.42 19.17 -13.96 2.56 [rand]
o=45 v=-0.014866: 101.36 3.99 -35.50 -23.16 -17.17 3.59 [rand]
o=46 v=-0.014839: 92.13 29.18 -38.24 -19.28 -27.13 15.13 [rand]
o=47 v=-0.014837: 118.75 0.25 -38.48 -11.98 -4.28 -19.20 [rand]
o=48 v=-0.014689: 79.05 -31.43 -28.33 -30.00 7.50 7.50 [grid]
o=49 v=-0.014620: 79.05 -31.43 -8.33 7.50 30.00 30.00 [grid]
o=50 v=-0.014505: 92.39 21.90 -28.33 -30.00 -7.50 -7.50 [grid]
o=51 v=-0.014461: 77.06 19.39 -38.24 -27.41 -42.10 -24.35 [rand]
o=52 v=-0.014396: 100.72 32.18 16.58 34.16 -13.44 -39.30 [rand]
o=53 v=-0.014281: 105.97 22.51 -36.15 -37.90 5.03 3.08 [rand]
o=54 v=-0.014091: 94.23 24.24 -33.03 -12.98 -30.57 5.92 [rand]
o=55 v=-0.014025: 92.39 21.90 -28.33 -30.00 7.50 7.50 [grid]
o=56 v=-0.013918: 95.77 18.13 -23.41 -23.24 -8.32 39.75 [rand]
o=57 v=-0.013631: 92.39 1.90 -28.33 -7.50 7.50 30.00 [grid]
o=58 v=-0.013613: 81.59 15.80 -31.72 -19.78 -42.00 -3.76 [rand]
o=59 v=-0.013359: 92.39 21.90 -28.33 -7.50 7.50 7.50 [grid]
o=60 v=-0.013329: 79.05 21.90 -28.33 -30.00 -30.00 -7.50 [grid]
o=61 v=-0.013268: 100.66 19.32 -31.59 -19.13 -33.53 22.48 [rand]
o=62 v=-0.013267: 79.90 -18.96 -31.30 -38.21 15.41 -13.51 [rand]
o=63 v=-0.013261: 65.47 22.51 -36.15 37.90 5.03 -3.08 [rand]
o=64 v=-0.013238: 92.13 29.18 -38.24 -19.28 -27.13 -15.13 [rand]
o=65 v=-0.013232: 81.12 -15.36 -33.24 -36.41 9.00 -22.11 [rand]
o=66 v=-0.013205: 102.49 16.01 -30.95 12.96 3.05 9.17 [rand]
o=67 v=-0.013094: 71.58 23.36 -25.12 27.82 2.77 -20.28 [rand]
o=68 v=-0.013012: 89.85 -25.34 -31.72 19.78 42.00 3.76 [rand]
o=69 v=-0.013012: 72.30 17.92 -33.00 4.37 -20.12 15.12 [rand]
o=70 v=-0.012997: 79.05 21.90 -28.33 30.00 7.50 -7.50 [grid]
o=71 v=-0.012965: 74.42 -38.90 -7.64 4.11 32.66 25.74 [rand]
o=72 v=-0.012890: 79.05 1.90 -28.33 -7.50 30.00 7.50 [grid]
o=73 v=-0.012859: 77.11 16.86 -30.37 14.86 -11.51 -26.00 [rand]
o=74 v=-0.012843: 112.39 -31.43 -28.33 30.00 7.50 7.50 [grid]
o=75 v=-0.012805: 92.13 29.18 -38.24 19.28 -27.13 15.13 [rand]
o=76 v=-0.012767: 79.05 -11.43 -28.33 30.00 7.50 7.50 [grid]
o=77 v=-0.012682: 81.59 -25.34 -31.72 -19.78 42.00 3.76 [rand]
o=78 v=-0.012654: 105.97 22.51 -36.15 -37.90 -5.03 3.08 [rand]
o=79 v=-0.012606: 71.58 23.36 -25.12 27.82 -2.77 -20.28 [rand]
o=80 v=-0.012585: 94.23 24.24 -33.03 -12.98 -30.57 -5.92 [rand]
o=81 v=-0.012548: 79.30 29.18 -38.24 19.28 -27.13 15.13 [rand]
o=82 v=-0.012494: 74.23 11.54 -35.42 19.17 -13.96 -2.56 [rand]
o=83 v=-0.012470: 99.14 17.92 -33.00 4.37 20.12 15.12 [rand]
o=84 v=-0.012428: 88.01 2.41 -33.36 -10.97 18.37 -5.04 [rand]
o=85 v=-0.012403: 70.43 16.12 -32.05 14.56 -29.40 15.92 [rand]
o=86 v=-0.012394: 72.01 -2.08 -35.96 41.38 -19.47 -31.64 [rand]
o=87 v=-0.012363: 70.08 3.99 -35.50 -23.16 -17.17 -3.59 [rand]
+ - A little optimization:*[#16333=-0.144252] *[#16334=-0.147572] *[#16337=-0.148384] *[#16338=-0.148441] *[#16339=-0.154] *[#16340=-0.155731] *[#16342=-0.155922] *[#16345=-0.157235] *[#16348=-0.157827] *[#16351=-0.158064] *[#16352=-0.158294] *[#16354=-0.1583] *[#16355=-0.158316] *[#16359=-0.158393] *[#16361=-0.158397] *[#16362=-0.158422] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.158422: 86.05 -3.69 -0.71 -0.53 -0.01 0.17 [grid] [f=43]
o= 1 v=-0.030286: 69.35 15.09 -32.08 20.10 3.38 -7.35 [rand] [f=54]
o= 2 v=-0.030579: 91.85 -10.39 -29.92 8.62 6.76 17.33 [grid] [f=45]
o= 3 v=-0.032514: 99.05 11.83 -39.51 -22.10 -44.19 30.85 [rand] [f=72]
o= 4 v=-0.037512: 76.14 8.40 -37.75 -16.42 10.21 8.29 [rand] [f=60]
o= 5 v=-0.025893: 69.60 15.14 -30.46 19.09 -2.29 -8.83 [rand] [f=42]
o= 6 v=-0.023991: 92.01 21.07 -31.89 -7.30 -30.44 27.78 [grid] [f=38]
o= 7 v=-0.026615: 100.33 10.03 -35.60 18.40 12.69 -1.78 [rand] [f=42]
o= 8 v=-0.026973: 95.13 19.70 -41.10 -27.03 -42.60 30.21 [rand] [f=42]
o= 9 v=-0.024128: 93.40 19.26 -39.07 -28.47 -42.88 -25.88 [rand] [f=36]
o=10 v=-0.038410: 77.24 7.80 -38.82 -14.57 8.31 0.64 [rand] [f=50]
o=11 v=-0.033223: 98.43 4.60 -32.33 16.52 7.26 0.86 [rand] [f=51]
o=12 v=-0.032746: 99.78 14.62 -32.39 -14.37 2.51 7.04 [rand] [f=58]
o=13 v=-0.024800: 81.50 18.56 -38.29 28.06 -42.29 24.54 [rand] [f=48]
o=14 v=-0.032143: 95.04 16.84 -31.22 -33.60 -9.46 35.60 [grid] [f=49]
o=15 v=-0.031507: 68.19 14.07 -34.37 17.99 1.10 6.71 [rand] [f=55]
o=16 v=-0.032234: 76.07 6.48 -38.34 -18.74 -9.19 -7.83 [rand] [f=61]
o=17 v=-0.023522: 97.01 16.11 -32.63 -34.91 -13.32 18.85 [rand] [f=48]
o=18 v=-0.028350: 101.20 8.83 -36.26 21.39 13.36 -4.73 [rand] [f=46]
o=19 v=-0.023595: 94.45 16.75 -30.48 -15.04 -11.81 30.94 [rand] [f=36]
o=20 v=-0.023454: 96.43 17.49 -33.04 -7.76 -34.53 20.18 [rand] [f=60]
o=21 v=-0.023295: 92.43 21.30 -30.94 -5.60 -9.38 34.34 [grid] [f=45]
o=22 v=-0.033790: 96.74 1.92 -32.08 6.63 7.13 6.63 [grid] [f=58]
o=23 v=-0.029880: 86.95 -28.44 -6.85 27.82 23.89 19.88 [rand] [f=46]
o=24 v=-0.044519: 79.40 6.23 -39.38 -20.85 1.50 1.29 [rand] [f=62]
o=25 v=-0.029850: 74.80 16.34 -34.93 17.90 0.83 8.94 [rand] [f=55]
o=26 v=-0.032403: 84.03 2.22 -34.35 -17.03 5.86 -10.73 [rand] [f=55]
o=27 v=-0.034188: 87.93 11.56 -34.57 -28.85 -12.98 36.49 [rand] [f=83]
o=28 v=-0.024616: 93.45 11.37 -39.03 19.23 -17.31 -2.31 [rand] [f=55]
o=29 v=-0.035684: 79.98 1.50 -33.36 -19.52 3.33 -8.04 [grid] [f=49]
o=30 v=-0.018851: 80.28 9.35 -31.87 42.82 15.64 13.11 [rand] [f=40]
o=31 v=-0.026218: 97.50 17.07 -33.85 -14.59 -32.05 17.79 [rand] [f=51]
o=32 v=-0.032083: 100.01 7.57 -34.75 -10.29 -5.61 5.76 [rand] [f=64]
o=33 v=-0.018674: 101.26 9.17 -35.41 -16.82 -33.31 11.89 [rand] [f=59]
o=34 v=-0.033591: 90.28 -9.07 -33.74 14.47 2.43 11.87 [grid] [f=76]
o=35 v=-0.028180: 93.10 21.13 -30.75 -32.01 -22.98 37.20 [grid] [f=56]
o=36 v=-0.026349: 96.35 1.83 -32.65 29.05 6.68 7.34 [grid] [f=55]
o=37 v=-0.021548: 78.85 -30.70 -9.94 28.07 28.11 27.14 [grid] [f=44]
o=38 v=-0.029556: 102.37 11.27 -33.62 -19.30 -1.51 7.70 [rand] [f=63]
o=39 v=-0.022030: 105.68 22.74 -32.11 -37.75 4.99 -3.11 [rand] [f=44]
o=40 v=-0.020972: 92.02 20.93 -27.79 7.06 -30.34 -0.63 [grid] [f=42]
o=41 v=-0.018336: 92.61 20.90 -28.75 -2.80 -30.25 -7.49 [grid] [f=40]
o=42 v=-0.018357: 52.43 -0.13 -38.28 16.47 -4.20 19.54 [rand] [f=47]
o=43 v=-0.025914: 78.94 12.36 -38.78 17.69 -6.87 8.24 [rand] [f=57]
o=44 v=-0.025162: 91.42 11.71 -41.00 19.66 -12.62 3.28 [rand] [f=57]
o=45 v=-0.030887: 102.65 6.32 -40.03 -22.17 -12.35 7.49 [rand] [f=66]
o=46 v=-0.024243: 93.48 21.80 -39.43 -16.78 -26.70 22.97 [rand] [f=61]
o=47 v=-0.023470: 120.54 5.39 -38.81 -14.46 -5.76 -21.28 [rand] [f=53]
o=48 v=-0.021578: 73.81 -32.32 -28.30 -29.44 7.71 7.73 [grid] [f=52]
o=49 v=-0.021961: 78.34 -33.30 -7.68 6.16 34.14 30.65 [grid] [f=66]
o=50 v=-0.018178: 93.25 22.48 -30.19 -31.31 -6.87 -8.85 [grid] [f=62]
o=51 v=-0.018119: 78.43 18.44 -41.00 -22.63 -43.59 -25.56 [rand] [f=43]
o=52 v=-0.023424: 100.99 25.03 14.71 33.25 -10.03 -44.80 [rand] [f=45]
o=53 v=-0.020405: 105.92 18.32 -36.18 -37.46 5.00 3.01 [rand] [f=41]
o=54 v=-0.018055: 93.93 23.48 -36.04 -16.10 -31.41 11.27 [rand] [f=49]
o=55 v=-0.024761: 100.05 21.71 -31.07 -27.71 1.29 11.50 [grid] [f=68]
o=56 v=-0.031859: 95.40 17.66 -28.25 -27.27 -10.00 40.59 [rand] [f=56]
o=57 v=-0.035021: 91.82 4.91 -37.18 -10.06 -1.30 31.71 [grid] [f=70]
o=58 v=-0.019102: 82.13 16.30 -31.71 -12.11 -39.60 -3.47 [rand] [f=44]
o=59 v=-0.022892: 94.86 19.49 -30.73 -13.32 2.59 10.66 [grid] [f=62]
o=60 v=-0.020388: 85.76 22.03 -30.45 -33.10 -36.83 -11.90 [grid] [f=53]
o=61 v=-0.027108: 94.48 19.09 -34.11 -17.68 -33.53 21.63 [rand] [f=63]
o=62 v=-0.022213: 79.17 -12.08 -33.72 -38.98 13.55 -12.54 [rand] [f=50]
o=63 v=-0.034719: 68.64 17.00 -32.33 30.83 7.17 -1.06 [rand] [f=45]
o=64 v=-0.017886: 92.65 28.42 -37.69 -18.59 -26.48 -9.13 [rand] [f=33]
o=65 v=-0.021171: 81.13 -14.70 -32.14 -33.29 9.42 -20.54 [rand] [f=62]
o=66 v=-0.034161: 96.96 1.66 -34.85 17.86 2.52 1.66 [rand] [f=63]
o=67 v=-0.023912: 73.95 19.98 -26.34 24.15 -2.71 -19.28 [rand] [f=71]
o=68 v=-0.016952: 90.41 -24.67 -32.43 18.47 44.46 3.70 [rand] [f=37]
o=69 v=-0.024032: 75.63 12.37 -32.22 8.42 -16.09 14.12 [rand] [f=57]
o=70 v=-0.031066: 71.21 19.33 -32.87 32.42 9.10 -5.60 [grid] [f=61]
o=71 v=-0.019020: 77.18 -38.21 -2.78 0.74 33.16 26.31 [rand] [f=41]
o=72 v=-0.019290: 80.73 -0.69 -26.64 -6.14 34.49 7.47 [grid] [f=47]
o=73 v=-0.024347: 74.73 13.50 -29.38 15.56 -10.27 -23.87 [rand] [f=46]
o=74 v=-0.017950: 112.31 -29.45 -29.03 35.47 7.46 6.91 [grid] [f=47]
o=75 v=-0.020444: 86.12 28.77 -41.50 23.48 -28.98 17.95 [rand] [f=58]
o=76 v=-0.033124: 80.54 -10.95 -33.26 32.11 6.59 6.30 [grid] [f=54]
o=77 v=-0.015899: 85.33 -24.93 -32.48 -20.26 43.69 3.84 [rand] [f=38]
o=78 v=-0.030005: 106.03 9.95 -41.66 -27.03 -7.73 9.63 [rand] [f=69]
o=79 v=-0.026739: 73.53 16.25 -28.27 23.69 -2.16 -15.59 [rand] [f=55]
o=80 v=-0.017988: 94.89 24.25 -33.63 -13.58 -25.98 -5.26 [rand] [f=34]
o=81 v=-0.031345: 81.08 16.15 -40.31 10.79 -17.65 4.48 [rand] [f=83]
o=82 v=-0.027903: 74.19 11.28 -39.81 17.71 -4.95 0.47 [rand] [f=60]
o=83 v=-0.019832: 98.61 12.51 -32.99 5.78 22.71 13.80 [rand] [f=53]
o=84 v=-0.018860: 83.48 1.82 -33.99 -10.93 18.05 -5.38 [rand] [f=46]
o=85 v=-0.015019: 71.66 16.11 -34.26 18.43 -29.41 12.20 [rand] [f=53]
o=86 v=-0.027905: 73.95 4.70 -39.15 37.53 -16.27 -34.51 [rand] [f=58]
o=87 v=-0.033629: 75.40 4.98 -38.77 -16.50 -8.30 -7.42 [rand] [f=59]
+ - saving # 0 for use with twobest
+ - saving #24 for use with twobest
+ - saving #10 for use with twobest
+ - saving # 4 for use with twobest
+ - saving #29 for use with twobest
+ - saving #57 for use with twobest
+ - saving #63 for use with twobest
+ - saving #27 for use with twobest
+ - saving #66 for use with twobest
+ - saving #22 for use with twobest
+ - saving #87 for use with twobest
+ - saving #34 for use with twobest
+ - saving #11 for use with twobest
+ - saving #76 for use with twobest
+ - saving #12 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #26 for use with twobest
+ - skip #16 for twobest: too close to set #87
+ - saving #14 for use with twobest
+ - saving #32 for use with twobest
+ - saving #56 for use with twobest
+ - saving #15 for use with twobest
+ - saving #81 for use with twobest
+ - saving #70 for use with twobest
+ - saving #45 for use with twobest
+ - saving # 2 for use with twobest
+ - saving # 1 for use with twobest
+ - saving #78 for use with twobest
+ - saving #23 for use with twobest
+ - saving #25 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=65 usiz=160
+ - retaining old weight image
+ - using 36247 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30267 total points stored in 62 'TOHD(17.3748)' bloks (0 duplicates)
*[#21082=-0.197662] *[#21090=-0.1991] *[#21102=-0.200976] *[#21103=-0.201636] *[#21105=-0.202284] *[#21106=-0.202753] *[#21107=-0.203598] *[#21112=-0.204986] *[#21114=-0.206642] *[#21117=-0.207141] *[#21121=-0.207211] *[#21124=-0.207306] *[#21125=-0.207497] *[#21128=-0.208208] *[#21133=-0.208437] *[#21136=-0.208481] *[#21138=-0.208673] *[#21139=-0.208929] *[#21140=-0.208983] *[#21143=-0.209168] *[#21145=-0.209264] *[#21146=-0.209448] *[#21147=-0.20962] *[#21150=-0.209825] *[#21151=-0.209942] *[#21152=-0.209968] *[#21153=-0.210044] *[#21154=-0.210198] *[#21156=-0.210244] *[#21159=-0.210289] *[#21163=-0.210314] *[#21167=-0.210331] *[#21168=-0.210375] *[#21169=-0.210454] *[#21170=-0.210603] *[#21171=-0.21079] *[#21172=-0.210856] *[#21175=-0.211025] *[#21176=-0.211239] *[#21177=-0.211464] *[#21179=-0.211569] *[#21180=-0.211716]
+ - param set #1 has cost=-0.211716 [o=0 t=0]
+ -- Parameters = 85.8736 -3.7203 -1.1165 -0.1440 0.2372 0.1380 0.9826 1.0187 0.9756 0.0020 0.0034 -0.0057
+ - param set #2 has cost=-0.037296 [o=24 t=1]
+ -- Parameters = 79.4038 6.1525 -39.4657 -20.8000 1.4321 1.3270 1.0037 0.9976 0.9984 0.0013 0.0001 0.0003
+ - param set #3 has cost=-0.039410 [o=10 t=2]
+ -- Parameters = 77.3703 3.8193 -38.3898 -13.9767 6.4991 0.2873 1.0003 0.9909 0.9816 -0.0068 -0.0038 0.0001
+ - param set #4 has cost=-0.037907 [o=4 t=3]
+ -- Parameters = 76.2762 6.2599 -37.2717 -16.0009 7.3976 2.9954 0.9933 0.9860 0.9811 -0.0013 0.0014 0.0035
+ - param set #5 has cost=-0.035582 [o=29 t=4]
+ -- Parameters = 80.0511 1.3002 -33.5832 -19.5638 3.2042 -8.1413 0.9906 0.9935 0.9948 0.0019 0.0015 0.0001
+ - param set #6 has cost=-0.029041 [o=57 t=5]
+ -- Parameters = 91.8047 4.9054 -37.1949 -10.0874 -1.2863 31.7045 0.9999 1.0007 1.0000 -0.0000 -0.0003 0.0011
+ - param set #7 has cost=-0.032430 [o=63 t=6]
+ -- Parameters = 69.1014 17.3819 -32.7390 30.6473 7.4616 -1.1620 1.0375 0.9956 0.9944 -0.0022 -0.0037 -0.0020
+ - param set #8 has cost=-0.029534 [o=27 t=7]
+ -- Parameters = 87.8382 11.4444 -34.6524 -28.9332 -13.1616 36.5855 1.0005 0.9991 1.0016 0.0002 -0.0001 0.0002
+ - param set #9 has cost=-0.032559 [o=66 t=8]
+ -- Parameters = 97.0781 1.7414 -34.4192 18.3540 2.6213 1.0100 0.9978 1.0016 1.0020 -0.0004 -0.0010 0.0002
+ - param set #10 has cost=-0.030224 [o=22 t=9]
+ -- Parameters = 96.7978 2.0082 -32.0708 6.6040 7.3226 6.5534 0.9999 1.0009 0.9993 0.0000 -0.0006 0.0005
++ 1476080 voxels in -source_automask+2
+ - param set #11 has cost=-0.032997 [o=87 t=10]
+ -- Parameters = 75.5291 4.5095 -39.2736 -15.7287 -9.0209 -7.6178 0.9906 0.9927 0.9859 -0.0023 0.0006 -0.0064
+ - param set #12 has cost=-0.028237 [o=34 t=11]
+ -- Parameters = 90.2251 -8.9606 -33.9962 14.3512 2.5899 11.8539 0.9975 1.0015 0.9936 0.0004 0.0007 -0.0001
+ - param set #13 has cost=-0.034561 [o=11 t=12]
+ -- Parameters = 100.4416 5.4224 -33.2486 14.8360 4.4159 -3.6787 0.9565 1.0019 0.9677 0.0194 0.0027 0.0003
+ - param set #14 has cost=-0.030330 [o=76 t=13]
+ -- Parameters = 80.5225 -11.1441 -33.0872 32.2208 6.5194 6.4566 1.0003 1.0330 0.9982 -0.0000 0.0002 0.0008
++ largeness ==> set -twobest 29
+ - param set #15 has cost=-0.031025 [o=12 t=14]
++ Zero-pad: ybot=4 ytop=2
++ Zero-pad: zbot=8 ztop=4
+ -- Parameters = 99.8077 14.6827 -32.3832 -14.3754 2.5485 7.0419 0.9999 1.0003 0.9996 0.0009 -0.0005 -0.0006
++ 36584 voxels [15.7%] in weight mask
++ Output dataset ./__tt_sub-02_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 36584
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - param set #16 has cost=-0.028256 [o=3 t=15]
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ -- Parameters = 99.0722 11.9683 -39.6707 -22.1199 -44.1971 30.5784 0.9904 0.9964 1.0014 -0.0008 -0.0005 0.0007
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 30.309 32.312 27.499 (index)
+ - param set #17 has cost=-0.031444 [o=26 t=16]
+ -- Parameters = 84.1486 2.8510 -34.2998 -17.0811 6.3874 -10.8582 0.9997 1.0052 0.9983 0.0005 0.0009 0.0016
+ - param set #18 has cost=-0.032296 [o=14 t=17]
+ -- Parameters = 94.8647 15.3124 -30.7010 -34.5351 -12.2349 33.9901 0.9809 0.9916 0.9875 0.0085 0.0067 0.0046
+ source center of mass = 90.077 103.579 111.760 (index)
+ source-target CM = 86.504 -0.102 -2.000 (xyz)
+ estimated center of mass shifts = 86.504 -0.102 -2.000
++ shift param auto-range: 25.8..147.2 -66.5..66.3 -67.5..63.5
+ - param set #19 has cost=-0.029231 [o=32 t=18]
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.504372 .. 96.504372 center = 86.504372
+ Range param#2 [y-shift] = -10.101776 .. 9.898224 center = -0.101776
+ Range param#3 [z-shift] = -12.000374 .. 7.999626 center = -2.000374
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ -- Parameters = 100.0976 7.5487 -34.6665 -10.3601 -5.7172 5.5827 0.9966 0.9971 1.0003 0.0007 -0.0002 -0.0014
+ - param set #20 has cost=-0.027248 [o=56 t=19]
+ -- Parameters = 95.2620 17.6821 -28.2161 -26.9709 -9.9747 40.4885 0.9999 0.9961 1.0058 -0.0005 -0.0002 0.0148
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.504372 .. 126.504372 center = 86.504372
+ Range param#2 [y-shift] = -40.101776 .. 39.898224 center = -0.101776
+ Range param#3 [z-shift] = -42.000374 .. 37.999626 center = -2.000374
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000090
++ Final parameter search ranges:
+ x-shift = 46.504 .. 126.504
+ y-shift = -40.102 .. 39.898
+ z-shift = -42.000 .. 38.000
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1476080 [out of 11534336] voxels
+ base mask has 49187 [out of 232960] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
+ - param set #21 has cost=-0.028699 [o=15 t=20]
+ -- Parameters = 70.8438 13.5995 -34.3672 19.1661 1.1120 6.1842 0.9873 1.0056 0.9918 -0.0007 -0.0009 0.0009
+ - param set #22 has cost=-0.030191 [o=81 t=21]
+ -- Parameters = 80.3682 16.1218 -40.4984 5.2683 -17.5434 4.6665 1.0038 1.0027 1.0046 0.0002 0.0003 0.0013
+ - param set #23 has cost=-0.029161 [o=70 t=22]
+ -- Parameters = 70.6914 19.0614 -32.4028 32.6704 6.9490 -5.4689 0.9962 0.9938 0.9914 -0.0010 0.0012 0.0007
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - param set #24 has cost=-0.030269 [o=45 t=23]
+ -- Parameters = 102.2308 7.2665 -40.7272 -21.9217 -16.2146 5.3088 0.9788 0.9701 0.9892 0.0006 -0.0042 -0.0002
+ - param set #25 has cost=-0.028067 [o=2 t=24]
+ -- Parameters = 91.0143 -10.9799 -29.9112 8.6364 4.5509 16.1745 1.0075 0.9829 0.9947 0.0009 0.0007 0.0011
+ - param set #26 has cost=-0.029001 [o=1 t=25]
+ -- Parameters = 69.2170 15.2484 -31.6682 20.2736 7.2107 -8.8623 0.9987 0.9760 0.9718 -0.0053 -0.0003 -0.0019
+ - param set #27 has cost=-0.035381 [o=78 t=26]
+ -- Parameters = 102.9123 9.5082 -41.2618 -24.8566 -9.7574 14.8442 0.9193 0.9519 0.9658 -0.0034 -0.0063 0.0049
+ - param set #28 has cost=-0.026624 [o=23 t=27]
+ -- Parameters = 86.3695 -28.5059 -6.7483 27.8640 23.0604 20.9073 0.9917 1.0019 1.0005 -0.0002 -0.0003 -0.0010
+ - param set #29 has cost=-0.028050 [o=25 t=28]
+ -- Parameters = 74.7571 16.2962 -34.9229 18.0407 0.7657 8.7864 0.9993 1.0012 0.9989 -0.0014 -0.0001 0.0009
*[#23800=-0.21189] *[#23803=-0.212332] *[#23804=-0.21257] *[#23805=-0.212798]
+ - param set #30 has cost=-0.212798 [o=-1 t=-1]
+ -- Parameters = 85.7854 -4.3457 -1.4276 0.0521 -0.7003 0.2827 0.9833 1.0218 0.9665 -0.0022 -0.0034 -0.0131
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0332
+ --- dist(#3,#1) = 0.462
+ --- dist(#4,#1) = 0.448
+ --- dist(#5,#1) = 0.475
+ --- dist(#6,#1) = 0.402
+ --- dist(#7,#1) = 0.498
+ --- dist(#8,#1) = 0.398
+ --- dist(#9,#1) = 0.473
+ --- dist(#10,#1) = 0.412
+ --- dist(#11,#1) = 0.391
+ --- dist(#12,#1) = 0.384
+ --- dist(#13,#1) = 0.411
+ --- dist(#14,#1) = 0.387
+ --- dist(#15,#1) = 0.396
+ --- dist(#16,#1) = 0.491
+ --- dist(#17,#1) = 0.383
+ --- dist(#18,#1) = 0.488
+ --- dist(#19,#1) = 0.415
+ --- dist(#20,#1) = 0.415
+ --- dist(#21,#1) = 0.387
+ --- dist(#22,#1) = 0.447
+ --- dist(#23,#1) = 0.378
+ --- dist(#24,#1) = 0.412
+ --- dist(#25,#1) = 0.483
+ --- dist(#26,#1) = 0.407
+ --- dist(#27,#1) = 0.356
+ --- dist(#28,#1) = 0.419
+ --- dist(#29,#1) = 0.447
+ --- dist(#30,#1) = 0.309
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=65 usiz=160
+ !source mask fill: ubot=56 usiz=154.5
+ - retaining old weight image
+ - using 36247 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30000 total points stored in 62 'TOHD(17.2644)' bloks (0 duplicates)
*[#23808=-0.238216] *[#23830=-0.238784] *[#23831=-0.239159] *[#23834=-0.239186] *[#23838=-0.23983] *[#23839=-0.239966] *[#23840=-0.240068] *[#23845=-0.240626] *[#23852=-0.241539] *[#23853=-0.241781] *[#23855=-0.241999] *[#23865=-0.242108] *[#23866=-0.242113] *[#23868=-0.242188] *[#23873=-0.242277] *[#23874=-0.242662] *[#23879=-0.242853] *[#23880=-0.242989] *[#23881=-0.243018] *[#23889=-0.243264] *[#23896=-0.243314] *[#23898=-0.243479] *[#23903=-0.243529] *[#23904=-0.243619] *[#23911=-0.243992] *[#23916=-0.244057]
+ - param set #1 has cost=-0.244057 [o=-1 t=-1]
+ -- Parameters = 85.7823 -3.9764 -1.0043 -0.1909 -0.3804 0.4195 0.9907 1.0221 0.9797 -0.0038 -0.0014 -0.0198
+ - copying weight image
+ - param set #2 has cost=-0.241586 [o=0 t=0]
+ - Search for coarse starting parameters
+ * Exit alignment setup routine
+ -- Parameters = 85.8663 -3.9158 -1.0972 -0.0026 -0.0575 0.0568 0.9922 1.0213 0.9766 -0.0007 0.0039 -0.0090
+ - using 36584 points from base image [use_all=2]
+ 30215 total points stored in 59 'TOHD(17.5558)' bloks (0 duplicates)
+ - param set #3 has cost=-0.034759 [o=10 t=2]
+ -- Parameters = 77.3643 3.8025 -38.4012 -13.9820 6.5007 0.2636 1.0007 0.9910 0.9814 -0.0068 -0.0037 0.0002
+ - number of free params = 6
+ - param set #4 has cost=-0.034220 [o=4 t=3]
+ -- Parameters = 76.5322 6.4447 -37.4824 -16.1404 7.1462 3.0586 0.9914 0.9843 0.9802 0.0076 0.0012 0.0037
+ - param set #5 has cost=-0.031789 [o=24 t=1]
+ -- Parameters = 79.3869 6.1870 -39.5127 -20.7715 1.4586 1.3168 1.0031 0.9999 0.9985 0.0013 -0.0006 0.0003
+ - param set #6 has cost=-0.033600 [o=29 t=4]
+ -- Parameters = 79.9923 1.4398 -33.6808 -19.6248 3.2791 -8.2094 0.9854 0.9954 0.9930 0.0094 0.0025 -0.0004
+ - param set #7 has cost=-0.029470 [o=78 t=26]
+ -- Parameters = 102.9961 9.8276 -41.2168 -24.7502 -9.3676 14.6004 0.9224 0.9503 0.9669 -0.0048 -0.0057 0.0140
+ - param set #8 has cost=-0.031245 [o=11 t=12]
+ -- Parameters = 100.5030 5.3826 -32.9218 14.9423 4.3678 -4.0327 0.9578 1.0019 0.9669 0.0198 0.0028 0.0003
+ - param set #9 has cost=-0.028861 [o=87 t=10]
+ -- Parameters = 75.5865 4.5088 -39.3753 -15.7410 -9.0704 -7.7341 0.9894 0.9917 0.9836 -0.0011 0.0021 -0.0074
+ - param set #10 has cost=-0.031736 [o=66 t=8]
+ -- Parameters = 96.0110 1.7467 -34.2637 17.7461 2.5286 0.1920 0.9870 0.9921 0.9878 -0.0037 -0.0042 -0.0037
+ - param set #11 has cost=-0.029725 [o=63 t=6]
+ -- Parameters = 69.0714 17.1591 -32.8921 30.7924 7.1112 -1.1700 1.0415 0.9916 0.9941 0.0068 -0.0047 -0.0011
+ - param set #12 has cost=-0.028590 [o=14 t=17]
+ -- Parameters = 95.1797 15.3871 -30.8876 -34.5273 -12.2645 33.8683 0.9735 0.9860 0.9841 0.0089 0.0100 0.0038
+ - param set #13 has cost=-0.029415 [o=26 t=16]
+ -- Parameters = 84.1463 2.8387 -34.2928 -17.0767 6.3879 -10.8691 0.9998 1.0053 0.9979 0.0005 0.0016 0.0011
+ - param set #14 has cost=-0.027029 [o=12 t=14]
+ -- Parameters = 99.7666 14.5935 -32.3996 -14.3128 2.5802 7.0727 0.9998 1.0020 0.9980 0.0019 -0.0023 0.0008
+ - param set #15 has cost=-0.026283 [o=76 t=13]
+ -- Parameters = 80.3341 -11.2973 -33.1938 32.2822 6.5394 6.5259 1.0011 1.0313 0.9968 -0.0006 0.0015 0.0007
+ - param set #16 has cost=-0.026267 [o=45 t=23]
+ -- Parameters = 101.9988 7.2035 -40.7731 -21.8777 -16.1365 5.4608 0.9817 0.9733 0.9871 0.0003 -0.0040 -0.0007
+ - param set #17 has cost=-0.028279 [o=22 t=9]
+ -- Parameters = 96.7930 1.9982 -32.1065 6.6558 7.2406 6.5366 1.0002 1.0009 0.9991 0.0003 -0.0005 0.0004
+ - param set #18 has cost=-0.031517 [o=81 t=21]
+ -- Parameters = 81.2648 15.0354 -40.7007 4.2094 -17.1101 4.4679 1.0069 0.9976 0.9821 -0.0010 -0.0031 -0.0006
+ - param set #19 has cost=-0.025711 [o=27 t=7]
+ -- Parameters = 87.8248 11.0537 -34.6766 -29.5445 -10.9695 37.2472 0.9951 0.9971 1.0001 -0.0010 -0.0006 -0.0012
+ - param set #20 has cost=-0.027659 [o=32 t=18]
+ -- Parameters = 100.3381 7.6064 -34.5891 -10.3651 -5.1608 5.5580 0.9966 0.9878 1.0031 -0.0012 0.0002 -0.0019
+ - param set #21 has cost=-0.029695 [o=70 t=22]
+ -- Parameters = 70.2884 16.9437 -31.8497 33.0430 6.5891 -4.7513 1.0205 0.9623 0.9827 0.0029 0.0112 0.0008
+ - param set #22 has cost=-0.021922 [o=57 t=5]
+ -- Parameters = 91.8379 4.8848 -37.2029 -10.0858 -1.2827 31.7383 0.9998 1.0006 1.0000 -0.0000 -0.0004 0.0011
+ - param set #23 has cost=-0.024451 [o=1 t=25]
+ -- Parameters = 68.8550 15.4534 -31.3802 20.1483 7.5450 -8.6261 0.9993 0.9758 0.9717 -0.0056 -0.0000 -0.0023
+ - param set #24 has cost=-0.027700 [o=15 t=20]
+ -- Parameters = 70.3758 14.5483 -34.0117 22.0494 1.9622 6.6715 0.9812 1.0027 1.0039 0.0002 -0.0009 0.0014
+ - param set #25 has cost=-0.024622 [o=3 t=15]
+ -- Parameters = 99.2374 11.8412 -39.5462 -21.7779 -44.3405 30.7002 0.9841 0.9947 1.0040 -0.0020 0.0007 0.0007
+ - param set #26 has cost=-0.027622 [o=34 t=11]
+ -- Parameters = 90.2214 -11.3593 -35.4524 8.3766 0.0823 13.1381 1.0044 0.9852 0.9644 0.0018 0.0032 0.0002
+ - param set #27 has cost=-0.030129 [o=2 t=24]
+ -- Parameters = 90.1843 -12.6452 -30.6926 9.5918 4.5613 15.0923 1.0125 0.9744 0.9855 0.0010 -0.0006 0.0011
+ - param set #28 has cost=-0.027130 [o=25 t=28]
+ -- Parameters = 74.6702 16.0229 -34.5812 17.9183 0.7937 8.8298 0.9951 0.9974 1.0022 -0.0012 -0.0003 0.0017
+ - param set #29 has cost=-0.024228 [o=56 t=19]
+ -- Parameters = 95.6352 17.0807 -28.5969 -26.7974 -10.5829 40.0246 0.9694 0.9828 0.9899 -0.0015 -0.0029 0.0151
+ - param set #30 has cost=-0.026045 [o=23 t=27]
+ -- Parameters = 84.8218 -28.7431 -6.9556 26.4042 22.9726 21.0220 0.9849 1.0008 1.0006 0.0009 -0.0008 0.0001
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0487
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.456
+ --- dist(#5,#1) = 0.408
+ --- dist(#6,#1) = 0.481
+ --- dist(#7,#1) = 0.416
+ --- dist(#8,#1) = 0.496
+ --- dist(#9,#1) = 0.399
+ --- dist(#10,#1) = 0.371
+ --- dist(#11,#1) = 0.399
+ --- dist(#12,#1) = 0.386
+ --- dist(#13,#1) = 0.503
+ --- dist(#14,#1) = 0.416
+ --- dist(#15,#1) = 0.48
+ --- dist(#16,#1) = 0.382
+ --- dist(#17,#1) = 0.389
+ --- dist(#18,#1) = 0.413
+ --- dist(#19,#1) = 0.42
+ --- dist(#20,#1) = 0.431
+ --- dist(#21,#1) = 0.42
+ --- dist(#22,#1) = 0.392
+ --- dist(#23,#1) = 0.402
+ --- dist(#24,#1) = 0.497
+ --- dist(#25,#1) = 0.31
+ --- dist(#26,#1) = 0.421
+ --- dist(#27,#1) = 0.488
+ --- dist(#28,#1) = 0.38
+ --- dist(#29,#1) = 0.44
+ --- dist(#30,#1) = 0.452
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=65 usiz=160
+ - retaining old weight image
+ - using 36247 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30038 total points stored in 63 'TOHD(17.1973)' bloks (0 duplicates)
*[#26870=-0.251311] *[#26892=-0.251615] *[#26894=-0.252052] *[#26895=-0.252529] *[#26897=-0.252728] *[#26899=-0.252812] *[#26900=-0.252949] *[#26901=-0.252972] *[#26902=-0.252985] *[#26903=-0.253122] *[#26904=-0.253247] *[#26907=-0.253362] *[#26910=-0.253406] *[#26913=-0.253623] *[#26920=-0.253719] *[#26921=-0.25381] *[#26922=-0.253868] *[#26925=-0.253943] *[#26926=-0.25404] *[#26927=-0.254103] *[#26928=-0.254167] *[#26929=-0.254184] *[#26934=-0.254248] *[#26937=-0.254318] *[#26940=-0.25466] *[#26941=-0.254821] *[#26942=-0.254854] *[#26951=-0.254953] *[#26955=-0.255093] *[#26956=-0.255242] *[#26963=-0.255275] *[#26972=-0.255313] *[#26976=-0.255339] *[#26979=-0.255363] *[#26980=-0.255378] *[#26988=-0.255413]
+ - param set #1 has cost=-0.255413 [o=-1 t=-1]
+ -- Parameters = 85.8750 -4.0688 -0.8970 -0.2495 -0.6040 0.1950 0.9935 1.0212 0.9771 -0.0105 0.0010 -0.0317
+ - param set #2 has cost=-0.255222 [o=0 t=0]
+ -- Parameters = 85.9845 -4.0432 -0.7949 -0.3680 -0.5875 -0.0309 0.9930 1.0195 0.9804 -0.0117 0.0072 -0.0288
+ - param set #3 has cost=-0.029607 [o=10 t=2]
+ -- Parameters = 77.3330 3.7400 -38.1765 -14.0977 6.5079 0.3764 1.0016 0.9925 0.9804 -0.0070 0.0018 -0.0002
+ - param set #4 has cost=-0.028526 [o=4 t=3]
+ -- Parameters = 76.5845 6.4197 -37.4816 -16.0930 7.0144 3.0146 0.9908 0.9834 0.9799 0.0131 0.0012 0.0038
+ - param set #5 has cost=-0.030208 [o=29 t=4]
+ -- Parameters = 80.0031 1.4205 -33.6958 -19.6550 3.2867 -8.2239 0.9853 0.9954 0.9929 0.0093 0.0025 -0.0004
+ - param set #6 has cost=-0.027229 [o=24 t=1]
+ -- Parameters = 79.3928 6.1858 -39.5952 -20.6257 1.6447 1.2581 1.0014 1.0081 0.9972 0.0007 -0.0012 0.0004
+ - param set #7 has cost=-0.028798 [o=66 t=8]
+ -- Parameters = 96.0372 2.0882 -34.2920 17.2826 2.7498 0.1803 0.9846 0.9988 0.9920 -0.0036 -0.0049 -0.0041
+ - param set #8 has cost=-0.029402 [o=81 t=21]
+ -- Parameters = 81.7000 14.7044 -40.7886 4.5173 -17.1908 4.5806 1.0116 0.9860 0.9807 -0.0037 -0.0039 -0.0018
+ - param set #9 has cost=-0.028437 [o=11 t=12]
+ -- Parameters = 100.4787 5.3414 -32.9088 15.0706 4.2862 -3.9919 0.9624 0.9993 0.9677 0.0199 0.0028 0.0008
+ - param set #10 has cost=-0.027313 [o=2 t=24]
+ -- Parameters = 89.9631 -12.5681 -30.7253 9.5585 4.6659 15.2663 1.0185 0.9762 0.9856 0.0001 -0.0002 0.0008
+ - param set #11 has cost=-0.025202 [o=63 t=6]
+ -- Parameters = 68.9062 16.8701 -32.9880 30.7197 7.2224 -1.0839 1.0416 0.9716 0.9927 0.0065 -0.0049 -0.0011
+ - param set #12 has cost=-0.024610 [o=70 t=22]
+ -- Parameters = 70.1939 16.9033 -32.0769 33.1289 6.4620 -4.7010 1.0209 0.9628 0.9802 0.0035 0.0094 0.0017
+ - param set #13 has cost=-0.024539 [o=78 t=26]
+ -- Parameters = 103.0684 9.7956 -41.0687 -24.7673 -9.3139 14.5054 0.9214 0.9503 0.9674 -0.0053 -0.0060 0.0138
+ - param set #14 has cost=-0.026027 [o=26 t=16]
+ -- Parameters = 84.1430 2.8310 -34.2967 -17.0779 6.3930 -10.8819 0.9999 1.0055 0.9978 0.0005 0.0015 0.0011
+ - param set #15 has cost=-0.025112 [o=87 t=10]
+ -- Parameters = 75.7266 4.4363 -39.2202 -15.8438 -9.1852 -8.0000 0.9873 0.9882 0.9837 0.0006 0.0042 -0.0080
+ - param set #16 has cost=-0.025192 [o=14 t=17]
+ -- Parameters = 95.1534 15.3266 -30.8907 -34.6087 -12.3411 33.8811 0.9729 0.9856 0.9846 0.0145 0.0101 0.0040
+ - param set #17 has cost=-0.025286 [o=22 t=9]
+ -- Parameters = 96.8633 1.9202 -32.0888 6.6220 7.1530 6.5086 0.9992 1.0008 0.9996 -0.0000 -0.0004 0.0011
+ - param set #18 has cost=-0.023730 [o=15 t=20]
+ -- Parameters = 70.3057 14.5895 -34.2406 22.0394 2.1720 6.6711 0.9802 1.0036 1.0013 0.0007 -0.0022 0.0029
+ - param set #19 has cost=-0.023631 [o=32 t=18]
+ -- Parameters = 100.4152 7.7533 -34.4805 -10.3798 -5.0955 5.4212 0.9951 0.9859 1.0052 -0.0007 0.0006 -0.0022
+ - param set #20 has cost=-0.026488 [o=34 t=11]
+ -- Parameters = 89.9109 -10.9302 -36.3299 8.0323 0.1715 13.0375 0.9911 0.9839 0.9539 0.0029 -0.0008 -0.0014
+ - param set #21 has cost=-0.025710 [o=25 t=28]
+ -- Parameters = 74.7761 16.0666 -34.5398 17.9324 1.0952 8.7630 0.9967 0.9761 1.0017 -0.0008 -0.0010 0.0013
+ - param set #22 has cost=-0.023329 [o=12 t=14]
+ -- Parameters = 99.7687 14.5075 -32.5438 -14.4133 2.6752 7.1660 0.9991 1.0085 0.9939 0.0036 -0.0018 0.0012
+ - param set #23 has cost=-0.021950 [o=76 t=13]
+ -- Parameters = 80.2883 -11.3077 -33.1460 32.4022 6.5306 6.6403 1.0011 1.0302 0.9984 0.0006 0.0011 0.0005
+ - param set #24 has cost=-0.022690 [o=45 t=23]
+ -- Parameters = 101.9252 7.1582 -40.8989 -21.8177 -16.0946 5.4748 0.9833 0.9740 0.9848 0.0004 0.0017 -0.0006
+ - param set #25 has cost=-0.022535 [o=23 t=27]
+ -- Parameters = 84.9013 -28.8441 -6.9070 26.3435 22.8799 21.0003 0.9813 0.9976 1.0024 -0.0010 -0.0003 0.0003
+ - param set #26 has cost=-0.019424 [o=27 t=7]
+ -- Parameters = 87.8294 11.0414 -34.6302 -29.4960 -11.0203 37.2879 0.9949 0.9964 1.0015 0.0049 -0.0008 -0.0009
+ - param set #27 has cost=-0.021469 [o=3 t=15]
+ -- Parameters = 99.2357 11.8658 -39.5413 -21.6338 -44.4072 30.8577 0.9829 0.9933 1.0042 -0.0042 -0.0010 -0.0010
+ - param set #28 has cost=-0.020528 [o=1 t=25]
+ -- Parameters = 68.7270 15.5653 -31.0707 20.3887 7.6012 -8.6576 0.9944 0.9766 0.9728 -0.0059 0.0048 -0.0027
+ - param set #29 has cost=-0.022276 [o=56 t=19]
+ -- Parameters = 95.7599 16.7036 -29.0202 -26.5490 -10.9095 37.8798 0.9701 0.9857 0.9929 -0.0013 -0.0022 0.0156
+ - param set #30 has cost=-0.017713 [o=57 t=5]
+ -- Parameters = 91.8026 5.0903 -37.2217 -10.0734 -1.2333 31.7567 0.9999 0.9987 0.9996 -0.0002 -0.0003 0.0011
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0279
+ --- dist(#3,#1) = 0.41
+ --- dist(#4,#1) = 0.466
+ --- dist(#5,#1) = 0.499
+ --- dist(#6,#1) = 0.417
+ --- dist(#7,#1) = 0.457
+ --- dist(#8,#1) = 0.4
+ --- dist(#9,#1) = 0.373
+ --- dist(#10,#1) = 0.484
+ --- dist(#11,#1) = 0.443
+ --- dist(#12,#1) = 0.417
+ --- dist(#13,#1) = 0.421
+ --- dist(#14,#1) = 0.39
+ --- dist(#15,#1) = 0.401
+ --- dist(#16,#1) = 0.382
+ --- dist(#17,#1) = 0.479
+ --- dist(#18,#1) = 0.39
+ --- dist(#19,#1) = 0.502
+ --- dist(#20,#1) = 0.417
+ --- dist(#21,#1) = 0.42
+ --- dist(#22,#1) = 0.396
+ --- dist(#23,#1) = 0.5
+ --- dist(#24,#1) = 0.31
+ --- dist(#25,#1) = 0.419
+ --- dist(#26,#1) = 0.403
+ --- dist(#27,#1) = 0.487
+ --- dist(#28,#1) = 0.377
+ --- dist(#29,#1) = 0.422
+ --- dist(#30,#1) = 0.454
+ - Total coarse refinement net CPU time = 0.0 s; 9054 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
#Script is running (command trimmed):
3dinfo ./__tt_sub-01_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/__tt_sub-01_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ !source mask fill: ubot=65 usiz=160
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 29469 total points stored in 62 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.253161 * [o=-1 t=-1]
+ -- Parameters = 85.8750 -4.0688 -0.8970 -0.2495 -0.6040 0.1950 0.9935 1.0212 0.9771 -0.0105 0.0010 -0.0317
+ - cost(#2)=-0.252994 [o=0 t=0]
+ -- Parameters = 85.9845 -4.0432 -0.7949 -0.3680 -0.5875 -0.0309 0.9930 1.0195 0.9804 -0.0117 0.0072 -0.0288
+ - cost(#3)=-0.028934 [o=29 t=4]
+ -- Parameters = 80.0031 1.4205 -33.6958 -19.6550 3.2867 -8.2239 0.9853 0.9954 0.9929 0.0093 0.0025 -0.0004
+ - cost(#4)=-0.028557 [o=10 t=2]
+ -- Parameters = 77.3330 3.7400 -38.1765 -14.0977 6.5079 0.3764 1.0016 0.9925 0.9804 -0.0070 0.0018 -0.0002
+ - cost(#5)=-0.025908 [o=81 t=21]
+ -- Parameters = 81.7000 14.7044 -40.7886 4.5173 -17.1908 4.5806 1.0116 0.9860 0.9807 -0.0037 -0.0039 -0.0018
+ - cost(#6)=-0.028722 [o=66 t=8]
+ -- Parameters = 96.0372 2.0882 -34.2920 17.2826 2.7498 0.1803 0.9846 0.9988 0.9920 -0.0036 -0.0049 -0.0041
+ - cost(#7)=-0.027012 [o=4 t=3]
+ -- Parameters = 76.5845 6.4197 -37.4816 -16.0930 7.0144 3.0146 0.9908 0.9834 0.9799 0.0131 0.0012 0.0038
+ - cost(#8)=-0.027560 [o=11 t=12]
+ -- Parameters = 100.4787 5.3414 -32.9088 15.0706 4.2862 -3.9919 0.9624 0.9993 0.9677 0.0199 0.0028 0.0008
+ - cost(#9)=-0.025875 [o=2 t=24]
+ -- Parameters = 89.9631 -12.5681 -30.7253 9.5585 4.6659 15.2663 1.0185 0.9762 0.9856 0.0001 -0.0002 0.0008
+ - cost(#10)=-0.026038 [o=24 t=1]
+ -- Parameters = 79.3928 6.1858 -39.5952 -20.6257 1.6447 1.2581 1.0014 1.0081 0.9972 0.0007 -0.0012 0.0004
+ - cost(#11)=-0.025894 [o=34 t=11]
+ -- Parameters = 89.9109 -10.9302 -36.3299 8.0323 0.1715 13.0375 0.9911 0.9839 0.9539 0.0029 -0.0008 -0.0014
+ - cost(#12)=-0.025662 [o=26 t=16]
+ -- Parameters = 84.1430 2.8310 -34.2967 -17.0779 6.3930 -10.8819 0.9999 1.0055 0.9978 0.0005 0.0015 0.0011
+ - cost(#13)=-0.023823 [o=25 t=28]
+ -- Parameters = 74.7761 16.0666 -34.5398 17.9324 1.0952 8.7630 0.9967 0.9761 1.0017 -0.0008 -0.0010 0.0013
+ - cost(#14)=-0.024896 [o=22 t=9]
+ -- Parameters = 96.8633 1.9202 -32.0888 6.6220 7.1530 6.5086 0.9992 1.0008 0.9996 -0.0000 -0.0004 0.0011
+ - cost(#15)=-0.023463 [o=63 t=6]
+ -- Parameters = 68.9062 16.8701 -32.9880 30.7197 7.2224 -1.0839 1.0416 0.9716 0.9927 0.0065 -0.0049 -0.0011
+ - cost(#16)=-0.023772 [o=14 t=17]
+ -- Parameters = 95.1534 15.3266 -30.8907 -34.6087 -12.3411 33.8811 0.9729 0.9856 0.9846 0.0145 0.0101 0.0040
+ - cost(#17)=-0.023740 [o=87 t=10]
+ -- Parameters = 75.7266 4.4363 -39.2202 -15.8438 -9.1852 -8.0000 0.9873 0.9882 0.9837 0.0006 0.0042 -0.0080
+ - cost(#18)=-0.023615 [o=70 t=22]
+ -- Parameters = 70.1939 16.9033 -32.0769 33.1289 6.4620 -4.7010 1.0209 0.9628 0.9802 0.0035 0.0094 0.0017
+ - cost(#19)=-0.022488 [o=78 t=26]
+ -- Parameters = 103.0684 9.7956 -41.0687 -24.7673 -9.3139 14.5054 0.9214 0.9503 0.9674 -0.0053 -0.0060 0.0138
+ - cost(#20)=-0.022027 [o=15 t=20]
+ -- Parameters = 70.3057 14.5895 -34.2406 22.0394 2.1720 6.6711 0.9802 1.0036 1.0013 0.0007 -0.0022 0.0029
+ - cost(#21)=-0.022102 [o=32 t=18]
+ -- Parameters = 100.4152 7.7533 -34.4805 -10.3798 -5.0955 5.4212 0.9951 0.9859 1.0052 -0.0007 0.0006 -0.0022
+ - cost(#22)=-0.021824 [o=12 t=14]
+ -- Parameters = 99.7687 14.5075 -32.5438 -14.4133 2.6752 7.1660 0.9991 1.0085 0.9939 0.0036 -0.0018 0.0012
+ - cost(#23)=-0.021088 [o=45 t=23]
+ -- Parameters = 101.9252 7.1582 -40.8989 -21.8177 -16.0946 5.4748 0.9833 0.9740 0.9848 0.0004 0.0017 -0.0006
+ - cost(#24)=-0.019439 [o=23 t=27]
+ -- Parameters = 84.9013 -28.8441 -6.9070 26.3435 22.8799 21.0003 0.9813 0.9976 1.0024 -0.0010 -0.0003 0.0003
+ - cost(#25)=-0.020328 [o=56 t=19]
+ -- Parameters = 95.7599 16.7036 -29.0202 -26.5490 -10.9095 37.8798 0.9701 0.9857 0.9929 -0.0013 -0.0022 0.0156
+ - cost(#26)=-0.019267 [o=76 t=13]
+ -- Parameters = 80.2883 -11.3077 -33.1460 32.4022 6.5306 6.6403 1.0011 1.0302 0.9984 0.0006 0.0011 0.0005
+ - cost(#27)=-0.019889 [o=3 t=15]
+ -- Parameters = 99.2357 11.8658 -39.5413 -21.6338 -44.4072 30.8577 0.9829 0.9933 1.0042 -0.0042 -0.0010 -0.0010
+ - cost(#28)=-0.018973 [o=1 t=25]
+ -- Parameters = 68.7270 15.5653 -31.0707 20.3887 7.6012 -8.6576 0.9944 0.9766 0.9728 -0.0059 0.0048 -0.0027
+ - cost(#29)=-0.018473 [o=27 t=7]
+ -- Parameters = 87.8294 11.0414 -34.6302 -29.4960 -11.0203 37.2879 0.9949 0.9964 1.0015 0.0049 -0.0008 -0.0009
+ - cost(#30)=-0.017367 [o=57 t=5]
+ -- Parameters = 91.8026 5.0903 -37.2217 -10.0734 -1.2333 31.7567 0.9999 0.9987 0.9996 -0.0002 -0.0003 0.0011
+ - cost(#31)=-0.184042 [o=-2 t=-2]
+ -- Parameters = 85.7192 -4.7679 -1.6647 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ Forming automask
+ Fixed clip level = 347.017487
+ -num_rtb 99 ==> refine all 31 cases
+ Used gradual clip level = 336.052002 .. 362.471405
+ Number voxels above clip level = 38060
+ Clustering voxels ...
+ Largest cluster has 37782 voxels
+ - cost(#1)=-0.253988 * [o=-1 t=-1]
+ -- Parameters = 85.8651 -3.9740 -0.8436 -0.2531 -0.7855 0.2793 0.9931 1.0178 0.9760 -0.0085 0.0024 -0.0347
+ - cost(#2)=-0.253662 [o=0 t=0]
+ -- Parameters = 85.9780 -3.9673 -0.8145 -0.3458 -0.6019 0.0386 0.9942 1.0187 0.9776 -0.0123 0.0076 -0.0300
+ - cost(#3)=-0.029081 [o=29 t=4]
+ -- Parameters = 80.0167 1.3990 -33.6683 -19.7076 3.3114 -8.2401 0.9831 0.9957 0.9929 0.0100 0.0014 -0.0005
+ Clustering voxels ...
+ Largest cluster has 37296 voxels
+ Filled 298 voxels in small holes; now have 37594 voxels
+ Clustering voxels ...
+ Largest cluster has 37594 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 126246 voxels
+ Mask now has 37594 voxels
++ 37594 voxels in the mask [out of 163840: 22.95%]
++ first 10 x-planes are zero [from L]
++ last 10 x-planes are zero [from R]
++ first 2 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 5 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
+ - cost(#4)=-0.028654 [o=10 t=2]
+ -- Parameters = 77.3701 3.7907 -38.2140 -14.1253 6.4696 0.3288 1.0023 0.9925 0.9803 -0.0137 0.0018 0.0002
+ - cost(#5)=-0.026777 [o=81 t=21]
+ -- Parameters = 81.9692 14.5431 -41.0561 5.0366 -17.4943 4.6737 1.0145 0.9890 0.9881 -0.0032 -0.0043 -0.0024
+ - cost(#6)=-0.028780 [o=66 t=8]
+ -- Parameters = 96.0748 2.0948 -34.2916 17.3132 2.7689 0.1456 0.9854 0.9980 0.9916 -0.0036 -0.0049 -0.0043
+ - cost(#7)=-0.027112 [o=4 t=3]
+ -- Parameters = 76.6310 6.3971 -37.4413 -16.1191 7.0182 2.9906 0.9911 0.9837 0.9798 0.0145 0.0021 0.0037
#++ Applying threshold of 844.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/844.000000))'
+ - cost(#8)=-0.028788 [o=11 t=12]
+ -- Parameters = 99.4677 4.7591 -33.4903 16.0414 4.7541 -4.7535 0.9579 0.9913 0.9675 0.0229 -0.0023 0.0011
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - cost(#9)=-0.026669 [o=2 t=24]
+ -- Parameters = 89.9428 -12.3464 -30.6047 9.5369 4.6400 15.2616 1.0188 0.9792 0.9873 -0.0074 -0.0004 0.0005
+ - cost(#10)=-0.026424 [o=24 t=1]
+ -- Parameters = 79.3892 6.3359 -39.5860 -20.1813 1.8145 1.3244 0.9993 1.0100 0.9958 0.0019 -0.0011 0.0000
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-01_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-01_T1w_ns+orig -prefix ./sub-01_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-01_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
+ - cost(#11)=-0.026686 [o=34 t=11]
+ -- Parameters = 89.6822 -10.5191 -36.5934 7.6179 0.5552 13.2331 0.9960 0.9825 0.9512 0.0054 -0.0005 -0.0017
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - cost(#12)=-0.025990 [o=26 t=16]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
+ -- Parameters = 83.9891 2.7257 -34.2250 -17.2188 6.6464 -10.8170 0.9990 1.0085 0.9985 0.0005 0.0006 0.0014
++ Source dataset: ./__tt_sub-01_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
+ - cost(#13)=-0.023915 [o=25 t=28]
+ -- Parameters = 74.8379 16.0762 -34.5736 17.8611 1.0585 8.7806 0.9986 0.9766 1.0018 -0.0011 -0.0018 0.0012
+ - cost(#14)=-0.024989 [o=22 t=9]
+ -- Parameters = 96.9030 1.8287 -32.0167 6.5877 7.2032 6.5307 0.9987 1.0007 0.9995 0.0018 0.0000 0.0009
+ - cost(#15)=-0.023598 [o=63 t=6]
+ -- Parameters = 68.7846 16.8162 -32.9431 30.5875 7.1739 -1.0367 1.0437 0.9723 0.9931 0.0065 -0.0050 -0.0010
+ - cost(#16)=-0.026296 [o=14 t=17]
+ -- Parameters = 95.4134 15.0857 -30.7224 -34.9143 -12.1185 34.4150 0.9748 0.9780 1.0130 0.0169 0.0098 0.0060
+ - cost(#17)=-0.024199 [o=87 t=10]
+ -- Parameters = 75.6378 4.5250 -39.1938 -15.7224 -9.1151 -8.0567 0.9861 0.9879 0.9827 0.0011 0.0112 -0.0078
+ - cost(#18)=-0.023871 [o=70 t=22]
+ -- Parameters = 69.7978 17.2293 -31.8050 33.0910 6.8472 -4.5704 1.0270 0.9650 0.9807 0.0076 0.0061 0.0027
+ - cost(#19)=-0.023098 [o=78 t=26]
+ -- Parameters = 103.2358 9.8564 -40.6256 -24.8366 -9.1025 14.3938 0.9187 0.9517 0.9673 -0.0068 -0.0064 0.0145
+ - cost(#20)=-0.022097 [o=15 t=20]
+ -- Parameters = 70.2784 14.5685 -34.2173 22.0527 2.1525 6.6984 0.9798 1.0031 1.0019 0.0004 -0.0030 0.0038
+ - cost(#21)=-0.022246 [o=32 t=18]
+ -- Parameters = 100.5255 7.7437 -34.4544 -10.4219 -5.0968 5.4251 0.9951 0.9842 1.0056 -0.0006 0.0007 -0.0020
+ - cost(#22)=-0.021868 [o=12 t=14]
+ -- Parameters = 99.7759 14.4720 -32.5730 -14.4447 2.6697 7.1771 0.9992 1.0090 0.9935 0.0037 -0.0019 0.0012
+ - cost(#23)=-0.021145 [o=45 t=23]
+ -- Parameters = 101.9082 7.1523 -40.9041 -21.8551 -16.0982 5.5015 0.9831 0.9740 0.9848 0.0004 0.0019 -0.0014
+ - cost(#24)=-0.019519 [o=23 t=27]
+ -- Parameters = 84.8226 -28.9011 -6.8806 26.2852 22.8602 21.0193 0.9823 0.9970 1.0022 -0.0011 -0.0004 0.0008
+ - cost(#25)=-0.021616 [o=56 t=19]
+ -- Parameters = 95.8096 16.8026 -29.4516 -26.9141 -11.4907 38.3361 0.9711 0.9796 0.9951 -0.0018 -0.0017 0.0182
+ - cost(#26)=-0.019318 [o=76 t=13]
+ -- Parameters = 80.2677 -11.3174 -33.1575 32.4045 6.5112 6.6060 1.0017 1.0302 0.9984 0.0013 0.0010 0.0005
+ - cost(#27)=-0.020005 [o=3 t=15]
+ -- Parameters = 99.4163 11.8431 -39.4840 -21.6759 -44.4155 30.8084 0.9814 0.9936 1.0045 -0.0044 -0.0004 -0.0012
+ - cost(#28)=-0.019426 [o=1 t=25]
+ -- Parameters = 68.5242 15.3979 -31.1142 20.2512 7.6334 -8.4231 0.9927 0.9796 0.9719 -0.0068 0.0113 -0.0015
+ - cost(#29)=-0.018496 [o=27 t=7]
+ -- Parameters = 87.8340 11.0447 -34.6311 -29.5483 -11.0456 37.2661 0.9947 0.9963 1.0016 0.0050 -0.0007 -0.0001
+ - cost(#30)=-0.018647 [o=57 t=5]
+ -- Parameters = 92.1738 3.7303 -36.1056 -9.9395 -1.2157 33.1077 0.9977 1.0002 0.9968 -0.0016 0.0005 0.0023
+ - cost(#31)=-0.252853 [o=-2 t=-2]
+ -- Parameters = 85.9834 -3.9723 -0.7869 -0.4481 -0.4847 0.0833 0.9930 1.0207 0.9786 -0.0159 0.0055 -0.0265
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.253988
+ - Initial fine Parameters = 85.8651 -3.9740 -0.8436 -0.2531 -0.7855 0.2793 0.9931 1.0178 0.9760 -0.0085 0.0024 -0.0347
+ - Finalish cost = -0.254340 ; 476 funcs
+ - ini Finalish Parameters = 85.9194 -4.0162 -0.7897 -0.3149 -0.8523 0.2010 0.9941 1.0177 0.9760 -0.0108 0.0042 -0.0390
+ - Final cost = -0.254340 ; 266 funcs
+ Final fine fit Parameters:
x-shift= 85.9191 y-shift= -4.0158 z-shift= -0.7888 ... enorm= 86.0165 mm
z-angle= -0.3149 x-angle= -0.8517 y-angle= 0.2032 ... total= 0.9309 deg
x-scale= 0.9941 y-scale= 1.0177 z-scale= 0.9760 ... vol3D= 0.9874 = base bigger than source
y/x-shear= -0.0108 z/x-shear= 0.0042 z/y-shear= -0.0390
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 85.9191 -4.0158 -0.7888 -0.3149 -0.8517 0.2032 0.9941 1.0177 0.9760 -0.0108 0.0042 -0.0390
++ Unloading unneeded data
++ Output dataset ./sub-03_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-03_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 20.1
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/__tt_sub-03_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/sub-03_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-03_T1w_ns+orig sub-03_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-03_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-03_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-03_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-03_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o++o-.o-o++.oo.oooo+.+-+.+-.-o+o.-+++++-+----o$oo----+- + - best 88 costs found:
o= 0 v=-0.103375: 86.50 -0.10 -2.00 0.00 0.00 0.00 [grid]
o= 1 v=-0.032802: 119.53 -5.12 -38.82 -11.98 4.28 -19.20 [rand]
o= 2 v=-0.032759: 67.39 23.31 -35.30 37.88 13.88 18.42 [rand]
o= 3 v=-0.031198: 113.35 -13.20 -34.58 -39.07 28.37 -8.01 [rand]
o= 4 v=-0.028305: 111.47 -29.15 -37.45 17.69 14.58 -34.92 [rand]
o= 5 v=-0.027403: 88.80 -7.28 -33.69 -10.97 18.37 -5.04 [rand]
o= 6 v=-0.027226: 88.80 -7.28 -33.69 -10.97 18.37 5.04 [rand]
o= 7 v=-0.027089: 66.25 27.18 -36.49 37.90 5.03 3.08 [rand]
o= 8 v=-0.026566: 119.53 -5.12 -38.82 -11.98 -4.28 -19.20 [rand]
o= 9 v=-0.026444: 84.21 -7.28 -33.69 -10.97 18.37 -5.04 [rand]
o=10 v=-0.025807: 88.80 -7.28 -33.69 10.97 18.37 5.04 [rand]
o=11 v=-0.025345: 84.21 -7.28 -33.69 -10.97 18.37 5.04 [rand]
o=12 v=-0.024803: 93.17 6.56 -28.67 -7.50 30.00 30.00 [grid]
o=13 v=-0.023827: 75.01 16.21 -35.75 19.17 13.96 2.56 [rand]
o=14 v=-0.023515: 93.17 -6.77 -28.67 7.50 7.50 7.50 [grid]
o=15 v=-0.023070: 59.66 -13.20 -34.58 39.07 28.37 8.01 [rand]
o=16 v=-0.022594: 66.25 27.18 -36.49 37.90 5.03 -3.08 [rand]
o=17 v=-0.022556: 84.21 -7.28 -33.69 10.97 18.37 5.04 [rand]
o=18 v=-0.022489: 79.84 6.56 -28.67 -7.50 7.50 -7.50 [grid]
o=19 v=-0.022190: 103.27 -20.88 -31.28 -12.96 3.05 -9.17 [rand]
o=20 v=-0.022076: 113.17 6.56 -28.67 -30.00 7.50 7.50 [grid]
o=21 v=-0.022070: 93.17 -6.77 -28.67 -30.00 7.50 -7.50 [grid]
o=22 v=-0.021902: 107.94 -20.08 -25.62 -27.55 25.32 -7.37 [rand]
o=23 v=-0.021463: 84.21 7.08 -33.69 10.97 18.37 5.04 [rand]
o=24 v=-0.021380: 119.53 -5.12 -38.82 11.98 4.28 -19.20 [rand]
o=25 v=-0.021283: 115.52 -27.24 -35.61 7.44 27.42 -25.63 [rand]
o=26 v=-0.020887: 88.80 7.08 -33.69 10.97 18.37 5.04 [rand]
o=27 v=-0.020854: 65.06 19.88 -25.62 27.55 25.32 -7.37 [rand]
o=28 v=-0.020753: 54.30 28.94 34.14 40.41 -19.09 40.75 [rand]
o=29 v=-0.020681: 70.86 8.66 -35.83 23.16 17.17 -3.59 [rand]
o=30 v=-0.020517: 57.49 27.04 31.61 -7.44 -27.42 -25.63 [rand]
o=31 v=-0.020487: 59.84 6.56 -28.67 30.00 7.50 -7.50 [grid]
o=32 v=-0.020279: 69.73 -20.88 -31.28 12.96 -3.05 -9.17 [rand]
o=33 v=-0.020248: 84.21 7.08 -33.69 -10.97 18.37 -5.04 [rand]
o=34 v=-0.019670: 119.53 -5.12 -38.82 11.98 -4.28 -19.20 [rand]
o=35 v=-0.019349: 113.17 -6.77 -28.67 -30.00 7.50 -7.50 [grid]
o=36 v=-0.019262: 119.53 4.92 -38.82 -11.98 4.28 -19.20 [rand]
o=37 v=-0.019131: 93.17 -6.77 -28.67 -7.50 30.00 7.50 [grid]
o=38 v=-0.019004: 93.17 -26.77 -28.67 -7.50 7.50 -7.50 [grid]
o=39 v=-0.018855: 84.21 7.08 -33.69 -10.97 18.37 5.04 [rand]
o=40 v=-0.018693: 119.00 -27.09 -33.89 9.69 21.75 -40.77 [rand]
o=41 v=-0.018621: 103.30 18.36 -34.36 -10.10 -16.32 39.31 [rand]
o=42 v=-0.018510: 79.84 -6.77 -28.67 -7.50 7.50 -30.00 [grid]
o=43 v=-0.018413: 79.84 -6.77 -28.67 -7.50 7.50 -7.50 [grid]
o=44 v=-0.018369: 93.17 -6.77 -28.67 7.50 7.50 -7.50 [grid]
o=45 v=-0.018355: 59.57 -28.24 35.94 15.05 -4.71 12.78 [rand]
o=46 v=-0.018317: 79.84 -26.77 -28.67 7.50 30.00 -30.00 [grid]
o=47 v=-0.018118: 62.13 30.03 -35.45 27.62 21.02 5.11 [rand]
o=48 v=-0.018078: 69.73 20.67 -31.28 12.96 3.05 -9.17 [rand]
o=49 v=-0.017966: 59.84 -6.77 -28.67 30.00 30.00 -7.50 [grid]
o=50 v=-0.017929: 69.73 -20.88 -31.28 12.96 3.05 -9.17 [rand]
o=51 v=-0.017921: 77.90 -21.73 -30.71 14.86 11.51 26.00 [rand]
o=52 v=-0.017913: 113.17 6.56 -28.67 -30.00 -7.50 30.00 [grid]
o=53 v=-0.017815: 70.86 8.66 -35.83 23.16 17.17 3.59 [rand]
o=54 v=-0.017737: 69.73 -20.88 -31.28 12.96 3.05 9.17 [rand]
o=55 v=-0.017402: 93.17 6.56 -28.67 30.00 7.50 -7.50 [grid]
o=56 v=-0.017375: 49.91 27.20 29.24 25.97 -37.53 40.21 [rand]
o=57 v=-0.017368: 70.36 9.36 -26.35 20.99 15.73 -16.09 [rand]
o=58 v=-0.017275: 93.17 6.56 -28.67 -7.50 30.00 7.50 [grid]
o=59 v=-0.017211: 53.48 -5.12 -38.82 11.98 4.28 19.20 [rand]
o=60 v=-0.016938: 75.01 -16.41 -35.75 19.17 13.96 -2.56 [rand]
o=61 v=-0.016903: 69.71 18.36 30.36 10.10 -16.32 -39.31 [rand]
o=62 v=-0.016842: 115.78 -15.86 -31.86 13.45 19.96 -14.96 [rand]
o=63 v=-0.016810: 113.35 -13.20 -34.58 -39.07 28.37 8.01 [rand]
o=64 v=-0.016810: 79.84 -26.77 -28.67 7.50 7.50 -7.50 [grid]
o=65 v=-0.016427: 61.18 28.57 -25.99 42.33 -22.93 -32.02 [rand]
o=66 v=-0.016336: 93.17 6.56 -28.67 -7.50 7.50 -7.50 [grid]
o=67 v=-0.016323: 97.99 -16.41 -35.75 -19.17 13.96 2.56 [rand]
o=68 v=-0.016278: 93.17 -26.77 -8.67 7.50 30.00 7.50 [grid]
o=69 v=-0.016082: 65.06 -20.08 -25.62 27.55 25.32 7.37 [rand]
o=70 v=-0.016082: 80.09 -34.04 34.57 -19.28 -27.13 15.13 [rand]
o=71 v=-0.015999: 60.98 -31.44 -36.28 42.46 -41.07 -35.63 [rand]
o=72 v=-0.015997: 59.84 26.56 -28.67 30.00 -7.50 -30.00 [grid]
o=73 v=-0.015987: 93.17 -26.77 -28.67 -7.50 30.00 7.50 [grid]
o=74 v=-0.015984: 116.40 -35.11 -22.42 29.29 23.14 -9.37 [rand]
o=75 v=-0.015979: 75.01 16.21 -35.75 19.17 13.96 -2.56 [rand]
o=76 v=-0.015854: 72.51 -15.64 -37.68 14.61 39.00 28.71 [rand]
o=77 v=-0.015834: 119.53 4.92 -38.82 -11.98 -4.28 -19.20 [rand]
o=78 v=-0.015816: 64.51 29.33 -24.26 28.45 -9.40 -40.53 [rand]
o=79 v=-0.015762: 100.50 -15.64 33.68 -14.61 -39.00 -28.71 [rand]
o=80 v=-0.015757: 72.51 15.43 33.68 14.61 -39.00 -28.71 [rand]
o=81 v=-0.015662: 111.47 -29.15 33.45 17.69 -14.58 -34.92 [rand]
o=82 v=-0.015653: 95.11 -21.73 -30.71 -14.86 11.51 26.00 [rand]
o=83 v=-0.015559: 121.70 -34.79 -23.57 18.41 40.28 -7.49 [rand]
o=84 v=-0.015547: 57.52 15.16 -23.28 32.35 9.77 -6.77 [rand]
o=85 v=-0.015542: 119.53 4.92 -38.82 11.98 -4.28 -19.20 [rand]
o=86 v=-0.015509: 79.84 -6.77 -28.67 7.50 7.50 -30.00 [grid]
o=87 v=-0.015488: 69.73 20.67 -31.28 12.96 -3.05 -9.17 [rand]
Performing center alignment with @Align_Centers
++ 1523606 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=6 ytop=4
++ Zero-pad: zbot=8 ztop=3
++ 37594 voxels [15.6%] in weight mask
++ Output dataset ./__tt_sub-01_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 37594
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.930 34.452 27.782 (index)
+ source center of mass = 88.107 124.744 123.492 (index)
+ source-target CM = 86.505 -0.604 -5.138 (xyz)
+ estimated center of mass shifts = 86.505 -0.604 -5.138
++ shift param auto-range: 25.8..147.2 -70.9..69.7 -69.3..59.1
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.505249 .. 96.505249 center = 86.505249
+ Range param#2 [y-shift] = -10.604294 .. 9.395706 center = -0.604294
+ Range param#3 [z-shift] = -15.138100 .. 4.861900 center = -5.138100
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.505249 .. 126.505249 center = 86.505249
+ Range param#2 [y-shift] = -40.604294 .. 39.395706 center = -0.604294
+ Range param#3 [z-shift] = -45.138100 .. 34.861900 center = -5.138100
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000088
++ Final parameter search ranges:
+ x-shift = 46.505 .. 126.505
+ y-shift = -40.604 .. 39.396
+ z-shift = -45.138 .. 34.862
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1523606 [out of 11534336] voxels
+ base mask has 50457 [out of 241536] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ !source mask fill: ubot=29 usiz=173.5
+ - A little optimization:*[#16334=-0.113003] *[#16335=-0.116997] *[#16337=-0.132691] *[#16338=-0.136482] *[#16339=-0.138703] *[#16340=-0.139618] *[#16343=-0.147151] *[#16346=-0.1526] *[#16347=-0.152682] *[#16350=-0.153857] *[#16352=-0.156789] *[#16355=-0.15957] *[#16356=-0.159654] *[#16358=-0.160369] *[#16359=-0.160804] *[#16361=-0.160811] *[#16362=-0.161027] *[#16363=-0.161065] *[#16365=-0.161096] *[#16367=-0.161271] *[#16368=-0.161503] *[#16370=-0.161594] *[#16371=-0.161626] *[#16372=-0.161677] *[#16373=-0.1618] *[#16375=-0.16201] *[#16376=-0.162159] *[#16377=-0.162353] *[#16381=-0.162622] *[#16383=-0.16263] *[#16386=-0.162645] *[#16388=-0.162657] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.162657: 87.09 0.69 0.27 -1.44 2.08 0.68 [grid] [f=69]
o= 1 v=-0.045172: 119.60 -4.73 -37.77 -6.08 6.03 -17.29 [rand] [f=74]
o= 2 v=-0.052059: 66.34 24.14 -37.73 40.14 10.76 12.50 [rand] [f=54]
o= 3 v=-0.034277: 112.69 -13.66 -30.66 -39.46 28.87 -7.90 [rand] [f=33]
o= 4 v=-0.029619: 111.10 -29.25 -37.09 17.62 14.61 -34.36 [rand] [f=38]
o= 5 v=-0.037097: 87.01 -5.42 -32.81 -11.42 12.24 -1.61 [rand] [f=54]
o= 6 v=-0.030794: 88.61 -7.00 -32.75 -6.87 19.24 4.83 [rand] [f=51]
o= 7 v=-0.052209: 69.01 26.15 -37.00 39.66 12.28 4.78 [rand] [f=80]
o= 8 v=-0.039285: 119.41 -8.96 -37.47 -8.72 3.46 -19.43 [rand] [f=57]
o= 9 v=-0.032833: 87.29 -7.59 -32.55 -12.17 16.85 -3.87 [rand] [f=59]
o=10 v=-0.034647: 86.53 -3.63 -34.19 4.78 14.66 6.73 [rand] [f=74]
o=11 v=-0.034729: 88.51 -7.61 -33.51 -13.63 14.67 3.69 [rand] [f=44]
o=12 v=-0.032831: 92.66 6.50 -30.67 -9.24 24.21 29.08 [grid] [f=55]
o=13 v=-0.044784: 65.17 17.40 -34.42 19.34 13.54 7.66 [rand] [f=51]
o=14 v=-0.034147: 95.59 -5.49 -32.40 9.28 8.22 6.55 [grid] [f=48]
o=15 v=-0.032005: 59.24 -9.40 -34.24 38.46 26.80 9.61 [rand] [f=58]
o=16 v=-0.055139: 70.36 25.59 -35.92 36.42 11.89 4.60 [rand] [f=72]
o=17 v=-0.028132: 87.88 -7.04 -34.15 11.10 18.50 5.29 [rand] [f=42]
o=18 v=-0.035762: 79.02 5.28 -26.72 -9.04 14.53 -6.10 [grid] [f=53]
o=19 v=-0.029068: 103.99 -21.38 -30.74 -16.12 3.04 -8.30 [rand] [f=73]
o=20 v=-0.030704: 112.12 9.94 -31.53 -31.28 7.19 6.41 [grid] [f=70]
o=21 v=-0.030108: 94.04 -2.19 -32.33 -29.85 12.95 -8.07 [grid] [f=46]
o=22 v=-0.027249: 108.43 -18.76 -24.56 -26.93 25.86 -1.61 [rand] [f=52]
o=23 v=-0.028820: 83.94 6.29 -36.99 10.84 15.21 9.24 [rand] [f=67]
o=24 v=-0.034537: 113.00 -4.88 -38.20 10.11 1.38 -18.09 [rand] [f=49]
o=25 v=-0.027676: 119.40 -26.98 -35.26 7.69 27.32 -25.86 [rand] [f=39]
o=26 v=-0.028821: 96.87 6.37 -34.95 10.65 18.04 5.99 [rand] [f=54]
o=27 v=-0.026565: 65.89 18.69 -28.16 25.07 21.96 -9.89 [rand] [f=69]
o=28 v=-0.031677: 47.26 25.85 32.18 37.50 -24.07 41.56 [rand] [f=49]
o=29 v=-0.034179: 63.44 8.56 -34.11 28.10 12.05 -2.36 [rand] [f=56]
+ - copying weight image
o=30 v=-0.028788: 58.28 22.34 32.96 -8.40 -27.94 -24.59 [rand] [f=40]
+ - using 37594 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
o=31 v=-0.032051: 58.62 12.23 -28.63 31.45 10.56 -10.26 [grid] [f=50]
o=32 v=-0.025133: 73.44 -19.97 -32.11 12.80 -2.16 -8.46 [rand] [f=46]
o=33 v=-0.042186: 83.72 3.63 -34.67 -17.05 7.89 3.10 [rand] [f=71]
+ 30887 total points stored in 59 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
o=34 v=-0.032138: 113.36 -4.62 -38.82 9.69 -2.13 -17.41 [rand] [f=60]
o=35 v=-0.032647: 114.21 -7.22 -31.31 -25.63 7.00 -11.70 [grid] [f=54]
o=36 v=-0.044604: 119.80 -3.92 -36.46 -9.16 7.53 -13.03 [rand] [f=55]
o=37 v=-0.039579: 91.39 -16.33 -28.13 -9.66 31.78 8.59 [grid] [f=64]
o=38 v=-0.025362: 92.08 -23.60 -28.68 -6.60 2.83 -7.77 [grid] [f=44]
o=39 v=-0.042159: 81.86 4.38 -37.76 -14.77 2.28 -0.92 [rand] [f=61]
o=40 v=-0.029260: 118.35 -29.44 -34.21 15.40 23.18 -29.72 [rand] [f=59]
o=41 v=-0.025683: 102.13 18.45 -35.38 -9.13 -12.09 39.33 [rand] [f=34]
o=42 v=-0.026349: 78.22 -9.17 -34.68 -9.14 8.25 -30.38 [grid] [f=52]
o=43 v=-0.029326: 75.86 -7.76 -29.34 -7.11 7.94 -6.53 [grid] [f=47]
o=44 v=-0.029918: 92.89 -5.85 -31.91 6.88 8.11 -2.08 [grid] [f=48]
o=45 v=-0.022671: 63.26 -27.92 36.33 13.35 -4.23 12.92 [rand] [f=58]
o=46 v=-0.020880: 80.03 -27.65 -24.38 6.42 30.23 -30.06 [grid] [f=54]
o=47 v=-0.051698: 69.48 23.42 -35.72 32.07 13.04 3.71 [rand] [f=66]
o=48 v=-0.042812: 67.92 17.81 -29.76 23.15 8.16 -2.99 [rand] [f=64]
o=49 v=-0.027585: 61.29 -6.19 -29.34 37.39 29.79 -5.46 [grid] [f=54]
o=50 v=-0.026936: 73.08 -19.75 -30.65 13.68 1.28 -10.10 [rand] [f=83]
o=51 v=-0.022348: 73.97 -22.52 -31.66 15.29 10.79 25.58 [rand] [f=46]
o=52 v=-0.035062: 112.70 4.21 -34.35 -34.32 -8.47 31.06 [grid] [f=41]
o=53 v=-0.042413: 61.83 12.90 -33.99 22.97 15.72 1.69 [rand] [f=72]
o=54 v=-0.023111: 67.26 -20.08 -31.17 12.30 2.68 8.62 [rand] [f=68]
o=55 v=-0.023736: 90.75 6.56 -31.38 29.98 11.06 -4.03 [grid] [f=83]
o=56 v=-0.036892: 50.65 27.00 32.48 31.72 -29.61 36.88 [rand] [f=46]
o=57 v=-0.027632: 65.12 11.07 -22.14 20.83 19.11 -15.72 [rand] [f=55]
o=58 v=-0.023384: 93.98 5.90 -30.14 -8.39 27.83 15.03 [grid] [f=46]
o=59 v=-0.022990: 50.63 -5.18 -39.25 10.93 3.97 17.83 [rand] [f=45]
o=60 v=-0.036163: 79.52 -18.21 -30.99 14.21 21.94 -4.99 [rand] [f=63]
o=61 v=-0.020956: 70.23 17.43 31.34 9.21 -18.34 -34.22 [rand] [f=63]
o=62 v=-0.026468: 116.83 -22.04 -31.65 7.23 19.48 -15.50 [rand] [f=47]
o=63 v=-0.027933: 113.45 -12.69 -27.79 -38.26 25.47 6.65 [rand] [f=44]
o=64 v=-0.026956: 80.15 -22.65 -27.07 8.02 6.33 -7.04 [grid] [f=50]
o=65 v=-0.025116: 57.95 28.09 -30.35 40.82 -24.23 -29.68 [rand] [f=55]
o=66 v=-0.034499: 92.52 5.24 -33.74 -8.03 6.47 -1.89 [grid] [f=40]
o=67 v=-0.029250: 94.24 -14.97 -31.91 -17.10 14.77 5.24 [rand] [f=64]
o=68 v=-0.020770: 93.60 -30.98 -8.45 8.54 29.91 6.59 [grid] [f=60]
o=69 v=-0.033861: 64.22 -14.01 -31.28 25.46 28.67 11.86 [rand] [f=50]
o=70 v=-0.022919: 80.64 -33.98 37.99 -20.93 -26.81 14.23 [rand] [f=56]
o=71 v=-0.019120: 61.65 -34.58 -39.31 42.10 -43.53 -36.54 [rand] [f=38]
o=72 v=-0.028378: 59.25 19.45 -29.08 39.28 -8.61 -30.71 [grid] [f=53]
o=73 v=-0.040590: 90.22 -18.61 -28.00 -6.33 29.13 8.84 [grid] [f=75]
o=74 v=-0.025207: 124.16 -37.37 -25.51 30.60 18.12 -12.55 [rand] [f=46]
o=75 v=-0.045007: 64.39 19.83 -35.46 25.62 13.68 10.42 [rand] [f=63]
o=76 v=-0.017271: 72.39 -15.25 -38.46 14.88 39.11 28.61 [rand] [f=42]
o=77 v=-0.031405: 120.58 -2.99 -39.18 -11.11 -1.89 -18.75 [rand] [f=76]
o=78 v=-0.024646: 65.08 25.59 -25.67 28.64 -9.51 -40.55 [rand] [f=49]
o=79 v=-0.022289: 101.04 -13.12 30.84 -7.42 -43.59 -28.14 [rand] [f=53]
o=80 v=-0.023150: 72.85 13.52 37.99 7.49 -40.41 -32.34 [rand] [f=60]
o=81 v=-0.019296: 111.12 -33.19 33.32 17.42 -14.28 -35.43 [rand] [f=36]
o=82 v=-0.043019: 88.98 -20.09 -29.42 -3.29 24.68 4.74 [rand] [f=70]
o=83 v=-0.021272: 121.25 -35.36 -19.27 16.44 41.21 -7.12 [rand] [f=60]
o=84 v=-0.035711: 58.01 14.93 -32.63 29.66 16.94 -3.22 [rand] [f=83]
o=85 v=-0.041535: 118.27 -4.58 -38.19 2.68 4.53 -18.21 [rand] [f=71]
o=86 v=-0.018354: 78.64 -7.08 -32.56 5.23 7.13 -30.27 [grid] [f=55]
o=87 v=-0.029125: 68.02 20.27 -31.31 18.46 1.49 -7.25 [rand] [f=71]
+ - saving # 0 for use with twobest
+ - saving #16 for use with twobest
+ - skip # 7 for twobest: too close to set #16
+ - saving # 2 for use with twobest
+ - skip #47 for twobest: too close to set #16
+ - saving # 1 for use with twobest
+ - saving #75 for use with twobest
+ - saving #13 for use with twobest
+ - skip #36 for twobest: too close to set # 1
+ - saving #82 for use with twobest
+ - saving #48 for use with twobest
+ - saving #53 for use with twobest
+ - saving #33 for use with twobest
+ - saving #39 for use with twobest
+ - saving #85 for use with twobest
+ - skip #73 for twobest: too close to set #82
+ - skip #37 for twobest: too close to set #73
+ - saving # 8 for use with twobest
+ - saving # 5 for use with twobest
+ - saving #56 for use with twobest
+ - saving #60 for use with twobest
+ - saving #18 for use with twobest
+ - saving #84 for use with twobest
+ - saving #52 for use with twobest
+ - saving #11 for use with twobest
+ - saving #10 for use with twobest
+ - saving #24 for use with twobest
+ - saving #66 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #29 for use with twobest
+ - saving #14 for use with twobest
+ - saving #69 for use with twobest
+ - saving # 9 for use with twobest
+ - saving #12 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=56 usiz=154.5
+ - retaining old weight image
+ - using 36584 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30268 total points stored in 61 'TOHD(17.3748)' bloks (0 duplicates)
*[#21358=-0.1975] *[#21369=-0.198158] *[#21387=-0.19934] *[#21390=-0.200457] *[#21391=-0.200469] *[#21397=-0.200671] *[#21398=-0.20124] *[#21401=-0.201905] *[#21406=-0.202971] *[#21410=-0.203339] *[#21411=-0.204989] *[#21412=-0.206032] *[#21413=-0.206965] *[#21414=-0.207495] *[#21417=-0.208247] *[#21418=-0.20851] *[#21421=-0.209251] *[#21422=-0.20942] *[#21423=-0.210656] *[#21426=-0.21118] *[#21427=-0.211258] *[#21428=-0.211775] *[#21429=-0.212482] *[#21432=-0.212989] *[#21433=-0.213542] *[#21434=-0.213803] *[#21437=-0.214108] *[#21442=-0.214626] *[#21449=-0.214931] *[#21452=-0.215859] *[#21453=-0.215948] *[#21454=-0.216383]
+ - param set #1 has cost=-0.216383 [o=0 t=0]
+ -- Parameters = 86.6831 0.3250 -0.0372 -0.6042 1.0144 0.6163 0.9754 1.0108 0.9777 0.0081 0.0107 -0.0177
+ - param set #2 has cost=-0.044223 [o=16 t=1]
+ -- Parameters = 70.2449 25.5184 -35.9114 36.4608 11.8691 4.5877 1.0016 1.0012 0.9999 0.0010 -0.0007 -0.0006
+ - param set #3 has cost=-0.045265 [o=2 t=2]
+ -- Parameters = 66.1566 23.6383 -38.2751 40.1868 10.3822 12.8581 0.9992 1.0008 0.9967 0.0011 -0.0010 -0.0005
+ - param set #4 has cost=-0.036790 [o=1 t=3]
+ -- Parameters = 119.3970 -4.7322 -37.7619 -6.7073 6.2867 -17.3936 1.0012 0.9988 0.9985 -0.0002 -0.0004 -0.0011
+ - param set #5 has cost=-0.035717 [o=75 t=4]
+ -- Parameters = 64.2770 19.5078 -35.6353 25.1971 13.3418 10.5538 1.0047 0.9989 1.0001 -0.0003 -0.0016 -0.0007
+ - param set #6 has cost=-0.037451 [o=13 t=5]
+ -- Parameters = 65.2737 17.4353 -34.5827 19.8397 13.5597 7.2686 0.9999 0.9949 0.9847 0.0004 -0.0006 0.0172
+ - param set #7 has cost=-0.032035 [o=82 t=6]
+ -- Parameters = 88.9254 -20.1146 -29.6879 -3.2167 24.6666 4.7886 0.9978 0.9978 0.9999 -0.0016 0.0006 0.0126
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-03_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
+ - param set #8 has cost=-0.034500 [o=48 t=7]
+ -- Parameters = 67.3156 18.0014 -29.8839 23.2557 7.8038 -2.6375 1.0040 0.9977 1.0022 0.0001 -0.0005 -0.0015
+ - param set #9 has cost=-0.032630 [o=53 t=8]
+ -- Parameters = 61.8040 12.8555 -33.9823 23.2533 15.7519 1.5205 0.9982 1.0010 0.9977 -0.0002 0.0014 -0.0013
+ - param set #10 has cost=-0.033250 [o=33 t=9]
+ -- Parameters = 83.7046 3.6480 -34.6809 -17.5786 7.6924 2.9043 1.0010 1.0035 0.9991 -0.0000 0.0002 0.0000
+ - param set #11 has cost=-0.034201 [o=39 t=10]
+ -- Parameters = 81.7103 4.3864 -37.9280 -14.7049 2.0979 -0.8996 0.9918 0.9948 1.0009 -0.0019 0.0005 -0.0009
+ - param set #12 has cost=-0.034103 [o=85 t=11]
+ -- Parameters = 118.2771 -4.5312 -38.1823 2.6510 4.5308 -18.2111 0.9999 0.9998 0.9998 0.0001 0.0001 0.0001
+ - param set #13 has cost=-0.031435 [o=8 t=12]
+ -- Parameters = 119.9406 -8.8760 -37.5239 -8.6131 3.7565 -19.1502 0.9965 1.0020 1.0097 -0.0000 0.0017 -0.0002
+ - param set #14 has cost=-0.027380 [o=5 t=13]
+ -- Parameters = 86.9422 -5.5173 -32.8254 -11.4186 12.1964 -1.5828 0.9998 1.0008 1.0001 0.0004 0.0006 -0.0006
+ - param set #15 has cost=-0.034684 [o=56 t=14]
+ -- Parameters = 50.9207 26.5828 33.0730 32.4755 -30.0917 36.7923 1.0048 0.9936 0.9987 0.0015 0.0015 0.0157
+ - param set #16 has cost=-0.028289 [o=60 t=15]
+ -- Parameters = 80.3202 -18.0686 -30.9873 13.7674 26.2174 -4.3151 1.0005 0.9990 0.9974 0.0007 0.0005 0.0002
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-03_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-03_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-03_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-03_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-03_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-03_T1w_ns_shft+tlrc.HEAD doesn't exist!
+ - param set #17 has cost=-0.030369 [o=18 t=16]
+ -- Parameters = 78.7563 4.9026 -26.7867 -8.7469 14.2377 -6.1675 0.9959 1.0008 1.0002 0.0120 0.0009 -0.0003
+ - param set #18 has cost=-0.028586 [o=84 t=17]
+ -- Parameters = 57.9419 15.0482 -32.3914 29.7544 17.1891 -3.0907 0.9930 1.0012 1.0009 0.0014 -0.0009 -0.0002
+ - param set #19 has cost=-0.031417 [o=52 t=18]
+ -- Parameters = 112.0966 3.9817 -34.8897 -34.4093 -7.9639 31.3480 0.9985 1.0042 0.9959 0.0002 0.0013 0.0140
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - param set #20 has cost=-0.026129 [o=11 t=19]
+ -- Parameters = 88.5581 -7.5950 -33.6988 -13.5529 15.0664 3.4357 0.9983 1.0020 0.9973 0.0011 -0.0010 0.0130
+ - param set #21 has cost=-0.026032 [o=10 t=20]
+ -- Parameters = 86.8910 -3.7122 -33.9972 5.3736 14.5635 6.4952 0.9971 1.0002 0.9999 0.0112 -0.0010 0.0004
+ - param set #22 has cost=-0.028343 [o=24 t=21]
+ -- Parameters = 113.2083 -4.9862 -38.3268 10.3640 0.9812 -18.7059 0.9924 0.9945 1.0055 -0.0007 0.0033 0.0015
+ - param set #23 has cost=-0.028711 [o=66 t=22]
+ -- Parameters = 92.2801 5.0943 -33.8652 -7.7928 6.3958 -1.9063 0.9985 1.0023 0.9979 0.0002 -0.0007 -0.0010
+ - param set #24 has cost=-0.028848 [o=3 t=23]
+ -- Parameters = 112.4810 -13.8448 -31.5142 -39.5188 28.6116 -8.7397 0.9914 1.0333 0.9988 -0.0005 0.0009 -0.0013
+ - param set #25 has cost=-0.027805 [o=29 t=24]
+ -- Parameters = 63.7116 8.3320 -34.3224 22.5670 11.9529 -2.3489 1.0022 1.0025 1.0032 0.0004 -0.0010 -0.0002
+ - param set #26 has cost=-0.028227 [o=14 t=25]
+ -- Parameters = 95.9094 -6.0533 -32.4679 8.9686 7.8156 6.5453 0.9977 0.9988 0.9988 0.0011 -0.0017 -0.0003
+ - param set #27 has cost=-0.028721 [o=69 t=26]
+ -- Parameters = 64.0170 -13.9774 -31.5337 25.4967 28.6335 12.1420 1.0021 0.9974 0.9955 -0.0005 0.0130 -0.0008
+ - param set #28 has cost=-0.028004 [o=9 t=27]
+ -- Parameters = 86.4560 -7.9715 -32.5231 -12.2010 11.5451 -2.9883 1.0012 0.9968 0.9805 0.0111 0.0012 0.0005
+ - param set #29 has cost=-0.023940 [o=12 t=28]
+ -- Parameters = 91.5939 6.2708 -24.9234 -9.4755 24.4521 30.4825 1.0078 0.9966 0.9796 0.0026 0.0028 0.0031
+ - param set #30 has cost=-0.205575 [o=-1 t=-1]
+ -- Parameters = 86.9995 -0.6612 -0.2034 -0.9183 1.2244 0.4380 0.9859 1.0521 0.9953 -0.0008 0.0121 -0.0052
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0593
+ --- dist(#3,#1) = 0.478
+ --- dist(#4,#1) = 0.448
+ --- dist(#5,#1) = 0.432
+ --- dist(#6,#1) = 0.472
+ --- dist(#7,#1) = 0.445
+ --- dist(#8,#1) = 0.447
+ --- dist(#9,#1) = 0.373
+ --- dist(#10,#1) = 0.474
+ --- dist(#11,#1) = 0.477
+ --- dist(#12,#1) = 0.433
+ --- dist(#13,#1) = 0.424
+ --- dist(#14,#1) = 0.371
+ --- dist(#15,#1) = 0.469
+ --- dist(#16,#1) = 0.436
+ --- dist(#17,#1) = 0.334
+ --- dist(#18,#1) = 0.432
+ --- dist(#19,#1) = 0.394
+ --- dist(#20,#1) = 0.423
+ --- dist(#21,#1) = 0.404
+ --- dist(#22,#1) = 0.479
+ --- dist(#23,#1) = 0.387
+ --- dist(#24,#1) = 0.405
+ --- dist(#25,#1) = 0.406
+ --- dist(#26,#1) = 0.429
+ --- dist(#27,#1) = 0.41
+ --- dist(#28,#1) = 0.421
+ --- dist(#29,#1) = 0.424
+ --- dist(#30,#1) = 0.332
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=56 usiz=154.5
+ - retaining old weight image
+ - using 36584 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30603 total points stored in 63 'TOHD(17.2644)' bloks (0 duplicates)
*[#24249=-0.230945] *[#24273=-0.231313] *[#24280=-0.231495] *[#24286=-0.231658] *[#24287=-0.231896] *[#24290=-0.231996] *[#24291=-0.232085] *[#24293=-0.232096] *[#24301=-0.232244] *[#24304=-0.232263] *[#24305=-0.232285] *[#24306=-0.232295] *[#24309=-0.232339] *[#24312=-0.232402] *[#24313=-0.232436] *[#24316=-0.232483] *[#24318=-0.23279] *[#24323=-0.232852] *[#24333=-0.232955] *[#24334=-0.23313] *[#24335=-0.233338] *[#24338=-0.233556] *[#24339=-0.233579] *[#24341=-0.233716] *[#24357=-0.233743]
+ - param set #1 has cost=-0.233743 [o=0 t=0]
+ -- Parameters = 86.7083 0.0970 0.0066 -0.6560 0.7684 0.3945 0.9828 1.0214 0.9795 -0.0009 0.0098 -0.0213
+ - param set #2 has cost=-0.229998 [o=-1 t=-1]
+ -- Parameters = 86.8311 -0.0796 -0.0525 -0.8458 1.1684 0.4678 0.9860 1.0282 0.9918 -0.0035 0.0103 -0.0119
+ - param set #3 has cost=-0.040040 [o=2 t=2]
+ -- Parameters = 66.0709 23.4179 -38.3479 40.0694 10.2088 12.9403 0.9949 1.0000 0.9961 0.0006 -0.0012 0.0080
+ - param set #4 has cost=-0.037308 [o=16 t=1]
+ -- Parameters = 69.9455 25.3146 -35.7219 36.4079 11.6869 4.4618 1.0001 0.9993 1.0086 0.0024 -0.0009 -0.0015
+ - param set #5 has cost=-0.030417 [o=13 t=5]
+ -- Parameters = 65.1010 17.7786 -34.6129 19.7639 13.7724 7.4129 0.9967 0.9905 0.9815 0.0017 -0.0012 0.0158
+ - param set #6 has cost=-0.029960 [o=1 t=3]
+ -- Parameters = 119.4330 -4.7654 -37.7168 -6.7443 6.2813 -17.4070 1.0006 0.9973 0.9989 -0.0002 -0.0005 -0.0010
+ - param set #7 has cost=-0.030098 [o=75 t=4]
+ -- Parameters = 64.2033 19.6452 -35.6762 25.1857 13.4890 10.4477 1.0069 0.9992 0.9981 -0.0007 -0.0024 -0.0007
+ - param set #8 has cost=-0.027799 [o=56 t=14]
+ -- Parameters = 50.7985 26.5607 32.9987 32.5229 -30.4898 35.9035 1.0065 0.9976 0.9924 0.0028 0.0055 0.0102
+ - param set #9 has cost=-0.029091 [o=48 t=7]
+ -- Parameters = 67.1294 18.0937 -29.8266 23.4030 7.6375 -2.5375 1.0004 0.9943 1.0036 -0.0009 -0.0015 -0.0033
+ - param set #10 has cost=-0.028479 [o=39 t=10]
+ -- Parameters = 81.8033 4.2790 -38.0123 -14.6925 2.1461 -1.0751 0.9923 0.9992 0.9997 -0.0013 0.0010 -0.0007
+ - param set #11 has cost=-0.028479 [o=85 t=11]
+ -- Parameters = 118.3099 -4.5322 -38.1955 2.5789 4.5225 -18.2208 1.0004 0.9995 1.0000 0.0003 -0.0001 0.0000
+ - param set #12 has cost=-0.027206 [o=33 t=9]
+ -- Parameters = 83.6800 3.8803 -34.6090 -17.4970 7.7161 2.9925 0.9997 1.0006 1.0002 0.0006 0.0007 0.0014
+ - param set #13 has cost=-0.025377 [o=53 t=8]
+ -- Parameters = 61.9101 12.9007 -34.1582 23.2052 15.7365 1.4793 0.9983 0.9994 0.9951 -0.0001 0.0012 -0.0012
+ - param set #14 has cost=-0.025103 [o=82 t=6]
+ -- Parameters = 89.8948 -20.7514 -30.0484 -3.8291 24.1973 4.2320 0.9981 0.9939 0.9898 -0.0072 0.0025 0.0192
+ - param set #15 has cost=-0.027899 [o=8 t=12]
+ -- Parameters = 119.1992 -8.0009 -37.7572 -5.4504 4.3637 -17.6152 1.0010 0.9864 1.0154 -0.0002 0.0023 0.0032
+ - param set #16 has cost=-0.025272 [o=52 t=18]
+ -- Parameters = 112.0758 3.9811 -34.9010 -34.6114 -8.0375 31.2941 1.0012 1.0041 0.9919 0.0007 0.0011 0.0129
+ - param set #17 has cost=-0.025175 [o=18 t=16]
+ -- Parameters = 78.7516 4.6176 -26.7892 -8.7131 14.0215 -6.2860 0.9958 1.0022 0.9997 0.0116 0.0087 0.0000
+ - param set #18 has cost=-0.023870 [o=3 t=23]
+ -- Parameters = 112.2062 -13.7368 -31.4210 -39.6818 28.7022 -8.8693 0.9884 1.0350 0.9991 -0.0018 0.0009 -0.0022
+ - param set #19 has cost=-0.024196 [o=69 t=26]
+ -- Parameters = 63.4036 -14.0366 -31.8824 25.4399 28.5785 12.5087 1.0122 1.0030 0.9901 0.0018 0.0104 -0.0023
+ - param set #20 has cost=-0.024252 [o=66 t=22]
+ -- Parameters = 92.0758 5.0869 -33.8748 -7.6138 6.3385 -1.9256 0.9982 1.0020 0.9990 0.0007 -0.0011 -0.0009
+ - param set #21 has cost=-0.027676 [o=84 t=17]
+ -- Parameters = 56.7514 14.9207 -31.1771 28.2174 18.4448 -1.8545 0.9687 0.9916 0.9768 0.0052 -0.0095 0.0013
+ - param set #22 has cost=-0.022233 [o=24 t=21]
+ -- Parameters = 113.2353 -4.9743 -38.3728 10.4200 1.0571 -18.8872 0.9876 0.9979 1.0062 -0.0020 0.0035 0.0022
+ - param set #23 has cost=-0.021348 [o=60 t=15]
+ -- Parameters = 80.5292 -18.1304 -31.0037 13.7924 26.1545 -4.3874 1.0002 0.9990 0.9970 0.0007 0.0004 0.0002
+ - param set #24 has cost=-0.023026 [o=14 t=25]
+ -- Parameters = 95.9424 -6.0181 -32.6696 8.9274 7.6029 6.8064 0.9937 0.9959 0.9982 0.0010 -0.0023 -0.0021
+ - param set #25 has cost=-0.022628 [o=9 t=27]
+ -- Parameters = 86.2913 -7.9522 -32.5745 -12.2791 11.5169 -2.9509 1.0003 0.9987 0.9804 0.0118 0.0022 0.0091
+ - param set #26 has cost=-0.022881 [o=29 t=24]
+ -- Parameters = 63.9106 8.3394 -34.5373 22.5319 11.8328 -2.2982 0.9956 0.9987 1.0091 0.0017 -0.0023 0.0000
+ - param set #27 has cost=-0.021681 [o=5 t=13]
+ -- Parameters = 86.7262 -5.5396 -32.8841 -11.6543 12.2711 -1.2931 1.0068 1.0023 0.9968 0.0001 0.0004 -0.0003
+ - param set #28 has cost=-0.020842 [o=11 t=19]
+ -- Parameters = 88.5374 -7.5342 -33.5630 -13.5232 15.1536 3.4409 0.9985 1.0011 0.9984 0.0012 -0.0012 0.0131
+ - param set #29 has cost=-0.020825 [o=10 t=20]
+ -- Parameters = 87.1201 -4.1423 -33.9799 5.3244 14.6539 6.5055 0.9960 0.9987 0.9975 0.0205 -0.0010 0.0003
+ - param set #30 has cost=-0.019040 [o=12 t=28]
+ -- Parameters = 91.6398 6.6350 -24.7315 -9.5572 24.6619 30.3399 1.0047 0.9921 0.9753 0.0018 0.0029 0.0110
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0423
+ --- dist(#3,#1) = 0.479
+ --- dist(#4,#1) = 0.447
+ --- dist(#5,#1) = 0.433
+ --- dist(#6,#1) = 0.446
+ --- dist(#7,#1) = 0.472
+ --- dist(#8,#1) = 0.373
+ --- dist(#9,#1) = 0.478
+ --- dist(#10,#1) = 0.475
+ --- dist(#11,#1) = 0.472
+ --- dist(#12,#1) = 0.449
+ --- dist(#13,#1) = 0.39
+ --- dist(#14,#1) = 0.433
+ --- dist(#15,#1) = 0.427
+ --- dist(#16,#1) = 0.436
+ --- dist(#17,#1) = 0.335
+ --- dist(#18,#1) = 0.376
+ --- dist(#19,#1) = 0.424
+ --- dist(#20,#1) = 0.399
+ --- dist(#21,#1) = 0.434
+ --- dist(#22,#1) = 0.408
+ --- dist(#23,#1) = 0.432
+ --- dist(#24,#1) = 0.407
+ --- dist(#25,#1) = 0.48
+ --- dist(#26,#1) = 0.411
+ --- dist(#27,#1) = 0.388
+ --- dist(#28,#1) = 0.42
+ --- dist(#29,#1) = 0.425
+ --- dist(#30,#1) = 0.333
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=56 usiz=154.5
+ - retaining old weight image
+ - using 36584 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30158 total points stored in 62 'TOHD(17.1973)' bloks (0 duplicates)
*[#27426=-0.257652] *[#27447=-0.257991] *[#27448=-0.258597] *[#27451=-0.258827] *[#27452=-0.258894] *[#27455=-0.25895] *[#27456=-0.258973] *[#27459=-0.259054] *[#27464=-0.259196] *[#27469=-0.259248] *[#27471=-0.259345] *[#27473=-0.259415] *[#27474=-0.259492] *[#27477=-0.259498] *[#27480=-0.259509] *[#27481=-0.259554] *[#27482=-0.259589] *[#27483=-0.259608] *[#27488=-0.259741] *[#27491=-0.259962] *[#27492=-0.259968] *[#27496=-0.260026] *[#27509=-0.260054] *[#27510=-0.260072] *[#27512=-0.260152] *[#27514=-0.260164] *[#27515=-0.260253] *[#27518=-0.260285] *[#27531=-0.260286] *[#27533=-0.260369] *[#27536=-0.260418] *[#27539=-0.260433]
+ - param set #1 has cost=-0.260433 [o=0 t=0]
+ -- Parameters = 86.7391 0.2599 -0.0294 -0.7628 0.4225 0.6636 0.9885 1.0151 0.9774 -0.0025 0.0043 -0.0299
+ - param set #2 has cost=-0.259653 [o=-1 t=-1]
+ -- Parameters = 86.6847 0.1486 -0.0739 -0.6643 0.4843 0.5306 0.9896 1.0200 0.9794 -0.0033 0.0069 -0.0265
+ - param set #3 has cost=-0.034830 [o=2 t=2]
+ -- Parameters = 66.2123 23.5093 -38.3808 39.8093 9.9964 12.9610 0.9948 0.9829 0.9933 0.0012 -0.0017 0.0072
+ - param set #4 has cost=-0.031428 [o=16 t=1]
+ -- Parameters = 70.0367 25.2737 -35.6587 36.3199 11.6337 4.4118 1.0004 0.9959 1.0091 0.0030 -0.0009 -0.0020
+ - param set #5 has cost=-0.026253 [o=13 t=5]
+ -- Parameters = 64.9775 19.1412 -34.6954 20.0127 14.8358 7.9163 0.9854 0.9900 0.9682 0.0011 -0.0002 0.0166
+ - param set #6 has cost=-0.024210 [o=75 t=4]
+ -- Parameters = 64.1843 19.8875 -35.5525 25.1687 13.6845 10.4226 1.0066 0.9985 0.9987 -0.0014 -0.0028 -0.0008
+ - param set #7 has cost=-0.026345 [o=1 t=3]
+ -- Parameters = 119.6281 -4.6363 -37.7501 -6.3316 6.5138 -17.2004 0.9997 0.9976 0.9949 -0.0005 0.0039 -0.0019
+ - param set #8 has cost=-0.022759 [o=48 t=7]
+ -- Parameters = 67.1825 18.1954 -29.8499 23.3147 7.7080 -2.4611 0.9996 0.9933 1.0034 -0.0016 -0.0015 -0.0032
+ - param set #9 has cost=-0.024308 [o=85 t=11]
+ -- Parameters = 118.4666 -4.5245 -38.3397 2.4266 4.4473 -18.2752 0.9997 0.9983 1.0002 0.0006 0.0000 0.0004
+ - param set #10 has cost=-0.025652 [o=39 t=10]
+ -- Parameters = 81.3263 4.3148 -37.2612 -14.2136 2.7303 -0.7687 0.9937 1.0087 1.0013 0.0002 0.0033 -0.0004
+ - param set #11 has cost=-0.024645 [o=8 t=12]
+ -- Parameters = 119.3091 -7.9336 -37.8074 -5.5422 4.3781 -17.7279 0.9970 0.9813 1.0150 -0.0002 0.0072 0.0033
+ - param set #12 has cost=-0.022977 [o=56 t=14]
+ -- Parameters = 50.7745 26.6202 32.9973 32.4598 -30.5752 35.8697 1.0087 0.9958 0.9930 0.0031 0.0062 0.0099
+ - param set #13 has cost=-0.024151 [o=84 t=17]
+ -- Parameters = 56.9201 15.8319 -31.2056 28.0291 18.6459 -1.6768 0.9659 0.9732 0.9728 0.0029 -0.0104 0.0004
+ - param set #14 has cost=-0.026840 [o=33 t=9]
+ -- Parameters = 82.7826 4.1891 -34.7917 -16.3925 6.7573 2.3442 0.9923 0.9858 0.9897 0.0003 0.0021 0.0014
+ - param set #15 has cost=-0.021254 [o=53 t=8]
+ -- Parameters = 62.0441 12.8825 -34.1527 22.8447 15.6465 1.8500 0.9966 0.9790 0.9967 0.0008 0.0007 -0.0001
+ - param set #16 has cost=-0.022670 [o=52 t=18]
+ -- Parameters = 112.1598 4.0064 -35.1184 -34.5497 -8.1465 31.1941 1.0041 1.0007 0.9887 0.0008 0.0072 0.0121
+ - param set #17 has cost=-0.024009 [o=18 t=16]
+ -- Parameters = 78.5408 4.1929 -26.4729 -8.6572 13.8287 -5.8158 0.9937 1.0018 0.9844 0.0203 0.0087 0.0004
+ - param set #18 has cost=-0.022541 [o=82 t=6]
+ -- Parameters = 89.9216 -20.6565 -29.7497 -3.8335 24.2854 4.1633 0.9928 0.9922 0.9918 -0.0073 0.0026 0.0189
+ - param set #19 has cost=-0.021303 [o=66 t=22]
+ -- Parameters = 90.5092 4.5347 -33.9519 -6.7415 5.5820 -1.1301 0.9992 1.0101 1.0077 -0.0041 0.0017 0.0090
+ - param set #20 has cost=-0.022569 [o=69 t=26]
+ -- Parameters = 63.6270 -14.0523 -31.9087 25.4494 28.5313 12.4582 1.0084 1.0046 0.9895 0.0024 0.0101 -0.0027
+ - param set #21 has cost=-0.021419 [o=3 t=23]
+ -- Parameters = 112.2073 -13.7212 -31.2241 -39.7042 28.7034 -8.8775 0.9884 1.0352 0.9988 -0.0020 0.0013 -0.0026
+ - param set #22 has cost=-0.019750 [o=14 t=25]
+ -- Parameters = 95.9926 -6.1628 -32.6906 9.0920 7.6291 6.6283 0.9951 0.9981 0.9959 0.0010 -0.0024 -0.0020
+ - param set #23 has cost=-0.018469 [o=29 t=24]
+ -- Parameters = 63.9089 8.3338 -34.5391 22.5400 11.8490 -2.3021 0.9947 0.9980 1.0086 0.0016 -0.0023 0.0000
+ - param set #24 has cost=-0.020131 [o=9 t=27]
+ -- Parameters = 86.2745 -7.7472 -32.6625 -12.5530 11.5188 -2.6840 0.9996 0.9959 0.9790 0.0119 0.0017 0.0154
+ - param set #25 has cost=-0.018908 [o=24 t=21]
+ -- Parameters = 113.2497 -4.9876 -38.3468 10.4201 1.0477 -18.9932 0.9860 0.9976 1.0064 -0.0020 0.0032 0.0022
+ - param set #26 has cost=-0.018480 [o=5 t=13]
+ -- Parameters = 86.6693 -5.5611 -32.9794 -11.6371 12.2803 -1.2798 1.0081 1.0028 0.9952 -0.0009 0.0015 0.0001
+ - param set #27 has cost=-0.021916 [o=60 t=15]
+ -- Parameters = 80.6329 -19.0266 -30.6308 12.0679 25.7186 -4.8097 0.9826 1.0053 1.0035 -0.0003 0.0006 0.0018
+ - param set #28 has cost=-0.017958 [o=11 t=19]
+ -- Parameters = 88.5670 -7.4674 -33.4751 -13.5005 15.1288 3.4030 1.0004 0.9994 1.0005 0.0070 -0.0008 0.0131
+ - param set #29 has cost=-0.017558 [o=10 t=20]
+ -- Parameters = 87.0521 -4.2052 -34.0472 5.1087 14.6295 6.4657 0.9964 1.0008 0.9914 0.0278 -0.0027 -0.0007
+ - param set #30 has cost=-0.018518 [o=12 t=28]
+ -- Parameters = 91.5202 7.0339 -24.7444 -9.6096 24.8147 30.3037 1.0040 0.9968 0.9706 0.0016 0.0029 0.0178
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0154 XXX
+ --- dist(#3,#1) = 0.445
+ --- dist(#4,#1) = 0.435
+ --- dist(#5,#1) = 0.472
+ --- dist(#6,#1) = 0.433
+ --- dist(#7,#1) = 0.465
+ --- dist(#8,#1) = 0.472
+ --- dist(#9,#1) = 0.479
+ --- dist(#10,#1) = 0.444
+ --- dist(#11,#1) = 0.39
+ --- dist(#12,#1) = 0.331
+ --- dist(#13,#1) = 0.45
+ --- dist(#14,#1) = 0.373
+ --- dist(#15,#1) = 0.439
+ --- dist(#16,#1) = 0.398
+ --- dist(#17,#1) = 0.372
+ --- dist(#18,#1) = 0.383
+ --- dist(#19,#1) = 0.433
+ --- dist(#20,#1) = 0.424
+ --- dist(#21,#1) = 0.427
+ --- dist(#22,#1) = 0.408
+ --- dist(#23,#1) = 0.408
+ --- dist(#24,#1) = 0.479
+ --- dist(#25,#1) = 0.329
+ --- dist(#26,#1) = 0.412
+ --- dist(#27,#1) = 0.431
+ --- dist(#28,#1) = 0.418
+ --- dist(#29,#1) = 0.425
+ - cast out 1 parameter set for being too close to best set
+ - Total coarse refinement net CPU time = 0.0 s; 9506 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=56 usiz=154.5
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 30 cases
+ 30509 total points stored in 64 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.255452 * [o=0 t=0]
+ -- Parameters = 86.7391 0.2599 -0.0294 -0.7628 0.4225 0.6636 0.9885 1.0151 0.9774 -0.0025 0.0043 -0.0299
+ - cost(#2)=-0.030767 [o=2 t=2]
+ -- Parameters = 66.2123 23.5093 -38.3808 39.8093 9.9964 12.9610 0.9948 0.9829 0.9933 0.0012 -0.0017 0.0072
+ - cost(#3)=-0.027489 [o=16 t=1]
+ -- Parameters = 70.0367 25.2737 -35.6587 36.3199 11.6337 4.4118 1.0004 0.9959 1.0091 0.0030 -0.0009 -0.0020
+ - cost(#4)=-0.025288 [o=33 t=9]
+ -- Parameters = 82.7826 4.1891 -34.7917 -16.3925 6.7573 2.3442 0.9923 0.9858 0.9897 0.0003 0.0021 0.0014
+ - cost(#5)=-0.023277 [o=1 t=3]
+ -- Parameters = 119.6281 -4.6363 -37.7501 -6.3316 6.5138 -17.2004 0.9997 0.9976 0.9949 -0.0005 0.0039 -0.0019
+ - cost(#6)=-0.023117 [o=13 t=5]
+ -- Parameters = 64.9775 19.1412 -34.6954 20.0127 14.8358 7.9163 0.9854 0.9900 0.9682 0.0011 -0.0002 0.0166
+ - cost(#7)=-0.024581 [o=39 t=10]
+ -- Parameters = 81.3263 4.3148 -37.2612 -14.2136 2.7303 -0.7687 0.9937 1.0087 1.0013 0.0002 0.0033 -0.0004
+ - cost(#8)=-0.021633 [o=8 t=12]
+ -- Parameters = 119.3091 -7.9336 -37.8074 -5.5422 4.3781 -17.7279 0.9970 0.9813 1.0150 -0.0002 0.0072 0.0033
+ - cost(#9)=-0.021493 [o=85 t=11]
+ -- Parameters = 118.4666 -4.5245 -38.3397 2.4266 4.4473 -18.2752 0.9997 0.9983 1.0002 0.0006 0.0000 0.0004
+ - cost(#10)=-0.021267 [o=75 t=4]
+ -- Parameters = 64.1843 19.8875 -35.5525 25.1687 13.6845 10.4226 1.0066 0.9985 0.9987 -0.0014 -0.0028 -0.0008
+ - cost(#11)=-0.021774 [o=84 t=17]
+ -- Parameters = 56.9201 15.8319 -31.2056 28.0291 18.6459 -1.6768 0.9659 0.9732 0.9728 0.0029 -0.0104 0.0004
+ - cost(#12)=-0.022507 [o=18 t=16]
+ -- Parameters = 78.5408 4.1929 -26.4729 -8.6572 13.8287 -5.8158 0.9937 1.0018 0.9844 0.0203 0.0087 0.0004
+ - cost(#13)=-0.019155 [o=56 t=14]
Center distance of 0.000008 mm
+ -- Parameters = 50.7745 26.6202 32.9973 32.4598 -30.5752 35.8697 1.0087 0.9958 0.9930 0.0031 0.0062 0.0099
+ - cost(#14)=-0.020245 [o=48 t=7]
+ -- Parameters = 67.1825 18.1954 -29.8499 23.3147 7.7080 -2.4611 0.9996 0.9933 1.0034 -0.0016 -0.0015 -0.0032
+ - cost(#15)=-0.021092 [o=52 t=18]
+ -- Parameters = 112.1598 4.0064 -35.1184 -34.5497 -8.1465 31.1941 1.0041 1.0007 0.9887 0.0008 0.0072 0.0121
+ - cost(#16)=-0.019877 [o=69 t=26]
+ -- Parameters = 63.6270 -14.0523 -31.9087 25.4494 28.5313 12.4582 1.0084 1.0046 0.9895 0.0024 0.0101 -0.0027
+ - cost(#17)=-0.020305 [o=82 t=6]
+ -- Parameters = 89.9216 -20.6565 -29.7497 -3.8335 24.2854 4.1633 0.9928 0.9922 0.9918 -0.0073 0.0026 0.0189
+ - cost(#18)=-0.021238 [o=60 t=15]
+ -- Parameters = 80.6329 -19.0266 -30.6308 12.0679 25.7186 -4.8097 0.9826 1.0053 1.0035 -0.0003 0.0006 0.0018
+ - cost(#19)=-0.019577 [o=3 t=23]
+ -- Parameters = 112.2073 -13.7212 -31.2241 -39.7042 28.7034 -8.8775 0.9884 1.0352 0.9988 -0.0020 0.0013 -0.0026
+ - cost(#20)=-0.019125 [o=66 t=22]
+ -- Parameters = 90.5092 4.5347 -33.9519 -6.7415 5.5820 -1.1301 0.9992 1.0101 1.0077 -0.0041 0.0017 0.0090
+ - cost(#21)=-0.019160 [o=53 t=8]
+ -- Parameters = 62.0441 12.8825 -34.1527 22.8447 15.6465 1.8500 0.9966 0.9790 0.9967 0.0008 0.0007 -0.0001
+ - cost(#22)=-0.020172 [o=9 t=27]
+ -- Parameters = 86.2745 -7.7472 -32.6625 -12.5530 11.5188 -2.6840 0.9996 0.9959 0.9790 0.0119 0.0017 0.0154
+ - cost(#23)=-0.018017 [o=14 t=25]
+ -- Parameters = 95.9926 -6.1628 -32.6906 9.0920 7.6291 6.6283 0.9951 0.9981 0.9959 0.0010 -0.0024 -0.0020
+ - cost(#24)=-0.016181 [o=24 t=21]
+ -- Parameters = 113.2497 -4.9876 -38.3468 10.4201 1.0477 -18.9932 0.9860 0.9976 1.0064 -0.0020 0.0032 0.0022
+ - cost(#25)=-0.014794 [o=12 t=28]
+ -- Parameters = 91.5202 7.0339 -24.7444 -9.6096 24.8147 30.3037 1.0040 0.9968 0.9706 0.0016 0.0029 0.0178
+ - cost(#26)=-0.018695 [o=5 t=13]
+ -- Parameters = 86.6693 -5.5611 -32.9794 -11.6371 12.2803 -1.2798 1.0081 1.0028 0.9952 -0.0009 0.0015 0.0001
+ - cost(#27)=-0.017638 [o=29 t=24]
+ -- Parameters = 63.9089 8.3338 -34.5391 22.5400 11.8490 -2.3021 0.9947 0.9980 1.0086 0.0016 -0.0023 0.0000
+ - cost(#28)=-0.017219 [o=11 t=19]
+ -- Parameters = 88.5670 -7.4674 -33.4751 -13.5005 15.1288 3.4030 1.0004 0.9994 1.0005 0.0070 -0.0008 0.0131
+ - cost(#29)=-0.017063 [o=10 t=20]
+ -- Parameters = 87.0521 -4.2052 -34.0472 5.1087 14.6295 6.4657 0.9964 1.0008 0.9914 0.0278 -0.0027 -0.0007
+ - cost(#30)=-0.178893 [o=-2 t=-2]
+ -- Parameters = 86.5044 -0.1018 -2.0004 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 30 cases
+ - cost(#1)=-0.255660 * [o=0 t=0]
+ -- Parameters = 86.7125 0.2669 -0.0021 -0.7024 0.4142 0.5794 0.9887 1.0147 0.9784 -0.0019 0.0047 -0.0307
+ - cost(#2)=-0.030808 [o=2 t=2]
+ -- Parameters = 66.1653 23.5076 -38.3757 39.8051 10.0069 12.9649 0.9950 0.9824 0.9929 0.0009 -0.0024 0.0078
+ - cost(#3)=-0.027550 [o=16 t=1]
+ -- Parameters = 70.0160 25.2465 -35.7171 36.2921 11.6333 4.4623 1.0002 0.9962 1.0086 0.0037 -0.0012 -0.0022
+ - cost(#4)=-0.025976 [o=33 t=9]
+ -- Parameters = 82.1383 3.9699 -34.8077 -15.6873 7.0210 2.3311 0.9829 0.9911 0.9915 0.0006 0.0053 0.0033
+ - cost(#5)=-0.023376 [o=1 t=3]
+ -- Parameters = 119.6504 -4.7668 -37.7110 -6.3066 6.5481 -17.3192 0.9990 0.9970 0.9954 0.0062 0.0040 -0.0018
+ - cost(#6)=-0.023429 [o=13 t=5]
+ -- Parameters = 64.9497 19.2244 -34.7972 20.2612 14.8086 7.7645 0.9852 0.9917 0.9652 0.0025 -0.0011 0.0169
+ - cost(#7)=-0.024689 [o=39 t=10]
+ -- Parameters = 81.2819 4.3621 -37.1324 -14.2265 2.8360 -0.7164 0.9936 1.0090 1.0028 -0.0004 0.0032 0.0003
+ - cost(#8)=-0.024009 [o=8 t=12]
+ -- Parameters = 119.6102 -5.3714 -37.9480 -6.4817 4.5304 -17.4562 0.9964 0.9829 1.0171 -0.0005 0.0070 0.0044
+ - cost(#9)=-0.021696 [o=85 t=11]
+ -- Parameters = 118.5756 -4.4539 -38.3775 2.4090 4.5177 -18.3318 0.9983 0.9982 0.9999 0.0004 -0.0001 0.0019
+ - cost(#10)=-0.021720 [o=75 t=4]
+ -- Parameters = 64.3571 20.2468 -35.4634 24.9826 13.7679 10.4626 1.0077 0.9983 1.0002 -0.0079 -0.0033 -0.0012
+ - cost(#11)=-0.021961 [o=84 t=17]
+ -- Parameters = 57.1021 16.0834 -31.1352 28.0267 18.6405 -1.7933 0.9645 0.9696 0.9760 0.0016 -0.0073 0.0016
+ - cost(#12)=-0.022563 [o=18 t=16]
+ -- Parameters = 78.5560 4.1947 -26.4685 -8.6348 13.8276 -5.8370 0.9928 1.0014 0.9845 0.0202 0.0089 0.0013
+ - cost(#13)=-0.019260 [o=56 t=14]
+ -- Parameters = 50.7450 26.6768 32.9984 32.5420 -30.6059 35.8321 1.0098 0.9924 0.9938 0.0049 0.0066 0.0102
+ - cost(#14)=-0.020298 [o=48 t=7]
+ -- Parameters = 67.2597 18.2385 -29.8468 23.2420 7.7532 -2.4245 0.9998 0.9926 1.0031 -0.0016 -0.0015 -0.0037
+ - cost(#15)=-0.021495 [o=52 t=18]
+ -- Parameters = 112.2651 4.2211 -35.3283 -34.2743 -8.3033 31.0150 1.0023 0.9906 0.9836 0.0015 0.0106 0.0095
+ - cost(#16)=-0.020361 [o=69 t=26]
+ -- Parameters = 63.7524 -13.8683 -32.0660 25.5860 28.5357 12.2220 1.0114 1.0052 0.9866 0.0006 0.0075 -0.0124
+ - cost(#17)=-0.022784 [o=82 t=6]
+ -- Parameters = 89.8447 -21.7826 -29.9994 -4.3826 23.7842 2.9962 0.9964 0.9920 0.9677 -0.0078 0.0027 0.0187
+ - cost(#18)=-0.021535 [o=60 t=15]
+ -- Parameters = 80.6030 -19.0204 -30.6881 12.0696 25.8116 -4.7863 0.9796 1.0076 1.0012 -0.0008 0.0004 0.0009
+ - cost(#19)=-0.019782 [o=3 t=23]
+ -- Parameters = 112.2654 -13.7246 -31.1781 -39.7658 28.6507 -8.8884 0.9889 1.0344 0.9988 -0.0019 0.0087 -0.0022
+ - cost(#20)=-0.019463 [o=66 t=22]
+ -- Parameters = 90.7233 4.4427 -33.9152 -6.7966 5.4483 -1.2778 0.9996 1.0122 1.0093 -0.0132 0.0017 0.0093
+ - cost(#21)=-0.019185 [o=53 t=8]
+ -- Parameters = 62.0352 12.8898 -34.1482 22.8480 15.6759 1.8424 0.9966 0.9789 0.9966 0.0017 0.0006 -0.0001
+ - cost(#22)=-0.020594 [o=9 t=27]
+ -- Parameters = 86.3741 -7.7429 -32.7742 -12.7298 11.5379 -2.5464 1.0004 0.9986 0.9777 0.0103 0.0095 0.0151
+ - cost(#23)=-0.018661 [o=14 t=25]
+ -- Parameters = 96.0254 -6.2801 -32.5617 8.8973 7.3887 6.8711 0.9919 1.0027 0.9997 0.0008 -0.0029 -0.0106
+ - cost(#24)=-0.016262 [o=24 t=21]
+ -- Parameters = 113.2998 -4.9742 -38.3496 10.4188 1.1154 -19.0046 0.9858 0.9981 1.0061 -0.0020 0.0031 0.0024
+ - cost(#25)=-0.018217 [o=12 t=28]
+ -- Parameters = 91.6982 9.9276 -22.3775 -9.9113 25.1579 30.5747 1.0043 0.9998 0.9681 0.0006 0.0026 0.0162
+ - cost(#26)=-0.019421 [o=5 t=13]
+ -- Parameters = 86.4514 -5.8654 -32.7965 -12.4374 12.1416 -0.6854 0.9958 1.0071 0.9953 -0.0008 0.0022 0.0013
+ - cost(#27)=-0.017925 [o=29 t=24]
+ -- Parameters = 64.1308 8.2562 -34.6042 22.3209 11.9257 -2.1655 0.9925 0.9949 1.0085 0.0034 -0.0026 -0.0001
Padding ...
+ - cost(#28)=-0.017992 [o=11 t=19]
+ -- Parameters = 87.7665 -7.8719 -33.6269 -12.7891 15.1244 3.5388 0.9966 0.9867 0.9961 0.0065 0.0013 0.0136
+ - cost(#29)=-0.017639 [o=10 t=20]
+ -- Parameters = 87.2614 -4.1945 -34.0504 4.9441 14.7488 6.2897 0.9942 1.0027 0.9890 0.0384 -0.0059 -0.0010
+ - cost(#30)=-0.252580 [o=-2 t=-2]
+ -- Parameters = 86.5889 0.1609 -0.0592 -0.4657 0.5906 1.0863 0.9877 1.0175 0.9726 0.0058 -0.0097 -0.0264
+ - case #1 [o=0 t=0] is now the best
+ - Initial cost = -0.255660
+ - Initial fine Parameters = 86.7125 0.2669 -0.0021 -0.7024 0.4142 0.5794 0.9887 1.0147 0.9784 -0.0019 0.0047 -0.0307
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - Finalish cost = -0.256033 ; 311 funcs
+ - ini Finalish Parameters = 86.7155 0.2521 0.0313 -0.7138 0.1308 0.5354 0.9890 1.0141 0.9770 -0.0026 0.0061 -0.0372
+ - Final cost = -0.256033 ; 268 funcs
+ Final fine fit Parameters:
x-shift= 86.7153 y-shift= 0.2521 z-shift= 0.0315 ... enorm= 86.7157 mm
z-angle= -0.7138 x-angle= 0.1311 y-angle= 0.5358 ... total= 0.9018 deg
x-scale= 0.9890 y-scale= 1.0141 z-scale= 0.9770 ... vol3D= 0.9799 = base bigger than source
y/x-shear= -0.0026 z/x-shear= 0.0061 z/y-shear= -0.0372
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.7153 0.2521 0.0315 -0.7138 0.1311 0.5358 0.9890 1.0141 0.9770 -0.0026 0.0061 -0.0372
++ Unloading unneeded data
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
++ Output dataset ./sub-02_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-02_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 23.2
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/__tt_sub-02_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/sub-02_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-02_T1w_ns+orig sub-02_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
Resampling ...
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-02_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-02_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-02_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-02_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - Test (64+191)*64 params [top5=*o+-.]:#oo*[#3=0.0126675] *..+o-o.-*[#28=-0.00260291] *o..o*[#60=-0.00541545] *-..*[#261=-0.0124944] *-*[#263=-0.013413] *.+..*[#272=-0.0135668] *.-*[#304=-0.0141442] *.*[#773=-0.0186251] *o-+-$-*[#5149=-0.0194544] **[#5179=-0.0254457] *-o.o.
+ - best 88 costs found:
o= 0 v=-0.025446: 111.47 -29.66 30.31 -17.69 -14.58 -34.92 [rand]
o= 1 v=-0.021106: 80.09 33.34 -41.71 19.28 -27.13 -15.13 [rand]
o= 2 v=-0.019649: 119.00 26.39 26.75 -9.69 21.75 -40.77 [rand]
o= 3 v=-0.019454: 111.47 28.45 -40.59 -17.69 -14.58 34.92 [rand]
o= 4 v=-0.018777: 69.71 17.86 -37.50 -10.10 -16.32 -39.31 [rand]
o= 5 v=-0.018625: 93.17 6.06 -31.80 30.00 7.50 7.50 [grid]
o= 6 v=-0.018542: 112.03 -31.94 29.14 -42.46 -41.07 -35.63 [rand]
o= 7 v=-0.017972: 99.67 37.28 -42.33 -14.03 -8.39 25.20 [rand]
o= 8 v=-0.017144: 93.17 -7.27 -31.80 30.00 7.50 7.50 [grid]
o= 9 v=-0.017013: 102.15 8.15 -38.97 23.16 17.17 -3.59 [rand]
o=10 v=-0.016933: 79.84 6.06 -31.80 -30.00 -7.50 7.50 [grid]
o=11 v=-0.016366: 88.80 -7.78 -36.83 -10.97 18.37 5.04 [rand]
o=12 v=-0.016075: 84.21 -7.78 -36.83 -10.97 18.37 -5.04 [rand]
o=13 v=-0.015985: 84.21 -7.78 -36.83 -10.97 18.37 5.04 [rand]
o=14 v=-0.015963: 119.00 -27.59 26.75 -9.69 -21.75 -40.77 [rand]
o=15 v=-0.015734: 115.52 -27.75 -38.75 7.44 27.42 -25.63 [rand]
o=16 v=-0.015518: 81.91 9.98 -36.71 -36.41 9.00 22.11 [rand]
o=17 v=-0.015310: 84.21 -7.78 -36.83 10.97 18.37 -5.04 [rand]
o=18 v=-0.015098: 113.17 26.06 -31.80 -30.00 -7.50 30.00 [grid]
o=19 v=-0.015080: 66.25 26.68 -39.62 37.90 5.03 -3.08 [rand]
o=20 v=-0.014624: 51.64 29.95 10.63 -11.68 29.04 39.38 [rand]
o=21 v=-0.014427: 67.39 22.81 -38.43 37.88 -13.88 -18.42 [rand]
o=22 v=-0.014352: 88.80 -7.78 -36.83 10.97 18.37 -5.04 [rand]
o=23 v=-0.014272: 92.92 -34.55 31.44 -19.28 -27.13 -15.13 [rand]
o=24 v=-0.014149: 80.09 33.34 -41.71 19.28 -27.13 15.13 [rand]
o=25 v=-0.014144: 79.84 -7.27 -31.80 -7.50 7.50 -7.50 [grid]
o=26 v=-0.013972: 73.34 -38.49 32.06 -14.03 -8.39 25.20 [rand]
o=27 v=-0.013950: 60.20 25.35 -22.93 15.82 -17.65 -37.25 [rand]
o=28 v=-0.013940: 84.21 6.58 -36.83 -10.97 18.37 5.04 [rand]
o=29 v=-0.013927: 79.84 6.06 -31.80 -30.00 7.50 7.50 [grid]
o=30 v=-0.013789: 64.51 28.83 -27.40 28.45 -9.40 -40.53 [rand]
o=31 v=-0.013685: 70.86 8.15 -38.97 -23.16 17.17 -3.59 [rand]
o=32 v=-0.013567: 79.84 -7.27 -31.80 -7.50 7.50 7.50 [grid]
o=33 v=-0.013413: 93.17 -7.27 -31.80 7.50 7.50 7.50 [grid]
o=34 v=-0.013271: 102.15 8.15 -38.97 23.16 17.17 3.59 [rand]
o=35 v=-0.013254: 113.44 27.53 32.81 -15.05 -4.71 -12.78 [rand]
o=36 v=-0.013176: 113.44 27.53 32.81 -15.05 4.71 -12.78 [rand]
o=37 v=-0.012950: 113.17 6.06 -31.80 30.00 7.50 30.00 [grid]
o=38 v=-0.012922: 111.88 -27.86 32.37 -41.23 -14.90 -41.72 [rand]
o=39 v=-0.012876: 79.84 -7.27 -31.80 -30.00 7.50 -7.50 [grid]
o=40 v=-0.012721: 66.25 26.68 -39.62 37.90 -5.03 -3.08 [rand]
o=41 v=-0.012705: 84.21 6.58 -36.83 -10.97 18.37 -5.04 [rand]
o=42 v=-0.012683: 119.00 -27.59 26.75 9.69 -21.75 -40.77 [rand]
o=43 v=-0.012657: 57.22 15.15 -35.00 13.45 19.96 -14.96 [rand]
o=44 v=-0.012649: 119.00 26.39 26.75 9.69 21.75 -40.77 [rand]
o=45 v=-0.012537: 98.00 -16.91 -38.89 -19.17 13.96 2.56 [rand]
o=46 v=-0.012494: 93.17 6.06 -31.80 7.50 7.50 7.50 [grid]
o=47 v=-0.012468: 69.71 -19.07 27.22 -10.10 -16.32 39.31 [rand]
o=48 v=-0.012432: 88.80 6.58 -36.83 -10.97 18.37 5.04 [rand]
o=49 v=-0.012315: 90.65 23.20 26.75 31.52 11.26 -33.63 [rand]
o=50 v=-0.012302: 118.71 28.44 -41.28 -40.41 -19.09 40.75 [rand]
o=51 v=-0.012277: 88.80 -7.78 -36.83 -10.97 18.37 -5.04 [rand]
o=52 v=-0.012059: 59.84 6.06 -31.80 7.50 7.50 7.50 [grid]
o=53 v=-0.011952: 118.71 -29.65 31.00 -40.41 -19.09 -40.75 [rand]
o=54 v=-0.011754: 82.36 23.20 26.75 -31.52 11.26 33.63 [rand]
o=55 v=-0.011751: 72.80 2.08 -39.43 -41.38 -19.47 31.64 [rand]
o=56 v=-0.011640: 73.34 37.28 32.06 -14.03 8.39 25.20 [rand]
o=57 v=-0.011617: 108.50 28.83 -27.40 -28.45 -9.40 40.53 [rand]
o=58 v=-0.011578: 54.30 28.44 31.00 40.41 19.09 40.75 [rand]
o=59 v=-0.011401: 88.80 6.58 -36.83 -10.97 18.37 -5.04 [rand]
o=60 v=-0.011354: 73.34 37.28 -42.33 14.03 -8.39 -25.20 [rand]
o=61 v=-0.011309: 122.12 -5.02 -42.01 -37.76 24.22 3.86 [rand]
o=62 v=-0.011260: 59.84 6.06 -31.80 -30.00 -30.00 30.00 [grid]
o=63 v=-0.011233: 53.48 4.41 31.68 11.98 4.28 19.20 [rand]
o=64 v=-0.011206: 88.80 -7.78 -36.83 10.97 18.37 5.04 [rand]
o=65 v=-0.011116: 49.91 -27.90 26.10 25.97 -37.53 40.21 [rand]
o=66 v=-0.011111: 75.02 15.70 -38.89 -19.17 13.96 2.56 [rand]
o=67 v=-0.011059: 53.30 4.92 12.70 27.00 -22.71 29.07 [rand]
o=68 v=-0.011018: 92.92 33.34 -41.71 19.28 -27.13 15.13 [rand]
o=69 v=-0.011006: 66.25 26.68 -39.62 37.90 5.03 3.08 [rand]
o=70 v=-0.010865: 64.04 28.55 -35.16 27.43 -11.56 -19.13 [rand]
o=71 v=-0.010857: 57.49 26.54 28.47 7.44 27.42 25.63 [rand]
o=72 v=-0.010705: 51.31 34.09 16.43 -18.41 40.28 7.49 [rand]
o=73 v=-0.010679: 111.83 28.07 -29.13 -42.33 -22.93 32.02 [rand]
o=74 v=-0.010664: 92.92 -34.55 -41.71 19.28 27.13 15.13 [rand]
o=75 v=-0.010656: 103.30 17.86 -37.50 -10.10 -16.32 39.31 [rand]
o=76 v=-0.010631: 79.84 6.06 -31.80 -7.50 7.50 -7.50 [grid]
o=77 v=-0.010626: 72.80 -3.29 -39.43 -41.38 -19.47 31.64 [rand]
o=78 v=-0.010618: 105.25 -37.00 27.91 -24.90 -4.74 -17.23 [rand]
o=79 v=-0.010567: 59.58 27.53 32.81 15.05 4.71 12.78 [rand]
o=80 v=-0.010553: 64.04 28.55 -35.16 27.43 11.56 -19.13 [rand]
o=81 v=-0.010429: 93.17 26.06 21.53 30.00 7.50 -30.00 [grid]
o=82 v=-0.010420: 96.56 22.30 16.61 23.24 -8.32 39.75 [rand]
o=83 v=-0.010405: 93.56 -20.42 -40.89 -25.15 27.05 27.90 [rand]
o=84 v=-0.010313: 79.84 -27.27 21.53 -30.00 -30.00 30.00 [grid]
o=85 v=-0.010254: 92.92 -34.55 -41.71 19.28 27.13 -15.13 [rand]
o=86 v=-0.010222: 77.99 -29.61 -36.51 -12.98 30.57 5.92 [rand]
o=87 v=-0.010193: 80.09 -34.55 31.44 -19.28 -27.13 15.13 [rand]
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Performing center alignment with @Align_Centers
Clipping -0.000100 1230.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-03_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-03_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/123.00=4.1e+02
+ - A little optimization:*[#16322=-0.0263551] *[#16324=-0.026653] *[#16329=-0.0266668] *[#16330=-0.0283642] *[#16339=-0.0286273] *[#16342=-0.0288497] *[#16343=-0.0288869] *[#16345=-0.0289987] *[#16346=-0.029476] *[#16350=-0.0296251] *[#16351=-0.0296529] *[#16354=-0.0296796] *[#16356=-0.0299392] *[#16357=-0.0300724] *[#16358=-0.0302129] *[#16359=-0.0302619] *[#16364=-0.0303326] *[#16367=-0.030376] *[#16370=-0.0303896] .*[#16387=-0.031782] *[#16397=-0.0318328] *[#16400=-0.0319649] *[#16401=-0.0322047] *[#16403=-0.0322139] *[#16404=-0.0323157] *[#16405=-0.0324078] *[#16408=-0.0325641] *[#16409=-0.0326751] *[#16410=-0.0327988] *[#16413=-0.0328508] ....*[#16599=-0.0329225] *[#16600=-0.0351681] *[#16602=-0.0355468] *[#16606=-0.0358748] *[#16609=-0.0360879] *[#16614=-0.0362597] *[#16615=-0.0363264] *[#16616=-0.0365211] *[#16617=-0.0369081] *[#16619=-0.0369621] *[#16622=-0.0371712] *[#16623=-0.0372721] *[#16626=-0.0373975] *[#16629=-0.0374203] *[#16630=-0.037422] *[#16632=-0.0374548] .*[#16672=-0.0375318] *[#16679=-0.0376069] *[#16680=-0.0376557] *[#16681=-0.037689] *[#16682=-0.0376913] *[#16689=-0.0377628] *[#16690=-0.0378209] *[#16697=-0.037879] ..................................................................................
+ - costs of the above after a little optimization:
o= 0 v=-0.030390: 112.76 -36.42 30.50 -17.23 -16.88 -36.63 [rand] [f=54]
o= 1 v=-0.032851: 80.09 34.47 -44.55 18.30 -20.51 -15.56 [rand] [f=43]
o= 2 v=-0.020514: 119.52 26.38 26.77 -10.24 21.96 -40.45 [rand] [f=29]
o= 3 v=-0.028398: 109.14 23.56 -38.13 -15.71 -14.60 31.85 [rand] [f=58]
o= 4 v=-0.023851: 70.21 21.11 -38.05 -8.07 -16.74 -39.61 [rand] [f=45]
o= 5 v=-0.037455: 96.37 -1.32 -34.01 24.40 8.09 12.14 [grid] [f=83]
*o= 6 v=-0.037879: 112.09 -37.80 34.85 -35.89 -33.65 -42.98 [rand] [f=62]
o= 7 v=-0.021925: 99.64 36.88 -42.59 -14.24 -3.88 25.83 [rand] [f=45]
o= 8 v=-0.032572: 96.42 -0.05 -36.38 29.96 12.82 5.33 [grid] [f=53]
o= 9 v=-0.030254: 95.46 3.51 -37.39 22.28 14.63 -1.55 [rand] [f=71]
o=10 v=-0.020158: 81.33 5.26 -33.67 -31.28 -5.79 6.56 [grid] [f=57]
o=11 v=-0.027627: 85.11 -10.10 -37.66 -3.23 13.99 3.92 [rand] [f=57]
o=12 v=-0.026288: 84.76 -10.68 -37.11 -0.13 11.62 -5.13 [rand] [f=53]
o=13 v=-0.028532: 84.29 -10.24 -37.51 -0.91 13.85 -0.38 [rand] [f=55]
o=14 v=-0.030712: 113.59 -29.05 26.36 -15.26 -19.12 -43.60 [rand] [f=46]
o=15 v=-0.024618: 113.44 -24.51 -42.74 -0.70 24.54 -17.53 [rand] [f=52]
o=16 v=-0.029867: 81.30 7.36 -39.29 -33.77 12.76 15.97 [rand] [f=67]
o=17 v=-0.027670: 84.22 -10.53 -37.74 -0.01 11.93 0.01 [rand] [f=63]
o=18 v=-0.026327: 110.26 25.76 -32.74 -26.98 -9.49 34.93 [grid] [f=68]
o=19 v=-0.018040: 67.11 26.04 -40.34 36.53 3.35 -7.22 [rand] [f=44]
o=20 v=-0.023465: 49.85 34.04 8.22 -9.76 33.18 35.58 [rand] [f=47]
o=21 v=-0.023928: 67.84 26.28 -38.04 31.48 -13.67 -19.65 [rand] [f=49]
o=22 v=-0.026325: 85.46 -7.24 -37.66 2.68 14.15 -2.93 [rand] [f=56]
o=23 v=-0.023619: 92.92 -34.36 34.66 -20.03 -26.34 -15.29 [rand] [f=53]
o=24 v=-0.020672: 81.02 33.61 -41.43 15.62 -28.22 12.28 [rand] [f=71]
o=25 v=-0.028710: 79.43 -4.58 -36.30 -5.08 5.62 -2.14 [grid] [f=66]
o=26 v=-0.029225: 72.33 -36.03 34.77 -21.51 -14.21 22.99 [rand] [f=55]
o=27 v=-0.024447: 61.79 28.84 -27.26 14.69 -17.00 -38.12 [rand] [f=54]
o=28 v=-0.025051: 80.30 1.57 -37.56 -10.68 12.11 1.83 [rand] [f=53]
o=29 v=-0.028396: 81.07 2.61 -36.50 -30.00 8.15 8.11 [grid] [f=52]
o=30 v=-0.018132: 65.81 31.00 -28.30 27.91 -10.51 -40.65 [rand] [f=43]
o=31 v=-0.020905: 76.20 7.29 -37.37 -22.44 16.80 -3.82 [rand] [f=48]
o=32 v=-0.029040: 82.93 -6.91 -39.05 -8.02 5.76 2.12 [grid] [f=63]
o=33 v=-0.020123: 93.04 0.07 -37.37 10.31 7.35 6.22 [grid] [f=45]
o=34 v=-0.027930: 97.07 5.70 -39.40 20.85 11.70 4.84 [rand] [f=50]
o=35 v=-0.021931: 112.68 23.18 34.82 -12.42 0.20 -17.39 [rand] [f=45]
o=36 v=-0.022917: 112.55 31.80 34.00 -14.20 5.20 -20.18 [rand] [f=52]
o=37 v=-0.016022: 110.97 6.26 -32.43 29.37 7.12 30.79 [grid] [f=45]
o=38 v=-0.036065: 110.62 -40.20 33.20 -34.52 -32.37 -36.57 [rand] [f=82]
o=39 v=-0.024958: 77.86 -6.21 -35.55 -32.44 2.92 -3.88 [grid] [f=52]
o=40 v=-0.019960: 67.80 26.30 -43.34 38.68 0.71 -4.64 [rand] [f=57]
o=41 v=-0.018515: 80.56 5.35 -37.74 -10.89 17.76 -4.56 [rand] [f=43]
o=42 v=-0.021989: 117.29 -28.03 31.49 11.27 -22.50 -36.36 [rand] [f=52]
o=43 v=-0.023219: 59.51 14.92 -37.91 13.79 19.38 -8.46 [rand] [f=74]
o=44 v=-0.027649: 116.67 22.78 30.94 9.14 15.65 -44.15 [rand] [f=58]
o=45 v=-0.023260: 95.33 -17.63 -40.32 -19.57 6.74 4.42 [rand] [f=66]
o=46 v=-0.021051: 94.56 3.93 -36.30 10.35 9.31 8.29 [grid] [f=44]
o=47 v=-0.024521: 72.40 -17.07 28.64 -17.80 -16.75 42.06 [rand] [f=71]
o=48 v=-0.022374: 85.74 2.12 -38.39 -10.65 18.86 2.60 [rand] [f=46]
o=49 v=-0.015637: 89.13 23.67 23.22 29.07 10.36 -35.62 [rand] [f=45]
o=50 v=-0.018001: 115.72 28.55 -40.74 -37.80 -21.67 38.24 [rand] [f=51]
o=51 v=-0.026030: 84.70 -7.24 -38.64 1.70 15.13 -0.77 [rand] [f=48]
o=52 v=-0.022752: 60.04 6.91 -34.66 8.57 7.39 4.79 [grid] [f=44]
o=53 v=-0.023449: 114.29 -29.93 29.40 -40.42 -19.78 -41.72 [rand] [f=41]
o=54 v=-0.018354: 82.51 24.56 27.24 -31.58 11.45 40.45 [rand] [f=46]
o=55 v=-0.026015: 72.43 1.36 -44.08 -40.54 -23.18 35.23 [rand] [f=58]
o=56 v=-0.014918: 71.92 36.48 32.56 -14.60 7.23 19.81 [rand] [f=41]
o=57 v=-0.022967: 110.43 28.35 -30.22 -29.68 -9.51 37.94 [rand] [f=60]
o=58 v=-0.018510: 53.65 30.28 34.12 41.22 21.50 38.42 [rand] [f=38]
o=59 v=-0.016903: 87.00 5.59 -36.13 -12.74 13.41 -4.38 [rand] [f=47]
o=60 v=-0.030165: 73.74 26.38 -41.90 13.71 -7.95 -25.22 [rand] [f=44]
o=61 v=-0.013027: 121.76 -4.89 -41.03 -37.77 23.93 3.58 [rand] [f=42]
o=62 v=-0.014211: 56.56 7.78 -32.42 -30.67 -30.07 28.77 [grid] [f=44]
o=63 v=-0.018476: 53.21 6.59 34.23 14.01 11.05 20.50 [rand] [f=42]
o=64 v=-0.021027: 87.33 -4.84 -39.92 7.05 17.40 4.65 [rand] [f=56]
o=65 v=-0.023185: 47.36 -17.54 34.48 31.11 -30.47 38.94 [rand] [f=55]
o=66 v=-0.021121: 75.54 11.39 -41.56 -20.99 13.91 3.65 [rand] [f=48]
o=67 v=-0.025549: 50.22 1.30 13.08 29.09 -26.94 35.20 [rand] [f=59]
o=68 v=-0.032479: 90.83 38.93 -39.01 26.80 -24.94 8.37 [rand] [f=69]
o=69 v=-0.017800: 66.39 26.38 -41.58 44.28 3.13 1.57 [rand] [f=44]
o=70 v=-0.031428: 70.23 26.59 -39.13 24.94 -11.30 -25.13 [rand] [f=59]
o=71 v=-0.020747: 56.19 30.43 29.51 17.36 32.68 23.71 [rand] [f=53]
o=72 v=-0.020749: 50.06 39.38 17.81 -16.24 38.47 13.34 [rand] [f=75]
o=73 v=-0.015511: 111.68 32.35 -29.16 -42.46 -22.92 31.62 [rand] [f=44]
o=74 v=-0.015480: 92.75 -30.35 -44.67 19.58 28.01 12.50 [rand] [f=43]
o=75 v=-0.029604: 108.74 25.97 -36.22 -14.50 -10.46 36.13 [rand] [f=77]
o=76 v=-0.022828: 78.41 -0.64 -37.55 -11.20 4.83 -8.73 [grid] [f=67]
o=77 v=-0.021268: 71.72 -0.36 -43.29 -41.83 -20.99 32.25 [rand] [f=63]
o=78 v=-0.021370: 105.98 -37.81 23.87 -19.34 -5.95 -19.96 [rand] [f=68]
o=79 v=-0.031133: 55.59 30.00 34.42 23.03 6.93 26.15 [rand] [f=50]
o=80 v=-0.018063: 65.39 22.24 -34.24 28.62 10.19 -17.09 [rand] [f=58]
o=81 v=-0.013161: 92.54 26.80 22.21 28.49 6.83 -33.44 [grid] [f=50]
o=82 v=-0.011025: 96.84 22.29 16.73 23.97 -8.60 40.08 [rand] [f=37]
o=83 v=-0.014779: 93.43 -16.08 -41.01 -25.67 26.85 23.77 [rand] [f=53]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
o=84 v=-0.015629: 79.48 -27.77 25.09 -28.57 -30.39 31.68 [grid] [f=48]
o=85 v=-0.021586: 93.13 -25.14 -44.16 15.37 26.85 -11.57 [rand] [f=83]
o=86 v=-0.020395: 75.88 -26.80 -44.22 -14.52 29.73 2.47 [rand] [f=50]
o=87 v=-0.024445: 70.91 -39.66 30.95 -18.49 -23.98 16.17 [rand] [f=67]
+ - saving # 6 for use with twobest
+ - saving # 5 for use with twobest
+ - saving #38 for use with twobest
+ - saving # 1 for use with twobest
+ - saving # 8 for use with twobest
+ - saving #68 for use with twobest
+ - saving #70 for use with twobest
+ - saving #79 for use with twobest
+ - saving #14 for use with twobest
+ - saving # 0 for use with twobest
+ - saving # 9 for use with twobest
+ - saving #60 for use with twobest
+ - saving #16 for use with twobest
+ - saving #75 for use with twobest
+ - saving #26 for use with twobest
+ - saving #32 for use with twobest
+ - skip #25 for twobest: too close to set #32
+ - saving #13 for use with twobest
+ - skip # 3 for twobest: too close to set #75
+ - saving #29 for use with twobest
+ - saving #34 for use with twobest
+ - skip #17 for twobest: too close to set #13
+ - saving #44 for use with twobest
+ - skip #11 for twobest: too close to set #13
+ - saving #18 for use with twobest
+ - skip #22 for twobest: too close to set #13
+ - skip #12 for twobest: too close to set #22
+ - skip #51 for twobest: too close to set #13
+ - saving #55 for use with twobest
+ - saving #67 for use with twobest
+ - saving #28 for use with twobest
+ - saving #39 for use with twobest
+ - saving #15 for use with twobest
+ - saving #47 for use with twobest
+ - saving #27 for use with twobest
+ - saving #87 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=29 usiz=173.5
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-02_T1w_ns_shft+orig
+ - retaining old weight image
+ - using 37594 points from base image [use_all=2]
+ deoblique
+ * Exit alignment setup routine
++ 3drefit processed 1 datasets
+ 31467 total points stored in 63 'TOHD(17.3748)' bloks (0 duplicates)
+ - param set #1 has cost=-0.031393 [o=6 t=0]
+ -- Parameters = 112.0792 -37.7271 34.8591 -35.8281 -33.5107 -42.9618 1.0009 1.0010 1.0046 -0.0002 -0.0003 -0.0002
+ - param set #2 has cost=-0.031567 [o=5 t=1]
+ -- Parameters = 96.5622 -0.8954 -33.7407 23.8779 8.0303 12.0393 1.0039 0.9910 0.9992 0.0015 0.0002 0.0121
+ - param set #3 has cost=-0.029430 [o=38 t=2]
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ -- Parameters = 110.3400 -40.3686 33.2115 -34.3376 -32.4957 -36.4576 0.9909 1.0040 0.9996 0.0017 0.0034 -0.0005
++ THD_rename_dataset_files: rename sub-02_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-02_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-02_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-02_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-02_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-02_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - param set #4 has cost=-0.028466 [o=1 t=3]
+ -- Parameters = 79.6117 34.6141 -44.4333 19.0884 -21.0032 -15.0516 0.9816 0.9631 1.0004 -0.0024 0.0100 0.0016
+ - param set #5 has cost=-0.030633 [o=8 t=4]
+ -- Parameters = 96.5187 0.6993 -36.1452 29.7006 11.3516 12.8700 0.9620 0.9815 0.9518 0.0041 -0.0057 -0.0166
+ - param set #6 has cost=-0.027500 [o=68 t=5]
+ -- Parameters = 90.6684 38.8004 -38.5626 26.7584 -25.6867 8.6190 0.9989 0.9873 1.0144 0.0013 0.0004 -0.0108
+ - param set #7 has cost=-0.028828 [o=70 t=6]
+ -- Parameters = 70.0875 26.5641 -39.4313 19.9777 -11.9594 -25.1207 0.9903 0.9990 0.9988 0.0003 0.0011 -0.0005
+ - param set #8 has cost=-0.026238 [o=79 t=7]
+ -- Parameters = 55.6278 29.6775 34.2153 23.2122 7.4376 26.3016 0.9973 1.0414 1.0009 0.0007 0.0001 -0.0011
+ - param set #9 has cost=-0.026295 [o=14 t=8]
+ -- Parameters = 113.3479 -29.0498 26.6883 -16.2297 -18.9653 -43.9397 1.0017 0.9949 1.0018 -0.0012 0.0121 -0.0001
+ - param set #10 has cost=-0.026903 [o=0 t=9]
+ -- Parameters = 112.7267 -36.4129 30.5490 -17.2371 -16.9208 -36.6507 1.0008 0.9994 0.9999 -0.0001 0.0126 0.0002
+ - param set #11 has cost=-0.024429 [o=9 t=10]
+ -- Parameters = 95.4455 3.3650 -37.3551 22.3621 14.4884 -1.4852 0.9980 1.0077 1.0011 0.0120 -0.0008 -0.0013
+ - param set #12 has cost=-0.028542 [o=60 t=11]
+ -- Parameters = 73.5452 26.3816 -42.9077 15.6025 -10.7886 -24.6658 1.0035 1.0039 0.9830 0.0032 0.0031 -0.0008
+ - param set #13 has cost=-0.025992 [o=16 t=12]
+ -- Parameters = 81.2263 7.7201 -39.1547 -33.3057 13.3318 15.9163 1.0077 1.0082 0.9981 0.0007 0.0003 0.0009
+ - param set #14 has cost=-0.023501 [o=75 t=13]
+ -- Parameters = 108.6346 25.9732 -36.2769 -14.5968 -10.4280 36.2018 1.0021 1.0007 0.9984 -0.0016 -0.0001 0.0021
+ - param set #15 has cost=-0.023587 [o=26 t=14]
+ -- Parameters = 72.9526 -36.3945 34.8444 -21.8064 -18.1304 21.5448 0.9848 1.0052 0.9818 -0.0009 0.0013 0.0028
+ - param set #16 has cost=-0.022797 [o=32 t=15]
+ -- Parameters = 83.0813 -7.0899 -39.1304 -8.0976 5.7134 2.1973 1.0020 0.9990 1.0001 -0.0006 -0.0012 -0.0004
+ - param set #17 has cost=-0.020583 [o=13 t=16]
+ -- Parameters = 84.2965 -10.3028 -37.5557 -1.0116 13.8498 -0.3659 0.9955 1.0010 0.9945 -0.0003 -0.0005 -0.0002
+ - param set #18 has cost=-0.025366 [o=29 t=17]
+ -- Parameters = 81.1529 3.6315 -36.4650 -34.2172 8.7573 7.0699 0.9876 0.9964 0.9906 -0.0019 -0.0021 -0.0017
+ - param set #19 has cost=-0.023916 [o=34 t=18]
+ -- Parameters = 97.5726 6.0262 -39.0841 20.7074 12.2625 5.2456 0.9998 0.9928 0.9908 -0.0010 0.0015 0.0018
+ - param set #20 has cost=-0.023736 [o=44 t=19]
+ -- Parameters = 117.5350 22.2079 31.3363 4.1007 15.2877 -43.2699 1.0046 1.0018 1.0053 -0.0013 -0.0009 -0.0029
+ - param set #21 has cost=-0.022827 [o=18 t=20]
+ -- Parameters = 110.2983 25.9444 -32.9114 -26.6899 -9.4244 35.0870 0.9983 1.0030 0.9967 -0.0000 0.0002 -0.0016
+ - param set #22 has cost=-0.019279 [o=55 t=21]
+ -- Parameters = 72.3642 1.4364 -43.8148 -40.5775 -23.0992 35.2553 1.0007 0.9976 1.0060 -0.0004 -0.0005 -0.0012
+ - param set #23 has cost=-0.019406 [o=67 t=22]
+ -- Parameters = 50.1056 1.3212 12.6470 29.3038 -27.0126 35.2315 0.9922 0.9995 0.9975 0.0010 -0.0004 0.0133
+ - param set #24 has cost=-0.018937 [o=28 t=23]
+ -- Parameters = 80.2547 1.4494 -37.7101 -10.6075 11.9926 1.7432 0.9986 1.0084 0.9906 0.0018 0.0002 -0.0013
+ - param set #25 has cost=-0.023252 [o=39 t=24]
+ -- Parameters = 78.5986 -7.0686 -35.3295 -31.5508 0.2464 -5.3140 0.9848 0.9849 0.9830 -0.0012 -0.0028 -0.0018
+ - param set #26 has cost=-0.021512 [o=15 t=25]
+ -- Parameters = 113.4210 -24.1197 -42.7046 -1.1970 24.0618 -17.8800 0.9797 0.9907 1.0029 0.0019 0.0004 0.0024
+ - param set #27 has cost=-0.024391 [o=47 t=26]
+ -- Parameters = 73.5287 -17.7607 29.5602 -19.3080 -16.5862 39.2818 0.9768 0.9515 0.9846 -0.0057 -0.0069 0.0036
+ - param set #28 has cost=-0.019764 [o=27 t=27]
+ -- Parameters = 61.9927 28.8052 -27.3654 14.6163 -16.9668 -38.1694 1.0010 0.9993 1.0008 0.0000 0.0008 -0.0001
+ - param set #29 has cost=-0.020665 [o=87 t=28]
+ -- Parameters = 70.1840 -40.0444 31.6543 -18.5269 -23.6147 15.7269 0.9965 1.0013 0.9933 -0.0002 0.0003 -0.0014
*[#23960=-0.260357] *[#23977=-0.272917] *[#23982=-0.280949] *[#23983=-0.281255] *[#23987=-0.283259] *[#23988=-0.285376] *[#23990=-0.293894] *[#23991=-0.296079] *[#23994=-0.296579] *[#23996=-0.303272] *[#23997=-0.305394] *[#23999=-0.306511] *[#24002=-0.307862] *[#24005=-0.309856] *[#24006=-0.310674] *[#24009=-0.311123] *[#24010=-0.311464] *[#24012=-0.311517] *[#24015=-0.311637] *[#24016=-0.311867] *[#24017=-0.311978] *[#24020=-0.312556] *[#24021=-0.313372] *[#24024=-0.313602] *[#24026=-0.313865] *[#24027=-0.314256] *[#24028=-0.314817] *[#24029=-0.315518] *[#24031=-0.315841] *[#24032=-0.316146] *[#24033=-0.316207] *[#24034=-0.316208] *[#24035=-0.316254] *[#24036=-0.316355] *[#24037=-0.316509] *[#24038=-0.316631] *[#24039=-0.316929] *[#24040=-0.316932] *[#24042=-0.316946] *[#24043=-0.317197] *[#24044=-0.317597] *[#24046=-0.317637] *[#24047=-0.317752] *[#24048=-0.317761] *[#24051=-0.317889] *[#24052=-0.318078] *[#24053=-0.318142] *[#24054=-0.318158] *[#24055=-0.318178]
+ - param set #30 has cost=-0.318178 [o=-1 t=-1]
+ -- Parameters = 86.2736 -1.0713 0.2741 -0.3638 -0.2556 -0.0692 0.9870 1.0233 0.9890 0.0009 0.0051 -0.0012
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.425
+ --- dist(#3,#1) = 0.477
+ --- dist(#4,#1) = 0.455
+ --- dist(#5,#1) = 0.491
+ --- dist(#6,#1) = 0.496
+ --- dist(#7,#1) = 0.54
+ --- dist(#8,#1) = 0.559
+ --- dist(#9,#1) = 0.498
+ --- dist(#10,#1) = 0.442
+ --- dist(#11,#1) = 0.487
+ --- dist(#12,#1) = 0.424
+ --- dist(#13,#1) = 0.493
+ --- dist(#14,#1) = 0.459
+ --- dist(#15,#1) = 0.47
+ --- dist(#16,#1) = 0.437
+ --- dist(#17,#1) = 0.492
+ --- dist(#18,#1) = 0.48
+ --- dist(#19,#1) = 0.442
+ --- dist(#20,#1) = 0.457
+ --- dist(#21,#1) = 0.445
+ --- dist(#22,#1) = 0.415
+ --- dist(#23,#1) = 0.493
+ --- dist(#24,#1) = 0.537
+ --- dist(#25,#1) = 0.487
+ --- dist(#26,#1) = 0.473
+ --- dist(#27,#1) = 0.423
+ --- dist(#28,#1) = 0.452
+ --- dist(#29,#1) = 0.551
+ --- dist(#30,#1) = 0.475
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=29 usiz=173.5
+ - retaining old weight image
+ - using 37594 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31192 total points stored in 63 'TOHD(17.2644)' bloks (0 duplicates)
*[#24057=-0.354015] *[#24085=-0.354034] *[#24091=-0.354531] *[#24092=-0.355316] *[#24093=-0.35629] *[#24096=-0.356441] *[#24099=-0.35681] *[#24102=-0.35702] *[#24103=-0.357146] *[#24108=-0.357319] *[#24113=-0.357434] *[#24114=-0.35745] *[#24118=-0.35788] *[#24120=-0.357889] *[#24121=-0.358006] *[#24124=-0.358185] *[#24133=-0.358224] *[#24134=-0.358235] *[#24138=-0.358258] *[#24139=-0.358301] *[#24140=-0.358337] *[#24141=-0.35841] *[#24142=-0.358488] *[#24145=-0.358506] *[#24146=-0.358507] *[#24147=-0.358535] *[#24148=-0.358572] *[#24151=-0.358631] *[#24152=-0.358698] *[#24153=-0.358721] *[#24154=-0.358769] *[#24155=-0.358784] *[#24159=-0.358788] *[#24162=-0.358817] *[#24163=-0.358864] *[#24166=-0.358876]
+ - param set #1 has cost=-0.358876 [o=-1 t=-1]
+ -- Parameters = 86.2059 -1.0470 0.3954 -0.3188 -0.1599 -0.0604 0.9876 1.0222 0.9677 0.0010 0.0071 -0.0041
+ - param set #2 has cost=-0.028253 [o=5 t=1]
+ -- Parameters = 96.6226 -0.9696 -33.6606 23.5353 8.0199 12.1317 1.0067 0.9904 0.9974 0.0009 0.0006 0.0125
+ - param set #3 has cost=-0.027526 [o=6 t=0]
+ -- Parameters = 112.0876 -37.7818 34.8613 -35.8319 -33.5357 -42.9533 1.0009 1.0020 1.0047 -0.0002 -0.0005 0.0005
+ - param set #4 has cost=-0.026825 [o=8 t=4]
+ -- Parameters = 96.5568 0.7261 -36.0472 29.7706 11.6107 12.6608 0.9616 0.9845 0.9547 0.0049 -0.0059 -0.0080
+ - param set #5 has cost=-0.025053 [o=38 t=2]
+ -- Parameters = 110.1802 -40.6040 33.3161 -34.3193 -32.9191 -36.9144 0.9916 1.0022 0.9975 0.0109 0.0038 -0.0007
+ - param set #6 has cost=-0.023520 [o=70 t=6]
+ -- Parameters = 70.1780 26.6238 -39.8021 19.2801 -11.7839 -25.4305 0.9824 0.9978 1.0085 -0.0000 0.0013 0.0005
+ - param set #7 has cost=-0.024606 [o=60 t=11]
+ -- Parameters = 73.4981 26.3564 -42.9607 15.5735 -10.8516 -24.6349 1.0028 1.0040 0.9869 0.0041 0.0026 -0.0015
+ - param set #8 has cost=-0.023303 [o=1 t=3]
+ -- Parameters = 79.4890 34.6036 -44.3009 19.2786 -20.9716 -14.8872 0.9802 0.9655 0.9991 -0.0023 0.0095 0.0020
+ - param set #9 has cost=-0.025835 [o=68 t=5]
+ -- Parameters = 90.1993 38.4139 -38.7248 30.1393 -25.4684 9.0833 1.0056 0.9803 1.0206 0.0020 -0.0002 -0.0107
+ - param set #10 has cost=-0.026228 [o=0 t=9]
+ -- Parameters = 112.2036 -37.8929 29.9032 -19.1316 -13.2939 -36.3915 0.9892 0.9670 1.0168 -0.0063 0.0055 -0.0045
+ - param set #11 has cost=-0.022055 [o=14 t=8]
+ -- Parameters = 113.2207 -29.0080 26.7598 -16.2490 -18.8878 -43.9288 0.9982 0.9954 1.0020 -0.0031 0.0117 0.0014
+ - param set #12 has cost=-0.022872 [o=79 t=7]
+ -- Parameters = 55.6011 29.7101 34.2114 23.2147 7.4555 26.3447 0.9972 1.0417 1.0008 0.0007 -0.0001 -0.0003
+ - param set #13 has cost=-0.022904 [o=16 t=12]
+ -- Parameters = 81.2257 7.6677 -39.2263 -33.4216 13.3326 16.0042 1.0078 1.0140 0.9959 0.0010 -0.0001 0.0011
+ - param set #14 has cost=-0.021127 [o=29 t=17]
+ -- Parameters = 81.0485 3.5993 -36.5449 -34.1662 8.6682 6.9365 0.9863 0.9968 0.9869 -0.0030 -0.0009 -0.0018
+ - param set #15 has cost=-0.020421 [o=9 t=10]
+ -- Parameters = 95.6238 3.3121 -37.1669 23.0454 14.3051 -1.7729 0.9909 1.0088 1.0072 0.0236 -0.0020 0.0006
+ - param set #16 has cost=-0.022356 [o=47 t=26]
+ -- Parameters = 73.7189 -17.9583 29.6332 -19.1371 -16.4170 38.9256 0.9678 0.9463 0.9832 -0.0063 0.0013 0.0039
+ - param set #17 has cost=-0.019520 [o=34 t=18]
+ -- Parameters = 97.5361 6.0123 -39.2250 20.6887 12.2904 5.3776 0.9992 0.9917 0.9888 -0.0006 0.0021 0.0026
Center distance of 0.000000 mm
+ - param set #18 has cost=-0.019845 [o=44 t=19]
+ -- Parameters = 117.4644 22.2320 31.2286 4.2090 15.1105 -43.2023 1.0090 1.0026 1.0054 -0.0015 0.0084 -0.0031
+ - param set #19 has cost=-0.020580 [o=26 t=14]
+ -- Parameters = 73.0005 -36.6965 34.8617 -21.5369 -18.0154 21.3029 0.9841 1.0062 0.9811 -0.0017 0.0010 0.0119
+ - param set #20 has cost=-0.020912 [o=75 t=13]
+ -- Parameters = 107.8665 25.7255 -36.1085 -14.5825 -10.4285 35.7443 1.0111 0.9810 0.9797 -0.0027 -0.0009 0.0007
+ - param set #21 has cost=-0.020099 [o=39 t=24]
+ -- Parameters = 78.3526 -7.0245 -35.1638 -31.1876 0.6116 -5.3473 0.9885 0.9853 0.9889 -0.0006 -0.0030 0.0064
+ - param set #22 has cost=-0.019406 [o=18 t=20]
+ -- Parameters = 110.3808 25.9039 -32.8701 -26.7415 -9.4763 34.9989 0.9934 1.0017 0.9971 -0.0016 0.0003 -0.0021
+ - param set #23 has cost=-0.018425 [o=32 t=15]
+ -- Parameters = 83.1150 -7.0181 -38.9994 -8.1942 5.5289 2.2218 1.0020 0.9952 1.0069 -0.0022 0.0003 -0.0014
+ - param set #24 has cost=-0.018961 [o=15 t=25]
+ -- Parameters = 113.2348 -24.1982 -42.2571 -1.6340 23.9661 -17.3153 0.9708 0.9955 1.0330 0.0056 0.0011 0.0046
+ - param set #25 has cost=-0.018841 [o=87 t=28]
+ -- Parameters = 70.4292 -40.5987 32.1142 -18.8175 -24.1341 10.9370 0.9823 0.9778 0.9887 0.0008 0.0005 0.0002
Padding ...
+ - param set #26 has cost=-0.017360 [o=13 t=16]
+ -- Parameters = 84.3779 -10.5994 -36.8407 -0.5873 13.4183 -0.3925 0.9980 1.0038 0.9891 0.0013 -0.0009 -0.0024
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - param set #27 has cost=-0.016828 [o=27 t=27]
+ -- Parameters = 62.0154 28.8539 -27.3180 14.6671 -17.0135 -38.2412 1.0012 0.9994 1.0007 -0.0009 0.0008 0.0001
+ - param set #28 has cost=-0.016927 [o=67 t=22]
+ -- Parameters = 50.0802 1.3253 12.5954 29.2092 -26.9162 35.1433 0.9913 1.0004 0.9986 0.0011 -0.0006 0.0218
+ - param set #29 has cost=-0.017138 [o=55 t=21]
+ -- Parameters = 73.1622 1.1644 -43.4688 -40.4065 -23.4359 35.3152 0.9974 0.9952 1.0229 -0.0005 0.0014 0.0002
+ - param set #30 has cost=-0.015568 [o=28 t=23]
+ -- Parameters = 83.3840 1.5159 -37.7447 -10.7621 12.0528 1.6729 1.0016 1.0089 0.9914 0.0012 0.0024 -0.0031
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.426
+ --- dist(#3,#1) = 0.477
+ --- dist(#4,#1) = 0.456
+ --- dist(#5,#1) = 0.461
+ --- dist(#6,#1) = 0.493
+ --- dist(#7,#1) = 0.494
+ --- dist(#8,#1) = 0.542
+ --- dist(#9,#1) = 0.502
+ --- dist(#10,#1) = 0.559
+ --- dist(#11,#1) = 0.495
+ --- dist(#12,#1) = 0.423
+ --- dist(#13,#1) = 0.433
+ --- dist(#14,#1) = 0.487
+ --- dist(#15,#1) = 0.462
+ --- dist(#16,#1) = 0.456
+ --- dist(#17,#1) = 0.446
+ --- dist(#18,#1) = 0.47
+ --- dist(#19,#1) = 0.444
+ --- dist(#20,#1) = 0.479
+ --- dist(#21,#1) = 0.495
+ --- dist(#22,#1) = 0.416
+ --- dist(#23,#1) = 0.533
+ --- dist(#24,#1) = 0.494
+ --- dist(#25,#1) = 0.492
+ --- dist(#26,#1) = 0.465
+ --- dist(#27,#1) = 0.548
+ --- dist(#28,#1) = 0.452
+ --- dist(#29,#1) = 0.424
+ --- dist(#30,#1) = 0.477
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
+ !source mask fill: ubot=29 usiz=173.5
+ - retaining old weight image
+ - using 37594 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31038 total points stored in 63 'TOHD(17.1973)' bloks (0 duplicates)
*[#27235=-0.376103] *[#27257=-0.376245] *[#27258=-0.376471] *[#27259=-0.376824] *[#27260=-0.377315] *[#27265=-0.377594] *[#27278=-0.377649] *[#27279=-0.377758] *[#27282=-0.377825] *[#27283=-0.377872] *[#27284=-0.377939] *[#27285=-0.378026] *[#27290=-0.378082] *[#27291=-0.378153] *[#27292=-0.378185] *[#27297=-0.378245] *[#27298=-0.378264] *[#27299=-0.378308] *[#27302=-0.378323] *[#27304=-0.378347] *[#27308=-0.378363] *[#27310=-0.378473] *[#27317=-0.378596] *[#27320=-0.37862] *[#27329=-0.378621] *[#27330=-0.378647] *[#27334=-0.378657] *[#27336=-0.378715] *[#27343=-0.378839] *[#27346=-0.378883] *[#27349=-0.378885] *[#27354=-0.378951]
+ - param set #1 has cost=-0.378951 [o=-1 t=-1]
+ -- Parameters = 86.1957 -0.9975 0.3693 -0.2527 -0.7206 -0.1287 0.9899 1.0226 0.9731 0.0024 0.0080 -0.0140
+ - param set #2 has cost=-0.024672 [o=5 t=1]
+ -- Parameters = 96.6628 -0.9456 -33.6972 23.4977 8.0287 12.0988 1.0067 0.9897 0.9956 0.0006 0.0014 0.0122
+ - param set #3 has cost=-0.024816 [o=6 t=0]
+ -- Parameters = 112.3583 -37.8816 34.8519 -35.7894 -33.8135 -43.1499 1.0004 1.0068 0.9918 0.0003 -0.0013 0.0006
Resampling ...
+ - param set #4 has cost=-0.022931 [o=8 t=4]
+ -- Parameters = 96.5279 0.6754 -35.8462 29.8396 11.5977 12.3999 0.9606 0.9845 0.9608 0.0053 -0.0052 -0.0021
+ - param set #5 has cost=-0.023778 [o=0 t=9]
+ -- Parameters = 112.1434 -37.9315 29.9747 -19.1573 -13.1316 -36.4627 0.9904 0.9666 1.0127 -0.0064 0.0052 -0.0044
+ - param set #6 has cost=-0.024569 [o=68 t=5]
+ -- Parameters = 90.0805 38.7333 -38.4809 30.5531 -25.3502 9.3891 1.0074 0.9584 1.0240 0.0010 0.0014 -0.0115
+ - param set #7 has cost=-0.021659 [o=38 t=2]
+ -- Parameters = 110.3401 -40.6012 33.3546 -34.3791 -32.9155 -36.9950 0.9895 1.0011 0.9968 0.0111 0.0093 -0.0009
+ - param set #8 has cost=-0.021586 [o=60 t=11]
+ -- Parameters = 73.5014 26.3280 -42.9750 15.5536 -10.8603 -24.6164 1.0030 1.0047 0.9865 0.0040 0.0025 -0.0014
+ - param set #9 has cost=-0.021212 [o=70 t=6]
+ -- Parameters = 70.3735 26.6134 -40.5861 18.8345 -11.9221 -25.7550 0.9828 0.9914 1.0050 -0.0013 0.0065 -0.0052
+ - param set #10 has cost=-0.020681 [o=1 t=3]
+ -- Parameters = 79.4664 34.5679 -44.4711 19.2693 -21.0485 -14.8759 0.9779 0.9674 0.9905 -0.0025 0.0089 0.0017
+ - param set #11 has cost=-0.019865 [o=16 t=12]
+ -- Parameters = 81.1725 7.7296 -39.2366 -33.5579 13.3712 16.0959 1.0116 1.0123 0.9951 0.0008 0.0002 0.0022
+ - param set #12 has cost=-0.019551 [o=79 t=7]
+ -- Parameters = 55.5284 29.7207 34.1705 23.2041 7.4842 26.4721 0.9966 1.0438 1.0005 0.0000 0.0005 0.0005
+ - param set #13 has cost=-0.021757 [o=47 t=26]
+ -- Parameters = 74.1714 -18.4852 29.5124 -19.7787 -16.7212 38.7845 0.9391 0.9387 0.9875 -0.0036 0.0018 0.0058
+ - param set #14 has cost=-0.020777 [o=14 t=8]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ -- Parameters = 113.0965 -29.0358 26.7540 -16.2813 -18.7497 -44.0268 0.9964 0.9923 1.0039 -0.0054 0.0181 0.0022
+ - param set #15 has cost=-0.017505 [o=29 t=17]
+ -- Parameters = 81.0034 3.4485 -36.5862 -34.1906 8.8652 7.0847 0.9833 0.9970 0.9830 0.0021 -0.0019 -0.0024
+ - param set #16 has cost=-0.018320 [o=75 t=13]
+ -- Parameters = 107.8936 26.0570 -36.1243 -14.5970 -10.4410 35.7767 1.0121 0.9792 0.9791 0.0035 -0.0009 0.0006
+ - param set #17 has cost=-0.018703 [o=26 t=14]
+ -- Parameters = 72.9411 -36.7741 34.8618 -21.5840 -18.4408 21.3678 0.9860 1.0029 0.9566 0.0072 0.0020 0.0124
+ - param set #18 has cost=-0.016705 [o=9 t=10]
+ -- Parameters = 95.8755 3.5949 -37.2745 23.2447 14.5449 -1.7914 0.9883 1.0144 1.0039 0.0227 0.0039 0.0005
+ - param set #19 has cost=-0.018298 [o=39 t=24]
+ -- Parameters = 78.3275 -7.2778 -35.0686 -30.8340 0.4001 -5.2855 0.9917 0.9807 0.9936 -0.0004 -0.0036 0.0089
+ - param set #20 has cost=-0.017919 [o=44 t=19]
+ -- Parameters = 117.6658 22.1995 31.4556 4.4921 14.9312 -43.1175 1.0234 0.9999 1.0001 -0.0019 0.0088 -0.0031
+ - param set #21 has cost=-0.017740 [o=34 t=18]
+ -- Parameters = 97.8673 5.9252 -39.4022 20.9917 12.4593 5.6541 1.0048 0.9887 0.9798 0.0082 0.0085 0.0065
+ - param set #22 has cost=-0.016833 [o=18 t=20]
+ -- Parameters = 110.1725 25.7703 -32.8100 -26.6956 -9.4768 34.8653 0.9923 1.0026 0.9992 -0.0023 0.0003 -0.0025
+ - param set #23 has cost=-0.016041 [o=15 t=25]
+ -- Parameters = 113.2377 -24.2793 -42.3134 -1.5938 24.1082 -17.0175 0.9664 0.9960 1.0333 0.0063 0.0004 0.0050
RMS[0] = 0.333679 0.165083 ITER = 9/50
0.333679
+ - param set #24 has cost=-0.016486 [o=87 t=28]
+ -- Parameters = 70.4937 -40.5492 32.0909 -18.6955 -24.1412 10.9360 0.9831 0.9784 0.9884 0.0010 0.0007 0.0006
+ - param set #25 has cost=-0.014772 [o=32 t=15]
+ -- Parameters = 83.0582 -6.9147 -38.9448 -8.3132 5.7068 2.2767 1.0069 0.9931 1.0100 -0.0017 -0.0007 -0.0034
Warping has converged.
+ - param set #26 has cost=-0.014997 [o=13 t=16]
+ -- Parameters = 84.4259 -10.6050 -36.8243 -0.5545 13.3830 -0.4547 0.9985 1.0042 0.9892 0.0015 -0.0011 -0.0028
+ - param set #27 has cost=-0.017566 [o=55 t=21]
+ -- Parameters = 73.6048 1.7693 -43.5022 -40.4062 -23.5654 36.0402 0.9657 1.0022 1.0588 0.0094 0.0084 0.0018
+ - param set #28 has cost=-0.015956 [o=67 t=22]
+ -- Parameters = 50.0529 1.3248 12.5498 29.2003 -26.8889 35.1372 0.9907 0.9999 0.9991 0.0010 -0.0006 0.0218
+ - param set #29 has cost=-0.015066 [o=27 t=27]
+ -- Parameters = 62.4831 28.1970 -27.6605 15.6787 -17.3253 -38.6343 0.9941 1.0022 0.9939 0.0043 0.0013 0.0003
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - param set #30 has cost=-0.014296 [o=28 t=23]
+ -- Parameters = 83.4746 1.4570 -37.7831 -13.5438 11.9701 1.5161 1.0021 1.0064 0.9818 0.0006 0.0034 -0.0031
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.478
+ --- dist(#3,#1) = 0.426
+ --- dist(#4,#1) = 0.497
+ --- dist(#5,#1) = 0.462
+ --- dist(#6,#1) = 0.453
+ --- dist(#7,#1) = 0.432
+ --- dist(#8,#1) = 0.495
+ --- dist(#9,#1) = 0.542
+ --- dist(#10,#1) = 0.512
+ --- dist(#11,#1) = 0.488
+ --- dist(#12,#1) = 0.561
+ --- dist(#13,#1) = 0.495
+ --- dist(#14,#1) = 0.423
+ --- dist(#15,#1) = 0.447
+ --- dist(#16,#1) = 0.456
+ --- dist(#17,#1) = 0.443
+ --- dist(#18,#1) = 0.478
+ --- dist(#19,#1) = 0.497
+ --- dist(#20,#1) = 0.548
+ --- dist(#21,#1) = 0.462
+ --- dist(#22,#1) = 0.415
+ --- dist(#23,#1) = 0.471
+ --- dist(#24,#1) = 0.494
+ --- dist(#25,#1) = 0.534
+ --- dist(#26,#1) = 0.452
+ --- dist(#27,#1) = 0.428
+ --- dist(#28,#1) = 0.465
+ --- dist(#29,#1) = 0.491
+ --- dist(#30,#1) = 0.477
+ - Total coarse refinement net CPU time = 0.0 s; 9451 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=29 usiz=173.5
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 30994 total points stored in 64 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.371851 * [o=-1 t=-1]
+ -- Parameters = 86.1957 -0.9975 0.3693 -0.2527 -0.7206 -0.1287 0.9899 1.0226 0.9731 0.0024 0.0080 -0.0140
+ - cost(#2)=-0.023779 [o=6 t=0]
+ -- Parameters = 112.3583 -37.8816 34.8519 -35.7894 -33.8135 -43.1499 1.0004 1.0068 0.9918 0.0003 -0.0013 0.0006
+ - cost(#3)=-0.021233 [o=5 t=1]
+ -- Parameters = 96.6628 -0.9456 -33.6972 23.4977 8.0287 12.0988 1.0067 0.9897 0.9956 0.0006 0.0014 0.0122
+ - cost(#4)=-0.021993 [o=68 t=5]
+ -- Parameters = 90.0805 38.7333 -38.4809 30.5531 -25.3502 9.3891 1.0074 0.9584 1.0240 0.0010 0.0014 -0.0115
+ - cost(#5)=-0.022545 [o=0 t=9]
+ -- Parameters = 112.1434 -37.9315 29.9747 -19.1573 -13.1316 -36.4627 0.9904 0.9666 1.0127 -0.0064 0.0052 -0.0044
+ - cost(#6)=-0.019564 [o=8 t=4]
+ -- Parameters = 96.5279 0.6754 -35.8462 29.8396 11.5977 12.3999 0.9606 0.9845 0.9608 0.0053 -0.0052 -0.0021
+ - cost(#7)=-0.020598 [o=47 t=26]
+ -- Parameters = 74.1714 -18.4852 29.5124 -19.7787 -16.7212 38.7845 0.9391 0.9387 0.9875 -0.0036 0.0018 0.0058
+ - cost(#8)=-0.019827 [o=38 t=2]
+ -- Parameters = 110.3401 -40.6012 33.3546 -34.3791 -32.9155 -36.9950 0.9895 1.0011 0.9968 0.0111 0.0093 -0.0009
+ - cost(#9)=-0.019709 [o=60 t=11]
+ -- Parameters = 73.5014 26.3280 -42.9750 15.5536 -10.8603 -24.6164 1.0030 1.0047 0.9865 0.0040 0.0025 -0.0014
+ - cost(#10)=-0.019739 [o=70 t=6]
+ -- Parameters = 70.3735 26.6134 -40.5861 18.8345 -11.9221 -25.7550 0.9828 0.9914 1.0050 -0.0013 0.0065 -0.0052
+ - cost(#11)=-0.020378 [o=14 t=8]
+ -- Parameters = 113.0965 -29.0358 26.7540 -16.2813 -18.7497 -44.0268 0.9964 0.9923 1.0039 -0.0054 0.0181 0.0022
+ - cost(#12)=-0.017940 [o=1 t=3]
+ -- Parameters = 79.4664 34.5679 -44.4711 19.2693 -21.0485 -14.8759 0.9779 0.9674 0.9905 -0.0025 0.0089 0.0017
+ - cost(#13)=-0.016945 [o=16 t=12]
+ -- Parameters = 81.1725 7.7296 -39.2366 -33.5579 13.3712 16.0959 1.0116 1.0123 0.9951 0.0008 0.0002 0.0022
+ - cost(#14)=-0.018322 [o=79 t=7]
+ -- Parameters = 55.5284 29.7207 34.1705 23.2041 7.4842 26.4721 0.9966 1.0438 1.0005 0.0000 0.0005 0.0005
+ - cost(#15)=-0.017098 [o=26 t=14]
+ -- Parameters = 72.9411 -36.7741 34.8618 -21.5840 -18.4408 21.3678 0.9860 1.0029 0.9566 0.0072 0.0020 0.0124
+ - cost(#16)=-0.017023 [o=75 t=13]
+ -- Parameters = 107.8936 26.0570 -36.1243 -14.5970 -10.4410 35.7767 1.0121 0.9792 0.9791 0.0035 -0.0009 0.0006
+ - cost(#17)=-0.016390 [o=39 t=24]
+ -- Parameters = 78.3275 -7.2778 -35.0686 -30.8340 0.4001 -5.2855 0.9917 0.9807 0.9936 -0.0004 -0.0036 0.0089
+ - cost(#18)=-0.016067 [o=44 t=19]
+ -- Parameters = 117.6658 22.1995 31.4556 4.4921 14.9312 -43.1175 1.0234 0.9999 1.0001 -0.0019 0.0088 -0.0031
+ - cost(#19)=-0.015797 [o=34 t=18]
+ -- Parameters = 97.8673 5.9252 -39.4022 20.9917 12.4593 5.6541 1.0048 0.9887 0.9798 0.0082 0.0085 0.0065
+ - cost(#20)=-0.015189 [o=55 t=21]
+ -- Parameters = 73.6048 1.7693 -43.5022 -40.4062 -23.5654 36.0402 0.9657 1.0022 1.0588 0.0094 0.0084 0.0018
+ - cost(#21)=-0.014910 [o=29 t=17]
+ -- Parameters = 81.0034 3.4485 -36.5862 -34.1906 8.8652 7.0847 0.9833 0.9970 0.9830 0.0021 -0.0019 -0.0024
+ - cost(#22)=-0.014831 [o=18 t=20]
+ -- Parameters = 110.1725 25.7703 -32.8100 -26.6956 -9.4768 34.8653 0.9923 1.0026 0.9992 -0.0023 0.0003 -0.0025
+ - cost(#23)=-0.014140 [o=9 t=10]
+ -- Parameters = 95.8755 3.5949 -37.2745 23.2447 14.5449 -1.7914 0.9883 1.0144 1.0039 0.0227 0.0039 0.0005
+ - cost(#24)=-0.016059 [o=87 t=28]
+ -- Parameters = 70.4937 -40.5492 32.0909 -18.6955 -24.1412 10.9360 0.9831 0.9784 0.9884 0.0010 0.0007 0.0006
+ - cost(#25)=-0.014013 [o=15 t=25]
+ -- Parameters = 113.2377 -24.2793 -42.3134 -1.5938 24.1082 -17.0175 0.9664 0.9960 1.0333 0.0063 0.0004 0.0050
+ - cost(#26)=-0.013060 [o=67 t=22]
+ -- Parameters = 50.0529 1.3248 12.5498 29.2003 -26.8889 35.1372 0.9907 0.9999 0.9991 0.0010 -0.0006 0.0218
+ - cost(#27)=-0.014405 [o=27 t=27]
+ -- Parameters = 62.4831 28.1970 -27.6605 15.6787 -17.3253 -38.6343 0.9941 1.0022 0.9939 0.0043 0.0013 0.0003
+ - cost(#28)=-0.013064 [o=13 t=16]
+ -- Parameters = 84.4259 -10.6050 -36.8243 -0.5545 13.3830 -0.4547 0.9985 1.0042 0.9892 0.0015 -0.0011 -0.0028
+ - cost(#29)=-0.013108 [o=32 t=15]
+ -- Parameters = 83.0582 -6.9147 -38.9448 -8.3132 5.7068 2.2767 1.0069 0.9931 1.0100 -0.0017 -0.0007 -0.0034
+ - cost(#30)=-0.012338 [o=28 t=23]
+ -- Parameters = 83.4746 1.4570 -37.7831 -13.5438 11.9701 1.5161 1.0021 1.0064 0.9818 0.0006 0.0034 -0.0031
+ - cost(#31)=-0.017540 [o=-2 t=-2]
+ -- Parameters = 86.5052 -0.6043 -5.1381 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.372702 * [o=-1 t=-1]
+ -- Parameters = 86.1767 -1.0089 0.3316 -0.2202 -0.7039 -0.1230 0.9912 1.0197 0.9743 0.0012 0.0076 -0.0146
+ - cost(#2)=-0.023867 [o=6 t=0]
+ -- Parameters = 112.3423 -37.8764 34.8526 -35.8333 -33.8581 -43.1990 0.9991 1.0081 0.9903 0.0001 -0.0013 0.0002
+ - cost(#3)=-0.021293 [o=5 t=1]
+ -- Parameters = 96.6537 -0.9739 -33.7720 23.5324 7.9909 12.1118 1.0068 0.9897 0.9953 0.0007 0.0020 0.0118
+ - cost(#4)=-0.022569 [o=68 t=5]
+ -- Parameters = 89.9926 38.7727 -38.3287 30.7456 -25.2137 9.4897 1.0121 0.9525 1.0322 -0.0085 0.0007 -0.0119
+ - cost(#5)=-0.022670 [o=0 t=9]
+ -- Parameters = 112.0438 -37.9442 30.0104 -19.1938 -13.1619 -36.5463 0.9909 0.9663 1.0103 -0.0129 0.0056 -0.0043
+ - cost(#6)=-0.020169 [o=8 t=4]
+ -- Parameters = 97.0138 1.4915 -36.2219 29.8334 11.0977 11.8960 0.9531 0.9750 0.9645 0.0048 -0.0059 0.0008
+ - cost(#7)=-0.020746 [o=47 t=26]
+ -- Parameters = 74.0782 -18.4853 29.6468 -19.8286 -16.7194 38.7711 0.9344 0.9382 0.9835 -0.0057 0.0012 0.0070
+ - cost(#8)=-0.019936 [o=38 t=2]
+ -- Parameters = 110.3211 -40.6039 33.4218 -34.3512 -32.9235 -37.0568 0.9864 0.9998 0.9973 0.0106 0.0092 -0.0009
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - cost(#9)=-0.019902 [o=60 t=11]
+ -- Parameters = 73.4993 26.3194 -42.8599 15.5031 -10.8614 -24.6973 1.0026 1.0046 1.0099 0.0038 0.0024 -0.0016
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
+ - cost(#10)=-0.021275 [o=70 t=6]
+ -- Parameters = 71.0899 26.1437 -40.3112 18.0232 -11.0931 -25.9051 0.9821 1.0011 1.0199 0.0020 0.0138 -0.0129
+ - cost(#11)=-0.020576 [o=14 t=8]
+ -- Parameters = 113.1728 -29.0738 26.7435 -16.2993 -18.7412 -43.9419 0.9944 0.9932 1.0057 -0.0061 0.0177 0.0025
+ - cost(#12)=-0.018654 [o=1 t=3]
+ -- Parameters = 79.5306 34.5479 -44.6702 19.5297 -21.1091 -14.7478 0.9732 0.9637 0.9780 -0.0018 -0.0025 0.0016
+ - cost(#13)=-0.017082 [o=16 t=12]
+ -- Parameters = 81.2807 7.7013 -39.2663 -33.5304 13.3789 15.9748 1.0136 1.0127 0.9950 0.0008 0.0007 0.0030
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - cost(#14)=-0.019670 [o=79 t=7]
+ -- Parameters = 55.2362 26.9078 32.6920 23.6065 6.9765 26.5738 0.9855 1.0438 1.0025 0.0004 0.0027 0.0010
+ - cost(#15)=-0.017618 [o=26 t=14]
+ -- Parameters = 73.0916 -36.8200 34.8585 -21.4264 -18.7484 20.9767 0.9864 1.0082 0.9326 0.0041 0.0004 0.0135
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - cost(#16)=-0.017254 [o=75 t=13]
+ -- Parameters = 107.8591 25.9989 -36.1458 -14.5102 -10.5105 35.7767 1.0131 0.9779 0.9788 0.0127 -0.0013 -0.0009
+ - cost(#17)=-0.016663 [o=39 t=24]
+ -- Parameters = 78.4135 -7.1335 -35.1923 -30.8315 0.3863 -5.2953 0.9938 0.9772 0.9911 -0.0003 -0.0048 0.0085
+ - cost(#18)=-0.018463 [o=44 t=19]
+ -- Parameters = 117.2054 21.7958 31.6366 4.1798 13.3150 -42.8293 1.0367 0.9844 0.9904 -0.0091 0.0078 -0.0011
+ - cost(#19)=-0.015872 [o=34 t=18]
+ -- Parameters = 97.8746 5.9800 -39.4999 21.0462 12.5047 5.6658 1.0047 0.9879 0.9763 0.0095 0.0083 0.0062
+ - cost(#20)=-0.015684 [o=55 t=21]
+ -- Parameters = 73.7723 1.8002 -43.5165 -40.3658 -23.7033 36.0304 0.9682 1.0041 1.0580 0.0162 0.0086 -0.0004
+ - cost(#21)=-0.014995 [o=29 t=17]
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
+ -- Parameters = 80.9251 3.3701 -36.5720 -34.1806 8.8282 7.1355 0.9823 0.9979 0.9844 0.0023 -0.0022 -0.0021
+ - cost(#22)=-0.014914 [o=18 t=20]
+ -- Parameters = 110.1408 25.7880 -32.8375 -26.7204 -9.4125 34.8342 0.9923 1.0028 0.9994 -0.0031 0.0011 -0.0028
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-03_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-03_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-03_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-03_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - cost(#23)=-0.014718 [o=9 t=10]
+ -- Parameters = 95.9261 3.8571 -37.4105 23.8495 14.5444 -1.9807 0.9755 1.0033 1.0193 0.0257 0.0041 -0.0004
+ - cost(#24)=-0.016115 [o=87 t=28]
+ -- Parameters = 70.5042 -40.5994 32.1316 -18.7438 -24.0996 10.9022 0.9831 0.9781 0.9881 0.0016 0.0005 0.0005
+ - cost(#25)=-0.014090 [o=15 t=25]
+ -- Parameters = 113.2660 -24.2754 -42.3073 -1.6221 24.0773 -17.0459 0.9636 0.9959 1.0356 0.0066 -0.0010 0.0050
+ - cost(#26)=-0.013339 [o=67 t=22]
+ -- Parameters = 49.9012 1.3336 12.3550 29.1900 -26.8506 35.1037 0.9846 0.9990 0.9949 -0.0050 -0.0009 0.0207
Clipping -0.000100 1326.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - cost(#27)=-0.014951 [o=27 t=27]
+ -- Parameters = 62.5493 28.2924 -27.4700 15.9069 -17.3638 -38.9253 0.9941 0.9997 0.9894 0.0026 0.0027 0.0004
+ - cost(#28)=-0.013810 [o=13 t=16]
+ -- Parameters = 84.2340 -10.8027 -36.7211 -0.5829 13.2703 -0.6337 0.9982 1.0054 0.9939 -0.0089 0.0000 -0.0033
+ - cost(#29)=-0.013388 [o=32 t=15]
+ -- Parameters = 83.0131 -7.0452 -38.8960 -8.2229 5.7418 2.0065 1.0110 0.9900 1.0154 -0.0022 -0.0001 0.0001
++ Output dataset ././__ats_tmp___pad15_pre.sub-03_T1w_ns+orig.BRIK
+ - cost(#30)=-0.012784 [o=28 t=23]
+ -- Parameters = 83.5685 1.1846 -37.6638 -13.4477 12.4991 1.3234 1.0063 1.0148 0.9736 0.0008 0.0026 -0.0023
+ - cost(#31)=-0.369675 [o=-2 t=-2]
+ -- Parameters = 86.1805 -1.1002 0.2862 -0.0253 -0.3479 -0.0295 0.9922 1.0237 0.9762 0.0086 0.0058 -0.0066
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.372702
+ - Initial fine Parameters = 86.1767 -1.0089 0.3316 -0.2202 -0.7039 -0.1230 0.9912 1.0197 0.9743 0.0012 0.0076 -0.0146
Unpadding ...
++ Output dataset ./__ats_tmp___rs_pre.sub-02_T1w_ns+tlrc.BRIK
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-02_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
+ - Finalish cost = -0.373597 ; 690 funcs
++ output dataset: ./__ats_tmp___upad15_pre.sub-03_T1w_ns+orig.BRIK
+ - ini Finalish Parameters = 86.1652 -0.8738 0.3592 -0.2053 -1.1470 -0.1502 0.9919 1.0185 0.9722 0.0025 0.0086 -0.0234
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - Final cost = -0.373598 ; 272 funcs
+ Final fine fit Parameters:
x-shift= 86.1650 y-shift= -0.8756 z-shift= 0.3614 ... enorm= 86.1702 mm
z-angle= -0.2058 x-angle= -1.1477 y-angle= -0.1491 ... total= 1.1753 deg
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
x-scale= 0.9918 y-scale= 1.0186 z-scale= 0.9723 ... vol3D= 0.9822 = base bigger than source
y/x-shear= 0.0025 z/x-shear= 0.0086 z/y-shear= -0.0234
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-03_T1w_ns+orig
++ image warp: parameters = 86.1650 -0.8756 0.3614 -0.2058 -1.1477 -0.1491 0.9918 1.0186 0.9723 0.0025 0.0086 -0.0234
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-03_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-03_T1w_ns+orig __ats_tmp___upad15_pre.sub-03_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-03_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-03_T1w_ns+tlrc.HEAD -> sub-03_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-03_T1w_ns+tlrc.BRIK -> sub-03_T1w_ns+tlrc.BRIK
Cleanup ...
++ Initial scale factor set to 0.30/117.00=3.9e+02
cat_matvec sub-03_T1w_ns+tlrc::WARP_DATA -I
++ Unloading unneeded data
if ( ! -f sub-03_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-03.r01.tcat+orig
++ Output dataset ./sub-01_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-01_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 26.3
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/__tt_sub-01_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/sub-01_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-01_T1w_ns+orig sub-01_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-03.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-01_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-01_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-01_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-01_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
+ Automask has 39096 voxels
+ 6008 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
Performing center alignment with @Align_Centers
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-01_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-01_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-01_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-01_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-01_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-01_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-01_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
RMS[0] = 0.34262 0.156252 ITER = 7/50
0.34262
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Center distance of 0.000008 mm
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Resampling ...
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-02_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-02_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-02_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-02_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-02_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-02_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-02_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-02_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-02_T1w_ns+orig __ats_tmp___upad15_pre.sub-02_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-02_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-02_T1w_ns+tlrc.HEAD -> sub-02_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-02_T1w_ns+tlrc.BRIK -> sub-02_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-02_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-02_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-02.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-02.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Automask has 39248 voxels
+ 6076 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1629.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-01_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-01_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/120.00=4e+02
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.016 pitch=-0.686 yaw=-0.068 dS=-0.647 dL=-0.033 dP=+0.070
++ Mean: roll=+0.057 pitch=+0.472 yaw=+0.062 dS=-0.125 dL=+0.198 dP=+0.296
++ Max : roll=+0.193 pitch=+0.955 yaw=+0.230 dS=+0.431 dL=+0.359 dP=+0.516
++ Max displacements (mm) for each sub-brick:
1.17(0.00) 1.17(0.04) 1.18(0.08) 1.17(0.13) 1.21(0.05) 1.23(0.04) 1.25(0.07) 1.25(0.06) 1.25(0.08) 1.29(0.06) 1.27(0.09) 1.33(0.14) 1.36(0.06) 1.29(0.07) 1.27(0.05) 1.32(0.08) 1.34(0.06) 1.38(0.07) 1.32(0.07) 1.32(0.06) 1.38(0.06) 1.31(0.13) 1.39(0.12) 1.36(0.09) 1.42(0.11) 1.37(0.09) 1.39(0.08) 1.36(0.08) 1.36(0.07) 1.41(0.07) 1.34(0.12) 1.48(0.19) 1.42(0.12) 1.48(0.11) 1.50(0.06) 1.50(0.04) 1.51(0.06) 1.51(0.03) 1.54(0.07) 1.52(0.06) 1.47(0.12) 1.52(0.09) 1.49(0.11) 1.51(0.14) 1.47(0.08) 1.46(0.06) 1.53(0.09) 1.50(0.08) 1.45(0.09) 1.45(0.10) 1.37(0.12) 1.43(0.08) 1.49(0.10) 1.42(0.09) 1.44(0.07) 1.43(0.12) 1.52(0.18) 1.56(0.05) 1.47(0.11) 1.67(0.22) 1.64(0.11) 1.60(0.14) 1.60(0.12) 1.41(0.44) 1.49(0.15) 1.47(0.22) 1.52(0.11) 0.71(0.89) 1.29(1.48) 1.05(0.35) 0.85(0.21) 0.53(0.80) 0.47(0.09) 0.55(0.14) 0.60(0.12) 0.52(0.09) 0.64(0.14) 0.77(0.16) 0.85(0.11) 0.81(0.12) 0.55(0.36) 0.62(0.23) 0.83(0.24) 0.63(0.20) 0.81(0.21) 0.90(0.20) 0.94(0.10) 0.91(0.08) 0.85(0.07) 0.83(0.10) 0.65(0.21) 0.75(0.19) 0.87(0.19) 0.86(0.13) 1.03(0.18) 0.92(0.11) 1.14(0.22) 1.13(0.08) 1.28(0.16) 1.29(0.08) 1.22(0.08) 1.14(0.09) 1.23(0.15) 1.19(0.08) 1.23(0.11) 1.13(0.16) 1.11(0.10) 1.05(0.11) 1.14(0.12) 1.15(0.06) 1.22(0.13) 1.27(0.06) 1.20(0.09) 1.24(0.07) 1.18(0.09) 1.25(0.10) 1.30(0.08) 1.24(0.09) 1.34(0.11) 1.30(0.07) 1.34(0.06) 1.39(0.07) 1.37(0.08) 1.40(0.07) 1.42(0.08) 1.45(0.06) 1.64(0.19) 1.63(0.06) 1.65(0.08) 1.68(0.05) 1.68(0.08) 1.75(0.10) 1.88(0.13) 1.80(0.12) 1.76(0.08) 1.74(0.10) 1.50(0.32) 1.31(0.29) 1.39(0.20) 1.49(0.12) 1.41(0.14) 1.50(0.11) 1.44(0.09) 1.50(0.08) 1.41(0.09) 1.30(1.12)
++ Max displacement in automask = 1.88 (mm) at sub-brick 132
++ Max delta displ in automask = 1.48 (mm) at sub-brick 68
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-03.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-03_T1w_ns+tlrc::WARP_DATA -I sub-03_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-03_T1w_ns+tlrc -input pb00.sub-03.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-03.r01.tcat+orig.HEAD
++ Base dataset: ./sub-03_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -3.451 -12.560 -40.570
+ shift search range is +/- = 57.780 69.336 57.780
+ 6.0% 18.1% 70.2%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
RMS[0] = 0.278413 0.171367 ITER = 7/50
0.278413
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.064 pitch=-0.321 yaw=-0.150 dS=-0.229 dL=-0.135 dP=-0.090
++ Mean: roll=+0.003 pitch=-0.039 yaw=-0.022 dS=-0.030 dL=+0.023 dP=+0.020
++ Max : roll=+0.057 pitch=+0.141 yaw=+0.137 dS=+0.171 dL=+0.173 dP=+0.138
++ Max displacements (mm) for each sub-brick:
0.75(0.00) 0.71(0.06) 0.62(0.09) 0.61(0.03) 0.63(0.05) 0.63(0.10) 0.65(0.12) 0.70(0.09) 0.63(0.07) 0.54(0.12) 0.57(0.13) 0.56(0.06) 0.53(0.06) 0.49(0.13) 0.52(0.12) 0.52(0.05) 0.53(0.05) 0.53(0.05) 0.54(0.09) 0.49(0.09) 0.54(0.09) 0.43(0.15) 0.48(0.10) 0.43(0.08) 0.45(0.06) 0.43(0.11) 0.47(0.05) 0.43(0.05) 0.48(0.09) 0.41(0.11) 0.48(0.10) 0.45(0.08) 0.40(0.08) 0.40(0.03) 0.41(0.08) 0.33(0.18) 0.34(0.06) 0.29(0.10) 0.30(0.06) 0.31(0.07) 0.31(0.06) 0.34(0.20) 0.33(0.27) 0.36(0.10) 0.35(0.04) 0.30(0.08) 0.33(0.13) 0.27(0.15) 0.27(0.04) 0.29(0.08) 0.28(0.08) 0.30(0.11) 0.27(0.13) 0.27(0.08) 0.28(0.04) 0.24(0.10) 0.22(0.06) 0.28(0.14) 0.25(0.17) 0.22(0.13) 0.22(0.07) 0.19(0.07) 0.24(0.12) 0.21(0.10) 0.22(0.06) 0.21(0.05) 0.19(0.07) 0.14(0.08) 0.17(0.11) 0.09(0.13) 0.10(0.06) 0.11(0.08) 0.08(0.07) 0.14(0.11) 0.07(0.18) 0.09(0.07) 0.18(0.24) 0.05(0.14) 0.09(0.05) 0.12(0.05) 0.06(0.09) 0.11(0.09) 0.00(0.11) 0.08(0.08) 0.04(0.06) 0.11(0.09) 0.10(0.10) 0.19(0.13) 0.16(0.13) 0.17(0.12) 0.10(0.16) 0.15(0.14) 0.15(0.05) 0.14(0.02) 0.18(0.05) 0.15(0.11) 0.14(0.07) 0.18(0.04) 0.13(0.06) 0.10(0.04) 0.09(0.04) 0.12(0.08) 0.14(0.05) 0.19(0.16) 0.12(0.12) 0.16(0.11) 0.11(0.11) 0.17(0.12) 0.16(0.10) 0.16(0.09) 0.14(0.09) 0.14(0.04) 0.19(0.06) 0.15(0.18) 0.20(0.16) 0.21(0.08) 0.27(0.07) 0.20(0.08) 0.23(0.07) 0.21(0.11) 0.23(0.08) 0.27(0.08) 0.32(0.07) 0.27(0.11) 0.25(0.07) 0.28(0.06) 0.29(0.10) 0.31(0.09) 0.30(0.06) 0.35(0.08) 0.29(0.08) 0.31(0.04) 0.26(0.18) 0.31(0.18) 0.30(0.05) 0.32(0.05) 0.31(0.05) 0.32(0.03) 0.34(0.12) 0.35(0.04) 0.41(0.16) 0.42(0.11) 0.43(0.08) 0.42(0.10) 0.44(0.10) 0.43(0.12)
++ Max displacement in automask = 0.75 (mm) at sub-brick 0
++ Max delta displ in automask = 0.27 (mm) at sub-brick 42
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-02.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Authored by: RW Cox
*+ WARNING: input 'a' is not used in the expression
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-01_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-01_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-01_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-01_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-01_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-01_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-01_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-01_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-01_T1w_ns+orig __ats_tmp___upad15_pre.sub-01_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-01_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-01_T1w_ns+tlrc.HEAD -> sub-01_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-01_T1w_ns+tlrc.BRIK -> sub-01_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-01_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-01_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-01.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-01.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 39709 voxels
+ 5833 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-02_T1w_ns+tlrc::WARP_DATA -I sub-02_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-02_T1w_ns+tlrc -input pb00.sub-02.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-02.r01.tcat+orig.HEAD
++ Base dataset: ./sub-02_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 4.087 0.458 -27.207
+ shift search range is +/- = 57.780 69.336 57.780
+ 7.1% 0.7% 47.1%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.9
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-03_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -4.335 -18.106 -35.069
+ shift search range is +/- = 57.780 69.336 57.780
+ 7.5% 26.1% 60.7%
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-02_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.6
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.934 -5.532 -24.833
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.6% 8.0% 43.0%
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.194 pitch=-0.073 yaw=-0.245 dS=-0.808 dL=-0.235 dP=-0.321
++ Mean: roll=-0.031 pitch=+0.095 yaw=-0.070 dS=-0.131 dL=-0.030 dP=-0.055
++ Max : roll=+0.085 pitch=+0.349 yaw=+0.097 dS=+0.101 dL=+0.117 dP=+0.052
++ Max displacements (mm) for each sub-brick:
0.48(0.00) 0.49(0.12) 0.52(0.05) 0.53(0.05) 0.50(0.07) 0.57(0.17) 0.48(0.13) 0.48(0.07) 0.52(0.08) 0.50(0.04) 0.48(0.04) 0.52(0.07) 0.48(0.11) 0.55(0.11) 0.50(0.08) 0.48(0.05) 0.50(0.06) 0.57(0.30) 0.41(0.27) 0.29(0.26) 0.29(0.05) 0.29(0.05) 0.27(0.06) 0.26(0.02) 0.31(0.11) 0.27(0.05) 0.32(0.09) 0.28(0.07) 0.31(0.09) 0.27(0.06) 0.27(0.03) 0.32(0.07) 0.26(0.08) 0.31(0.08) 0.23(0.10) 0.27(0.06) 0.12(0.19) 0.04(0.12) 0.05(0.05) 0.00(0.05) 0.05(0.05) 0.21(0.25) 0.07(0.16) 0.09(0.05) 0.05(0.08) 0.07(0.07) 0.06(0.08) 0.07(0.09) 0.07(0.08) 0.09(0.07) 0.06(0.05) 0.07(0.03) 0.07(0.04) 0.06(0.06) 0.07(0.04) 0.07(0.04) 0.08(0.07) 0.07(0.06) 0.09(0.12) 0.63(0.64) 0.45(0.26) 0.43(0.07) 0.37(0.08) 0.42(0.09) 0.37(0.08) 0.38(0.04) 0.44(0.07) 0.38(0.06) 0.45(0.11) 0.45(0.03) 0.42(0.04) 0.47(0.06) 0.41(0.08) 0.43(0.06) 0.41(0.07) 0.38(0.04) 0.48(0.12) 0.45(0.09) 0.46(0.07) 0.50(0.05) 0.40(0.11) 0.43(0.05) 0.48(0.06) 0.40(0.08) 0.52(0.13) 0.49(0.08) 0.48(0.11) 0.56(0.09) 0.45(0.12) 0.51(0.06) 0.52(0.04) 0.53(0.06) 0.60(0.09) 0.48(0.13) 0.55(0.08) 0.79(0.64) 0.53(0.46) 0.53(0.13) 0.60(0.12) 0.55(0.08) 0.63(0.12) 0.51(0.14) 0.53(0.06) 0.50(0.05) 0.50(0.05) 0.55(0.07) 0.53(0.06) 0.56(0.09) 0.62(0.07) 0.57(0.07) 0.64(0.10) 0.55(0.11) 0.62(0.08) 0.59(0.06) 0.57(0.04) 0.61(0.05) 0.57(0.09) 0.55(0.03) 0.64(0.10) 0.63(0.20) 0.67(0.24) 0.66(0.16) 0.71(0.13) 0.68(0.11) 0.70(0.08) 0.72(0.03) 0.69(0.04) 0.73(0.06) 0.70(0.06) 0.75(0.08) 0.72(0.07) 0.71(0.05) 0.78(0.09) 0.73(0.08) 0.75(0.08) 0.78(0.06) 0.81(0.10) 0.84(0.09) 0.76(0.10) 0.87(0.13) 0.88(0.12) 0.89(0.06) 0.86(0.05) 0.82(0.07) 0.88(0.09) 1.48(0.61)
++ Max displacement in automask = 1.48 (mm) at sub-brick 145
++ Max delta displ in automask = 0.64 (mm) at sub-brick 95
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-01.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-03.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-03.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 39026 voxels
+ 6033 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-01_T1w_ns+tlrc::WARP_DATA -I sub-01_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-01_T1w_ns+tlrc -input pb00.sub-01.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-01.r01.tcat+orig.HEAD
++ Base dataset: ./sub-01_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.745 -17.539 -10.712
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.3% 25.3% 18.5%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 6.0
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-02.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-02.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 39133 voxels
+ 6057 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.082 pitch=-0.607 yaw=-0.290 dS=-0.402 dL=-0.165 dP=-0.180
++ Mean: roll=-0.005 pitch=+0.247 yaw=-0.087 dS=-0.017 dL=-0.008 dP=+0.019
++ Max : roll=+0.157 pitch=+1.000 yaw=+0.133 dS=+0.464 dL=+0.119 dP=+0.290
++ Max displacements (mm) for each sub-brick:
1.21(0.00) 1.14(0.09) 1.04(0.14) 0.90(0.16) 0.83(0.13) 0.78(0.12) 0.77(0.14) 0.67(0.11) 0.64(0.12) 0.54(0.13) 0.42(0.21) 0.42(0.17) 0.53(0.17) 0.43(0.12) 0.43(0.05) 0.45(0.32) 0.34(0.19) 0.27(0.13) 1.03(1.04) 0.95(0.19) 0.81(0.20) 0.76(0.08) 0.76(0.04) 0.68(0.10) 0.71(0.22) 0.59(0.16) 0.51(0.10) 0.53(0.05) 0.39(0.16) 0.29(0.14) 0.20(0.15) 0.13(0.18) 0.14(0.12) 0.07(0.08) 0.14(0.08) 0.13(0.10) 0.09(0.10) 0.08(0.09) 0.00(0.08) 0.06(0.06) 0.05(0.06) 0.10(0.11) 0.09(0.09) 0.15(0.11) 0.24(0.11) 0.20(0.04) 0.22(0.12) 0.28(0.08) 0.34(0.08) 0.36(0.11) 0.45(0.12) 0.48(0.05) 0.49(0.09) 0.57(0.15) 0.58(0.06) 0.61(0.08) 0.60(0.02) 0.63(0.08) 0.63(0.06) 0.69(0.06) 0.81(0.15) 0.95(0.17) 0.79(0.24) 0.64(0.20) 0.86(0.25) 0.75(0.16) 0.81(0.09) 0.83(0.10) 0.81(0.27) 0.97(1.35) 0.77(0.34) 0.68(0.25) 0.63(0.15) 0.65(0.12) 0.64(0.18) 0.58(0.08) 0.63(0.14) 0.60(0.09) 0.68(0.12) 0.52(0.28) 0.32(0.24) 0.36(0.13) 0.37(0.14) 0.44(0.08) 0.39(0.07) 0.47(0.10) 0.52(0.10) 0.56(0.11) 0.61(0.08) 0.68(0.11) 0.70(0.15) 0.68(0.10) 0.74(0.06) 0.73(0.06) 0.79(0.07) 0.84(0.07) 0.87(0.11) 0.94(0.15) 1.10(0.17) 1.08(0.04) 0.93(0.15) 0.92(0.24) 1.00(0.29) 0.95(0.07) 1.02(0.08) 1.07(0.06) 1.01(0.07) 1.08(0.12) 0.95(0.67) 0.74(0.48) 1.14(0.42) 1.29(0.28) 1.31(0.13) 1.24(0.10) 1.23(0.11) 1.14(0.11) 1.03(0.16) 1.12(0.12) 1.06(0.10) 1.18(0.17) 1.22(0.22) 1.33(0.16) 1.34(0.10) 1.30(0.07) 1.38(0.08) 1.35(0.11) 1.38(0.23) 1.41(0.03) 1.29(0.14) 1.34(0.23) 1.47(0.19) 1.45(0.11) 1.45(0.13) 1.45(0.05) 1.43(0.05) 1.45(0.07) 1.46(0.02) 1.46(0.04) 1.52(0.06) 1.53(0.05) 1.44(0.10) 1.52(0.12) 1.54(0.09) 1.53(0.13) 1.61(0.08) 1.45(0.26)
++ Max displacement in automask = 1.61 (mm) at sub-brick 144
++ Max delta displ in automask = 1.35 (mm) at sub-brick 69
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-03.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.6
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-01_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 1.696 -24.906 -9.444
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.9% 35.9% 16.3%
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-03_T1w_ns+tlrc::WARP_DATA -I sub-03_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-03_T1w_ns+tlrc -input pb00.sub-03.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-03.r02.tcat+orig.HEAD
++ Base dataset: ./sub-03_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -3.283 -12.324 -40.685
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.7% 17.8% 70.4%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.4
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-01.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-01.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 39640 voxels
+ 5772 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.387 pitch=+0.076 yaw=+0.120 dS=+0.314 dL=-0.780 dP=-1.005
++ Mean: roll=-0.310 pitch=+0.475 yaw=+0.189 dS=+0.467 dL=-0.522 dP=-0.832
++ Max : roll=-0.064 pitch=+0.624 yaw=+0.248 dS=+0.608 dL=-0.171 dP=-0.520
++ Max displacements (mm) for each sub-brick:
0.76(0.00) 0.87(0.15) 0.94(0.17) 0.84(0.20) 0.97(0.14) 1.01(0.09) 1.06(0.08) 1.02(0.08) 0.99(0.10) 1.05(0.07) 1.06(0.04) 1.02(0.20) 0.94(0.20) 1.02(0.10) 1.11(0.09) 1.11(0.05) 1.21(0.15) 1.26(0.07) 1.21(0.11) 1.25(0.06) 1.23(0.06) 1.28(0.10) 1.20(0.15) 1.17(0.06) 1.27(0.14) 1.34(0.11) 1.40(0.15) 1.41(0.04) 1.43(0.10) 1.44(0.06) 1.39(0.08) 1.41(0.08) 1.42(0.14) 1.49(0.08) 1.38(0.11) 1.46(0.09) 1.46(0.05) 1.49(0.11) 1.54(0.06) 1.61(0.08) 1.60(0.04) 1.60(0.07) 1.58(0.05) 1.62(0.07) 1.63(0.11) 1.66(0.20) 1.65(0.05) 1.62(0.25) 1.59(0.15) 1.60(0.14) 1.65(0.06) 1.69(0.04) 1.71(0.09) 1.69(0.06) 1.72(0.04) 1.62(0.16) 1.60(0.12) 1.63(0.08) 1.63(0.06) 1.66(0.17) 1.69(0.07) 1.68(0.04) 1.70(0.08) 1.66(0.07) 1.69(0.06) 1.63(0.09) 1.67(0.06) 1.73(0.10) 1.72(0.13) 1.72(0.10) 1.81(0.15) 1.78(0.08) 1.75(0.19) 1.78(0.19) 1.83(0.05) 1.84(0.08) 1.83(0.06) 1.78(0.04) 1.83(0.05) 1.83(0.05) 1.79(0.10) 1.79(0.14) 1.76(0.10) 1.74(0.09) 1.70(0.09) 1.77(0.17) 1.75(0.05) 1.77(0.07) 1.77(0.08) 1.79(0.07) 1.70(0.13) 1.70(0.15) 1.70(0.06) 1.68(0.10) 1.70(0.05) 1.71(0.07) 1.66(0.19) 1.71(0.11) 1.70(0.08) 1.72(0.06) 1.72(0.05) 1.74(0.07) 1.70(0.16) 1.74(0.06) 1.73(0.11) 1.73(0.16) 1.73(0.18) 1.68(0.16) 1.73(0.10) 1.75(0.08) 1.76(0.04) 1.79(0.07) 1.81(0.10) 1.70(0.14) 1.80(0.14) 1.76(0.10) 1.78(0.04) 1.80(0.16) 1.78(0.11) 1.77(0.08) 1.82(0.10) 1.76(0.08) 1.79(0.04) 1.80(0.04) 1.78(0.04) 1.82(0.05) 1.76(0.21) 1.72(0.13) 1.74(0.06) 1.76(0.06) 1.75(0.03) 1.74(0.09) 1.76(0.08) 1.76(0.20) 1.80(0.10) 1.81(0.09) 1.76(0.11) 1.77(0.05) 1.84(0.08) 1.81(0.11) 1.81(0.08) 1.80(0.12) 1.78(0.08) 1.84(0.07) 1.83(0.06) 1.80(0.04)
++ Max displacement in automask = 1.84 (mm) at sub-brick 138
++ Max delta displ in automask = 0.25 (mm) at sub-brick 47
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-02.r02.tcat+orig -expr 1 -prefix rm.epi.all1
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.0
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-03_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -4.335 -18.106 -35.069
+ shift search range is +/- = 57.780 69.336 57.780
+ 7.5% 26.1% 60.7%
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-02_T1w_ns+tlrc::WARP_DATA -I sub-02_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-02_T1w_ns+tlrc -input pb00.sub-02.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-02.r02.tcat+orig.HEAD
++ Base dataset: ./sub-02_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 4.539 1.128 -27.695
+ shift search range is +/- = 57.780 69.336 57.780
+ 7.9% 1.6% 47.9%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.3
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-03.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-02_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.934 -5.532 -24.833
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.6% 8.0% 43.0%
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.158 pitch=+0.157 yaw=+0.034 dS=-0.653 dL=-0.309 dP=-0.817
++ Mean: roll=-0.073 pitch=+0.402 yaw=+0.110 dS=-0.502 dL=-0.054 dP=-0.725
++ Max : roll=+0.129 pitch=+0.654 yaw=+0.186 dS=-0.386 dL=+0.205 dP=-0.644
++ Max displacements (mm) for each sub-brick:
1.43(0.00) 1.34(0.13) 1.31(0.05) 1.30(0.04) 1.16(0.24) 1.14(0.16) 1.17(0.06) 1.17(0.10) 1.21(0.08) 1.16(0.07) 1.17(0.03) 1.16(0.04) 1.17(0.05) 1.18(0.06) 1.18(0.06) 1.17(0.07) 1.19(0.08) 1.21(0.07) 1.22(0.07) 1.22(0.03) 1.21(0.04) 1.23(0.04) 1.21(0.07) 1.23(0.02) 1.19(0.10) 1.18(0.11) 1.21(0.09) 1.24(0.10) 1.22(0.05) 1.21(0.07) 1.24(0.05) 1.35(0.24) 1.27(0.13) 1.30(0.07) 1.33(0.05) 1.37(0.10) 1.35(0.09) 1.31(0.06) 1.32(0.02) 1.28(0.07) 1.27(0.03) 1.31(0.06) 1.29(0.06) 1.27(0.04) 1.29(0.05) 1.26(0.03) 1.30(0.04) 1.30(0.06) 1.26(0.06) 1.26(0.05) 1.28(0.09) 1.27(0.03) 1.33(0.11) 1.29(0.09) 1.26(0.04) 1.29(0.03) 1.27(0.02) 1.27(0.05) 1.26(0.07) 1.31(0.09) 1.34(0.07) 1.33(0.08) 1.38(0.11) 1.39(0.06) 1.42(0.07) 1.43(0.03) 1.38(0.08) 1.47(0.13) 1.46(0.09) 1.45(0.02) 1.46(0.07) 1.43(0.12) 1.45(0.07) 1.52(0.10) 1.49(0.10) 1.53(0.13) 1.51(0.13) 1.55(0.19) 1.50(0.16) 1.51(0.10) 1.56(0.06) 1.52(0.08) 1.53(0.07) 1.58(0.06) 1.53(0.09) 1.58(0.15) 1.56(0.13) 1.53(0.03) 1.60(0.12) 1.58(0.06) 1.56(0.04) 1.59(0.04) 1.51(0.12) 1.52(0.10) 1.55(0.08) 1.52(0.04) 1.63(0.16) 1.55(0.16) 1.54(0.10) 1.57(0.04) 1.53(0.10) 1.57(0.10) 1.58(0.06) 1.53(0.06) 1.58(0.08) 1.54(0.09) 1.55(0.03) 1.59(0.06) 1.57(0.06) 1.58(0.11) 1.59(0.08) 1.58(0.07) 1.58(0.13) 1.55(0.09) 1.54(0.08) 1.53(0.09) 1.56(0.10) 1.55(0.09) 1.59(0.07) 1.60(0.06) 1.57(0.09) 1.58(0.04) 1.57(0.06) 1.65(0.12) 1.60(0.09) 1.58(0.06) 1.62(0.09) 1.66(0.06) 1.61(0.07) 1.60(0.06) 1.60(0.09) 1.62(0.07) 1.71(0.15) 1.60(0.23) 1.59(0.14) 1.60(0.09) 1.58(0.06) 1.66(0.16) 1.59(0.14) 1.63(0.14) 1.66(0.10) 1.67(0.07) 1.70(0.13) 1.80(0.12) 1.82(0.08) 1.80(0.03)
++ Max displacement in automask = 1.82 (mm) at sub-brick 144
++ Max delta displ in automask = 0.24 (mm) at sub-brick 31
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-01.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./pb01.sub-03.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-03.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-01_T1w_ns+tlrc::WARP_DATA -I sub-01_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-01_T1w_ns+tlrc -input pb00.sub-01.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.5
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-01.r02.tcat+orig.HEAD
++ Base dataset: ./sub-01_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.753 -16.928 -10.613
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.3% 24.4% 18.4%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-02.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb01.sub-03.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-03_T1w_ns+tlrc::WARP_DATA -I sub-03_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-03_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-03_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -3.267 -12.225 -40.624
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.7% 17.6% 70.3%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-03_T1w_ns+tlrc anat_final.sub-03
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-03+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-03+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -0.071 0.600 0.635
+ shift search range is +/- = 70.299 81.855 67.410
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.130118
sp = 0.323546
mi = 2.64436
crM = 0.0353931
nmi = 0.835478
je = 2.64436
hel = -0.102632
crA = 0.188406
crU = 0.198585
lss = 0.869882
lpc = 0.361136
lpa = 0.638864
lpc+ = 0.494552
lpa+ = 0.77228
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.9
++ ###########################################################
3dAllineate -source sub-03_T1w+orig -master anat_final.sub-03+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-03_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.6
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-01_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.7
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-03.r01.blur pb01.sub-03.r01.volreg+tlrc
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 1.696 -24.906 -9.444
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.9% 35.9% 16.3%
++ Output dataset ./pb01.sub-02.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-02.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-03.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-03.r02.blur pb01.sub-03.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-03.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-03.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-03.r01.blur+tlrc
++ Forming automask
+ Fixed clip level = 356.639801
+ Used gradual clip level = 326.405518 .. 374.955383
+ Number voxels above clip level = 69262
+ Clustering voxels ...
+ Largest cluster has 68709 voxels
+ Clustering voxels ...
+ Largest cluster has 68304 voxels
+ Filled 160 voxels in small holes; now have 68464 voxels
+ Filled 2 voxels in large holes; now have 68466 voxels
+ Clustering voxels ...
+ Largest cluster has 68466 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 203167 voxels
+ Mask now has 68466 voxels
++ 68466 voxels in the mask [out of 271633: 25.21%]
++ first 6 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 9 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-03.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-03.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 358.210419
+ Used gradual clip level = 328.055573 .. 376.247681
+ Number voxels above clip level = 69189
+ Clustering voxels ...
+ Largest cluster has 68623 voxels
+ Clustering voxels ...
+ Largest cluster has 68226 voxels
+ Filled 178 voxels in small holes; now have 68404 voxels
+ Filled 3 voxels in large holes; now have 68407 voxels
+ Clustering voxels ...
+ Largest cluster has 68407 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 203226 voxels
+ Mask now has 68407 voxels
++ 68407 voxels in the mask [out of 271633: 25.18%]
++ first 6 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 9 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-03
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 68545 survived, 203088 were zero
++ writing result full_mask.sub-03...
++ Output dataset ./full_mask.sub-03+tlrc.BRIK
3dresample -master full_mask.sub-03+tlrc -input sub-03_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-03
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.6
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 77716 survived, 193917 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-03...
++ Output dataset ./mask_anat.sub-03+tlrc.BRIK
++ Output dataset ./pb01.sub-02.r02.volreg+tlrc.BRIK
3dmask_tool -input full_mask.sub-03+tlrc mask_anat.sub-03+tlrc -inter -prefix mask_epi_anat.sub-03
end
cat_matvec -ONELINE sub-02_T1w_ns+tlrc::WARP_DATA -I sub-02_T1w_al_junk_mat.aff12.1D -I
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
3dAllineate -base sub-02_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 12063 clipped, 67099 survived, 192471 were zero
++ writing result mask_epi_anat.sub-03...
++ Output dataset ./mask_epi_anat.sub-03+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-03+tlrc mask_anat.sub-03+tlrc
tee out.mask_ae_overlap.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-02_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 4.105 0.430 -27.201
+ shift search range is +/- = 57.780 69.336 57.780
+ 7.1% 0.6% 47.1%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-02_T1w_ns+tlrc anat_final.sub-02
#A=./full_mask.sub-03+tlrc.BRIK B=./mask_anat.sub-03+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
68545 77716 79162 67099 1446 10617 2.1096 13.6613 1.0453 1.0038 1.1173
3ddot -dodice full_mask.sub-03+tlrc mask_anat.sub-03+tlrc
tee out.mask_ae_dice.txt
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-02+tlrc
tee out.allcostX.txt
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
0.917524
3dresample -master full_mask.sub-03+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-02+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-01.r01.volreg
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-03+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = 0.028 1.958 0.788
+ shift search range is +/- = 68.373 86.670 69.336
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
0.949783
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-03.r01.blur+tlrc
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.098481
sp = 0.305774
mi = 2.57849
crM = 0.0169835
nmi = 0.813651
je = 2.57849
hel = -0.124723
crA = 0.130738
crU = 0.141169
lss = 0.901519
lpc = 0.495823
lpa = 0.504177
lpc+ = 0.580243
lpa+ = 0.588596
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.8
++ ###########################################################
3dAllineate -source sub-02_T1w+orig -master anat_final.sub-02+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-02_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-03.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-03.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.3
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-02.r01.blur pb01.sub-02.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-02.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-02.r02.blur pb01.sub-02.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-02.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-02.r01.blur+tlrc
++ Output dataset ./pb01.sub-01.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-01.r02.volreg
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-02.r01.blur+tlrc
++ Forming automask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Fixed clip level = 366.194977
+ Used gradual clip level = 344.260895 .. 383.901855
+ Number voxels above clip level = 70414
+ Clustering voxels ...
+ Largest cluster has 69533 voxels
+ Clustering voxels ...
+ Largest cluster has 68992 voxels
+ Filled 115 voxels in small holes; now have 69107 voxels
+ Filled 1 voxels in large holes; now have 69108 voxels
+ Clustering voxels ...
+ Largest cluster has 69107 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202526 voxels
+ Mask now has 69107 voxels
++ 69107 voxels in the mask [out of 271633: 25.44%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 3 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-02.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-02.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 367.034882
+ Used gradual clip level = 343.943481 .. 384.986053
+ Number voxels above clip level = 70311
+ Clustering voxels ...
+ Largest cluster has 69471 voxels
+ Clustering voxels ...
+ Largest cluster has 68914 voxels
+ Filled 117 voxels in small holes; now have 69031 voxels
+ Clustering voxels ...
+ Largest cluster has 69030 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202603 voxels
+ Mask now has 69030 voxels
++ 69030 voxels in the mask [out of 271633: 25.41%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-02
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 69370 survived, 202263 were zero
++ writing result full_mask.sub-02...
++ Output dataset ./full_mask.sub-02+tlrc.BRIK
3dresample -master full_mask.sub-02+tlrc -input sub-02_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-02
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 77297 survived, 194336 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-02...
++ Output dataset ./mask_anat.sub-02+tlrc.BRIK
3dmask_tool -input full_mask.sub-02+tlrc mask_anat.sub-02+tlrc -inter -prefix mask_epi_anat.sub-02
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 11321 clipped, 67673 survived, 192639 were zero
++ writing result mask_epi_anat.sub-02...
++ Output dataset ./mask_epi_anat.sub-02+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-02+tlrc mask_anat.sub-02+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-02+tlrc.BRIK B=./mask_anat.sub-02+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
69370 77297 78994 67673 1697 9624 2.4463 12.4507 1.0872 0.9817 1.0720
3ddot -dodice full_mask.sub-02+tlrc mask_anat.sub-02+tlrc
tee out.mask_ae_dice.txt
++ Output dataset ./pb03.sub-03.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-03.r02.blur+tlrc
0.922812
3dresample -master full_mask.sub-02+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-02+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.958927
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-02.r01.blur+tlrc
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-03.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-03.r02.scale
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-02.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-02.r01.scale
++ Output dataset ./pb01.sub-01.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-01_T1w_ns+tlrc::WARP_DATA -I sub-01_T1w_al_junk_mat.aff12.1D -I
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
3dAllineate -base sub-01_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-01_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.746 -17.615 -10.729
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.3% 25.4% 18.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-01_T1w_ns+tlrc anat_final.sub-01
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-01+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-01+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -0.105 1.996 -4.467
+ shift search range is +/- = 68.373 79.929 69.336
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.152565
sp = 0.321396
mi = 2.65023
crM = 0.0415924
nmi = 0.843509
je = 2.65023
hel = -0.0964992
crA = 0.205041
crU = 0.22624
lss = 0.847435
lpc = 0.232077
lpa = 0.767923
lpc+ = 0.39217
lpa+ = 0.928017
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.8
++ ###########################################################
3dAllineate -source sub-01_T1w+orig -master anat_final.sub-01+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-01_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.5
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-01.r01.blur pb01.sub-01.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-01.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-01.r02.blur pb01.sub-01.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
++ Output dataset ./pb03.sub-03.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-01.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-01.r01.blur+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-01.r01.blur+tlrc
++ Forming automask
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-03
++ Output dataset ./pb03.sub-02.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-02.r02.blur+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 14
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-03_censor.1D -show_trs_uncensored encoded
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
+ Fixed clip level = 347.084930
+ Used gradual clip level = 332.422607 .. 361.193390
+ Number voxels above clip level = 69663
+ Clustering voxels ...
+ Largest cluster has 69472 voxels
+ Clustering voxels ...
+ Largest cluster has 69350 voxels
+ Filled 93 voxels in small holes; now have 69443 voxels
+ Clustering voxels ...
+ Largest cluster has 69442 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202191 voxels
+ Mask now has 69442 voxels
++ 69442 voxels in the mask [out of 271633: 25.56%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-03.r01.scale+tlrc.HEAD pb03.sub-03.r02.scale+tlrc.HEAD -censor motion_sub-03_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-03 -errts errts.sub-03 -bucket stats.sub-03
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-01.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-01.r02.blur+tlrc
++ Forming automask
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-03.r01.scale+tlrc.HEAD pb03.sub-03.r02.scale+tlrc.HEAD
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-02.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-02.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
+ Fixed clip level = 351.576874
+ Used gradual clip level = 338.083405 .. 365.682617
+ Number voxels above clip level = 69705
+ Clustering voxels ...
+ Largest cluster has 69525 voxels
+ Clustering voxels ...
+ Largest cluster has 69400 voxels
+ Filled 112 voxels in small holes; now have 69512 voxels
+ Clustering voxels ...
+ Largest cluster has 69512 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202121 voxels
+ Mask now has 69512 voxels
++ 69512 voxels in the mask [out of 271633: 25.59%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-01
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 69731 survived, 201902 were zero
++ writing result full_mask.sub-01...
++ Output dataset ./full_mask.sub-01+tlrc.BRIK
3dresample -master full_mask.sub-01+tlrc -input sub-01_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-01
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76259 survived, 195374 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-01...
++ Output dataset ./mask_anat.sub-01+tlrc.BRIK
3dmask_tool -input full_mask.sub-01+tlrc mask_anat.sub-01+tlrc -inter -prefix mask_epi_anat.sub-01
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ STAT automask has 263371 voxels (out of 271633 = 97.0%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (before censor) ; 278 (after)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 11994 clipped, 66998 survived, 192641 were zero
++ writing result mask_epi_anat.sub-01...
++ Output dataset ./mask_epi_anat.sub-01+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-01+tlrc mask_anat.sub-01+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-01+tlrc.BRIK B=./mask_anat.sub-01+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
69731 76259 78992 66998 2733 9261 3.9193 12.1441 1.0475 0.9399 1.1213
3ddot -dodice full_mask.sub-01+tlrc mask_anat.sub-01+tlrc
tee out.mask_ae_dice.txt
0.917844
3dresample -master full_mask.sub-01+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-01+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.956878
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-01.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-01.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-01.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-03.r01.scale+tlrc.HEAD pb03.sub-03.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-03_REML -Rvar stats.sub-03_REMLvar \
-Rfitts fitts.sub-03_REML -Rerrts errts.sub-03_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (278x20): 4.9813 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (278x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (278x18): 4.97649 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (278x12): 4.07135 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (278x6): 1.03158 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 2.98009e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.66 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=5.067
++ Job #2: processing voxels 67908 to 101861; elapsed time=5.081
++ Job #3: processing voxels 101862 to 135815; elapsed time=5.095
++ Job #4: processing voxels 135816 to 169769; elapsed time=5.108
++ Job #5: processing voxels 169770 to 203723; elapsed time=5.121
++ Job #6: processing voxels 203724 to 237677; elapsed time=5.134
++ Job #7: processing voxels 237678 to 271632; elapsed time=5.148
++ Job #0: processing voxels 0 to 33953; elapsed time=5.158
++ Output dataset ./pb03.sub-02.r02.scale+tlrc.BRIK
++ voxel loop:0123456789.0123456789.0123456789.0123456789.01234567++ Job #1 finished; elapsed time=9.159
8++ Job #7 finished; elapsed time=9.223
9.
++ Job #0 waiting for children to finish; elapsed time=9.244
++ Job #2 finished; elapsed time=9.254
++ Job #5 finished; elapsed time=9.260
++ Job #6 finished; elapsed time=9.262
++ Job #4 finished; elapsed time=9.291
++ Job #3 finished; elapsed time=9.293
++ Job #0 now finishing up; elapsed time=9.304
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
++ Smallest FDR q [0 Full_Fstat] = 4.19148e-10
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 0
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-02_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-02.r01.scale+tlrc.HEAD pb03.sub-02.r02.scale+tlrc.HEAD -censor motion_sub-02_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-02 -errts errts.sub-02 -bucket stats.sub-02
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-02.r01.scale+tlrc.HEAD pb03.sub-02.r02.scale+tlrc.HEAD
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
++ Output dataset ./pb03.sub-01.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-01.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Smallest FDR q [2 congruent#0_Tstat] = 1.7412e-05
++ Smallest FDR q [3 congruent_Fstat] = 1.74123e-05
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-01.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-01.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ STAT automask has 254352 voxels (out of 271633 = 93.6%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (no censoring)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.48011e-08
++ Smallest FDR q [6 incongruent_Fstat] = 2.4799e-08
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-02.r01.scale+tlrc.HEAD pb03.sub-02.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-02_REML -Rvar stats.sub-02_REMLvar \
-Rfitts fitts.sub-02_REML -Rerrts errts.sub-02_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (292x20): 5.94795 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (292x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (292x18): 5.93916 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (292x12): 3.34088 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (292x6): 1 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 5.62387e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.72 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=4.663
++ Job #2: processing voxels 67908 to 101861; elapsed time=4.678
++ Job #3: processing voxels 101862 to 135815; elapsed time=4.693
++ Job #4: processing voxels 135816 to 169769; elapsed time=4.709
++ Job #5: processing voxels 169770 to 203723; elapsed time=4.727
++ Job #6: processing voxels 203724 to 237677; elapsed time=4.743
++ Job #7: processing voxels 237678 to 271632; elapsed time=4.760
++ Job #0: processing voxels 0 to 33953; elapsed time=4.770
++ Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.0824997
++ Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.0824962
++ Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.0824997
++ Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.0824962
++ Wrote bucket dataset into ./stats.sub-03+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Output dataset ./pb03.sub-01.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
++ voxel loop:0123456789.0123456789.0123456789.0123456789.01234567++ Job #1 finished; elapsed time=9.304
++ Job #7 finished; elapsed time=9.359
8++ Job #2 finished; elapsed time=9.387
++ Job #5 finished; elapsed time=9.390
9.
++ Job #0 waiting for children to finish; elapsed time=9.447
++ Job #3 finished; elapsed time=9.534
++ Job #4 finished; elapsed time=9.548
++ Job #6 finished; elapsed time=9.553
++ Job #0 now finishing up; elapsed time=9.575
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
++ Smallest FDR q [0 Full_Fstat] = 1.28834e-10
++ Wrote 3D+time dataset into ./fitts.sub-03+tlrc.BRIK
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 6
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-01_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-01.r01.scale+tlrc.HEAD pb03.sub-01.r02.scale+tlrc.HEAD -censor motion_sub-01_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-01 -errts errts.sub-01 -bucket stats.sub-01
++ Smallest FDR q [2 congruent#0_Tstat] = 2.40858e-08
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-01.r01.scale+tlrc.HEAD pb03.sub-01.r02.scale+tlrc.HEAD
++ Smallest FDR q [3 congruent_Fstat] = 2.4085e-08
++ Wrote 3D+time dataset into ./errts.sub-03+tlrc.BRIK
++ Program finished; elapsed time=22.529
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 14 : 4.8%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 34 : 11.6%
final DF : 258 : 88.4%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-03.r01.scale+tlrc.HEAD pb03.sub-03.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-03_REML -Rvar stats.sub-03_REMLvar -Rfitts fitts.sub-03_REML -Rerrts errts.sub-03_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ STAT automask has 251719 voxels (out of 271633 = 92.7%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (before censor) ; 286 (after)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Smallest FDR q [5 incongruent#0_Tstat] = 1.80249e-09
++ Smallest FDR q [6 incongruent_Fstat] = 1.80249e-09
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ FDR automask has 263371 voxels (out of 271633 = 97.0%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (278x20): 4.9813 ++ VERY GOOD ++
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-01.r01.scale+tlrc.HEAD pb03.sub-01.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-01_REML -Rvar stats.sub-01_REMLvar \
-Rfitts fitts.sub-01_REML -Rerrts errts.sub-01_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (286x20): 5.56002 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (286x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (286x18): 5.53333 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (286x12): 4.07236 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (286x6): 1.03009 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 4.40903e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.68 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=2.922
++ Job #2: processing voxels 67908 to 101861; elapsed time=2.938
++ Job #3: processing voxels 101862 to 135815; elapsed time=2.953
++ Job #4: processing voxels 135816 to 169769; elapsed time=2.968
++ Job #5: processing voxels 169770 to 203723; elapsed time=2.983
++ Job #6: processing voxels 203724 to 237677; elapsed time=2.997
++ Job #7: processing voxels 237678 to 271632; elapsed time=3.012
++ Job #0: processing voxels 0 to 33953; elapsed time=3.022
+ masked off 22238 voxels for being all zero; 249395 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.30
+ X matrix: 79.011% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=278 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.32
+ average case bandwidth = 15.40
*+ WARNING: Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.999782 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.996785 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.999782 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.996785 ==> few true single voxel detections
++ Wrote bucket dataset into ./stats.sub-02+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ voxel loop:0123456789.0123456789.0123456789.0123456789.++ Job #1 finished; elapsed time=7.671
++ Job #7 finished; elapsed time=7.675
01234++ Job #6 finished; elapsed time=8.268
5++ Job #3 finished; elapsed time=8.504
6++ Job #5 finished; elapsed time=8.574
++ Job #2 finished; elapsed time=8.576
++ Wrote 3D+time dataset into ./fitts.sub-02+tlrc.BRIK
789.
++ Job #0 waiting for children to finish; elapsed time=8.864
++ Job #4 finished; elapsed time=8.960
++ Job #0 now finishing up; elapsed time=8.972
++ Smallest FDR q [0 Full_Fstat] = 1.49527e-11
++ Wrote 3D+time dataset into ./errts.sub-02+tlrc.BRIK
++ Program finished; elapsed time=22.157
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
++ Smallest FDR q [2 congruent#0_Tstat] = 6.62071e-10
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 0 : 0.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 20 : 6.8%
final DF : 272 : 93.2%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-02.r01.scale+tlrc.HEAD pb03.sub-02.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-02_REML -Rvar stats.sub-02_REMLvar -Rfitts fitts.sub-02_REML -Rerrts errts.sub-02_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ Smallest FDR q [3 congruent_Fstat] = 6.62055e-10
++ FDR automask has 254352 voxels (out of 271633 = 93.6%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (292x20): 5.94795 ++ VERY GOOD ++
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.03222e-11
++ Smallest FDR q [6 incongruent_Fstat] = 2.03206e-11
+ masked off 44070 voxels for being all zero; 227563 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.41
+ X matrix: 79.110% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=292 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.44
+ average case bandwidth = 16.10
++ Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 7.38077e-05
++ Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 7.38071e-05
++ Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 7.38077e-05
++ Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 7.38071e-05
++ Wrote bucket dataset into ./stats.sub-01+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Wrote 3D+time dataset into ./fitts.sub-01+tlrc.BRIK
++ Wrote 3D+time dataset into ./errts.sub-01+tlrc.BRIK
++ Program finished; elapsed time=20.589
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 6 : 2.1%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 26 : 8.9%
final DF : 266 : 91.1%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-01.r01.scale+tlrc.HEAD pb03.sub-01.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-01_REML -Rvar stats.sub-01_REMLvar -Rfitts fitts.sub-01_REML -Rerrts errts.sub-01_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.012345
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=21.46
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ FDR automask has 251719 voxels (out of 271633 = 92.7%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (286x20): 5.56002 ++ VERY GOOD ++
+ masked off 23715 voxels for being all zero; 247918 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.47
+ X matrix: 79.056% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=286 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.51
+ average case bandwidth = 15.84
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.01
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=20.46
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.012345
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=18.49
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=49.55
++ Output dataset ./stats.sub-03_REMLvar+tlrc.BRIK
++ Output dataset ./fitts.sub-03_REML+tlrc.BRIK
++ Output dataset ./errts.sub-03_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-03_REML+tlrc
++ Smallest FDR q [0 Full_Fstat] = 7.91926e-11
++ Smallest FDR q [2 congruent#0_Tstat] = 2.24712e-05
++ Smallest FDR q [3 congruent_Fstat] = 2.24712e-05
++ Smallest FDR q [5 incongruent#0_Tstat] = 9.69477e-08
++ Smallest FDR q [6 incongruent_Fstat] = 9.69477e-08
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=48.43
++ Output dataset ./stats.sub-02_REMLvar+tlrc.BRIK
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.379661 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.379661 ==> few true single voxel detections
++ Output dataset ./fitts.sub-02_REML+tlrc.BRIK
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.379661 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.379661 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-03_REML+tlrc
++ Output dataset ./stats.sub-03_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=61.47
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-03 pb03.sub-03.r01.scale+tlrc.HEAD pb03.sub-03.r02.scale+tlrc.HEAD
++ Output dataset ./errts.sub-02_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-02_REML+tlrc
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Smallest FDR q [0 Full_Fstat] = 1.04165e-10
++ elapsed time = 0.7 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-03+tlrc[0..65,69,72..143,146..162,165..213,216..252,256..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-03_REML+tlrc[0..65,69,72..143,146..162,165..213,216..252,256..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Smallest FDR q [2 congruent#0_Tstat] = 2.55006e-09
++ Smallest FDR q [3 congruent_Fstat] = 2.55007e-09
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-03
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-03+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-03_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.5772e-09
++ Smallest FDR q [6 incongruent_Fstat] = 2.5772e-09
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-03+tlrc rm.errts.unit+tlrc
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.999774 ==> few true single voxel detections
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 68545 voxels survive the mask
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.999774 ==> few true single voxel detections
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.137945
3dmaskave -quiet -mask full_mask.sub-03+tlrc errts.sub-03_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 68545 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-03_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-03_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-03_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=48.02
++ Output dataset ./stats.sub-01_REMLvar+tlrc.BRIK
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.999774 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.999774 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-02_REML+tlrc
++ Output dataset ./fitts.sub-01_REML+tlrc.BRIK
++ Output dataset ./stats.sub-02_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=59.27
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-02 pb03.sub-02.r01.scale+tlrc.HEAD pb03.sub-02.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: THD_Tcorr1D: 22238 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
++ Output dataset ./errts.sub-01_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-01_REML+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ elapsed time = 0.7 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-02+tlrc[0..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-03.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ Smallest FDR q [0 Full_Fstat] = 2.08062e-11
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..65,69,72..143 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-03+tlrc[0..65,69,72..143]
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-02_REML+tlrc[0..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
++ Smallest FDR q [2 congruent#0_Tstat] = 9.37983e-09
++ Smallest FDR q [3 congruent_Fstat] = 9.3798e-09
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-02
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-02+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-02_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Smallest FDR q [5 incongruent#0_Tstat] = 1.13772e-10
++ Smallest FDR q [6 incongruent_Fstat] = 1.13772e-10
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.79 mm
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-02+tlrc rm.errts.unit+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 69370 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.225595
3dmaskave -quiet -mask full_mask.sub-02+tlrc errts.sub-02_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.00690584
+++ 69370 voxels survive the mask
++ Smallest FDR q [9 incongruent-congruent_Fstat] = 0.00690584
3dTcorr1D -prefix corr_brain errts.sub-02_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-02_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-02_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
*+ WARNING: THD_Tcorr1D: 44070 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
++ Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.00690584
1dcat X.nocensor.xmat.1D[7]
++ Smallest FDR q [12 congruent-incongruent_Fstat] = 0.00690584
+ Added 9 FDR curves to dataset stats.sub-01_REML+tlrc
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
++ Output dataset ./stats.sub-01_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=58.77
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-01 pb03.sub-01.r01.scale+tlrc.HEAD pb03.sub-01.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-02.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-02+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69370
++ elapsed time = 0.8 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-01+tlrc[0..57,60..93,96..143,146..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ detrending start: 11 baseline funcs, 146 time points
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-01_REML+tlrc[0..57,60..93,96..143,146..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..162,165..213,216..252,256..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-03+tlrc[146..162,165..213,216..252,256..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-01
+ detrending done (0.00 CPU s thus far)
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-01+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-01_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ start ACF calculations out to radius = 17.57 mm
+ detrending done (0.00 CPU s thus far)
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-01+tlrc rm.errts.unit+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 69731 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
++ start ACF calculations out to radius = 17.62 mm
cat out.gcor.1D
-- GCOR = 0.0697554
3dmaskave -quiet -mask full_mask.sub-01+tlrc errts.sub-01_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 69731 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-01_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-01_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-01_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
*+ WARNING: THD_Tcorr1D: 23715 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-01.1D
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
mkdir files_ACF
-show_trs_run $run`
touch blur.epits.1D
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..57,60..93,96..143 == ) continue
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-01+tlrc[0..57,60..93,96..143]
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-02+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69731
++ Number of voxels in mask = 69370
++ detrending start: 11 baseline funcs, 140 time points
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.810233 3.34725 13.0921 8.48996
average epits ACF blurs: 0.810233 3.34725 13.0921 8.48996
echo 0.810233 3.34725 13.0921 8.48996 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..65,69,72..143 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-03+tlrc[0..65,69,72..143]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
++ start ACF calculations out to radius = 17.52 mm
++ start ACF calculations out to radius = 17.50 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.71 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.841405 3.32481 14.1278 8.37934
average epits ACF blurs: 0.841405 3.32481 14.1278 8.37934
echo 0.841405 3.32481 14.1278 8.37934 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-01+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69731
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-02+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69370
++ detrending start: 11 baseline funcs, 146 time points
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..162,165..213,216..252,256..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-03+tlrc[146..162,165..213,216..252,256..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
+ detrending done (0.00 CPU s thus far)
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.61 mm
++ start ACF calculations out to radius = 17.51 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.63 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.835226 3.35654 12.3908 8.40587
average epits ACF blurs: 0.835226 3.35654 12.3908 8.40587
echo 0.835226 3.35654 12.3908 8.40587 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-02+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69370
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..57,60..93,96..143 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-01+tlrc[0..57,60..93,96..143]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69731
++ detrending start: 11 baseline funcs, 146 time points
++ detrending start: 11 baseline funcs, 140 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.824311 3.35581 13.1607 8.46258
average errts ACF blurs: 0.824311 3.35581 13.1607 8.46258
echo 0.824311 3.35581 13.1607 8.46258 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..65,69,72..143 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-03_REML+tlrc[0..65,69,72..143]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.45 mm
++ start ACF calculations out to radius = 17.49 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.67 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.852865 3.32426 14.5555 8.35301
average errts ACF blurs: 0.852865 3.32426 14.5555 8.35301
echo 0.852865 3.32426 14.5555 8.35301 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-01+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-02_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69731
++ Number of voxels in mask = 69370
++ detrending start: 11 baseline funcs, 146 time points
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..162,165..213,216..252,256..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-03+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-03_REML+tlrc[146..162,165..213,216..252,256..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68545
*+ WARNING: removed 9 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 139 time points
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.47 mm
++ start ACF calculations out to radius = 17.58 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.59 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-02+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-02_REML+tlrc[146..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69370
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.843022 3.35601 12.4114 8.38415
average errts ACF blurs: 0.843022 3.35601 12.4114 8.38415
echo 0.843022 3.35601 12.4114 8.38415 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..57,60..93,96..143 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-01_REML+tlrc[0..57,60..93,96..143]
++ detrending start: 11 baseline funcs, 146 time points
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 69731
++ detrending start: 11 baseline funcs, 140 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.824937 3.35061 13.1355 8.44969
average err_reml ACF blurs: 0.824937 3.35061 13.1355 8.44969
echo 0.824937 3.35061 13.1355 8.44969 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-03 -dsets pb00.sub-03.r01.tcat+orig.HEAD pb00.sub-03.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-03+tlrc.HEAD -ss_review_dset out.ss_review.sub-03.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-03.txt
subject ID : sub-03
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-03+tlrc.HEAD
final stats dset : stats.sub-03_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 8
+ detrending done (0.00 CPU s thus far)
average motion (per TR) : 0.0976639
average censored motion : 0.0819588
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.81088
++ start ACF calculations out to radius = 17.42 mm
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.79726
++ start ACF calculations out to radius = 17.46 mm
average outlier frac (TR) : 0.00255359
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 139 139
num TRs per run (censored): 7 7
fraction censored per run : 0.0479452 0.0479452
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 278
degrees of freedom used : 20
degrees of freedom left : 258
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 14
censor fraction : 0.047945
num regs of interest : 2
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
num TRs per stim (orig) : 120 115
num TRs censored per stim : 7 5
fraction TRs censored : 0.058 0.043
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
echo 0 0 0 0 # err_reml FWHM blur estimates
++ Authored by: RWC et al.
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.855736 3.32213 14.7419 8.34399
average err_reml ACF blurs: 0.855736 3.32213 14.7419 8.34399
echo 0.855736 3.32213 14.7419 8.34399 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-02 -dsets pb00.sub-02.r01.tcat+orig.HEAD pb00.sub-02.r02.tcat+orig.HEAD
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-02+tlrc.HEAD -ss_review_dset out.ss_review.sub-02.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-01+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-01_REML+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
++ Number of voxels in mask = 69731
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-02.txt
subject ID : sub-02
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-02+tlrc.HEAD
final stats dset : stats.sub-02_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 0
ave mot per sresp (orig) : 0.104433 0.088395
ave mot per sresp (cens) : 0.082356 0.078794
++ detrending start: 11 baseline funcs, 146 time points
average motion (per TR) : 0.0641705
TSNR average : 122.576
global correlation (GCOR) : 0.137945
anat/EPI mask Dice coef : 0.917524
anat/templ mask Dice coef : 0.949783
average censored motion : 0.0641705
maximum F-stat (masked) : 42.0812
blur estimates (ACF) : 0.824937 3.35061 13.1355
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.86278
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.86278
average outlier frac (TR) : 0.000850754
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 37 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 868.000000 -func_range 868.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-03/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-03/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ start ACF calculations out to radius = 17.54 mm
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ Copy ulay to visualize (volumetric) within user's range:
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Ulay to be visualized within user range:
[0, 868.000000]
TRs censored : 0
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
censor fraction : 0.000000
num regs of interest : 2
++ Output dataset ./QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w/tmp_olay.nii
++ User-entered function range value value (868.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=6..56 y=5..61 z=0..34
++ 3dAutobox: output dataset = QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_i61rrTb4JIoVYBg9nm33pQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
++ Output dataset /tmp/3dcalc_XYZ_i61rrTb4JIoVYBg9nm33pQ+orig.BRIK
++ 101745 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ -1.283127 -3.054207 -33.1325
++ Will have the ref box central gapord: 7 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :519
[1] 2465822
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
ave mot per sresp (orig) : 0.064054 0.068270
ave mot per sresp (cens) : 0.064054 0.068270
TSNR average : 122.647
global correlation (GCOR) : 0.225595
anat/EPI mask Dice coef : 0.922812
anat/templ mask Dice coef : 0.958927
maximum F-stat (masked) : 45.5679
blur estimates (ACF) : 0.855736 3.32213 14.7419
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 38 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
echo average err_reml ACF blurs: 0.84362 3.35238 12.3604 8.3738
average err_reml ACF blurs: 0.84362 3.35238 12.3604 8.3738
echo 0.84362 3.35238 12.3604 8.3738 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-01 -dsets pb00.sub-01.r01.tcat+orig.HEAD pb00.sub-01.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-01+tlrc.HEAD -ss_review_dset out.ss_review.sub-01.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-01.txt
subject ID : sub-01
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-01+tlrc.HEAD
final stats dset : stats.sub-01_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 3
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w++ Writing palette image to QC_sub-03/media/qc_00_vorig_EPI.pbar.jpg
average motion (per TR) : 0.0601768
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-03/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 868.000000 -com SET_FUNC_RANGE 868.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ -1.283127 -3.054207 -33.1325 -com SAVE_JPEG sagittalimage QC_sub-03/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-03/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-03/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w
+* Removing temporary image directory 'QC_sub-03/media/__tmp_chauf_UFKQoxXOx9w'.
[1] Done Xvfb :519 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-03/media/qc_00_vorig_EPI*
average censored motion : 0.0561726
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 881.000000 -func_range 881.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-02/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-02/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-02/media/__tmp_chauf_F78SW5oqxEW
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.3322
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 881.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__tmp_chauf_F78SW5oqxEW/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__tmp_chauf_F78SW5oqxEW/tmp_olay.nii
++ User-entered function range value value (881.000000)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
max censored displacement : 1.3322
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
average outlier frac (TR) : 0.00147247
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=8..53 y=4..58 z=0..35
++ 3dAutobox: output dataset = QC_sub-02/media/__tmp_chauf_F78SW5oqxEW/ulay_box_0.nii
++ Output dataset ./QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Output dataset ./QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-02/media/__tmp_chauf_F78SW5oqxEW/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_cKOdnWUtSI-VwnLCja5UMg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ Output dataset /tmp/3dcalc_XYZ_cKOdnWUtSI-VwnLCja5UMg+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 91080 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 4.5 15.9578 -19.73494
++ Will have the ref box central gapord: 6 7 5
------------------- end of optionizing -------------------
++ Output dataset ./QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr/eac_3_cp.nii
-- trying to start Xvfb :305
[1] 2468874
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+ Fixed clip level = 122.572800
+ Used gradual clip level = 110.527199 .. 138.509995
+ Number voxels above clip level = 223362
+ Clustering voxels ...
+ Largest cluster has 222886 voxels
+ Clustering voxels ...
+ Largest cluster has 216015 voxels
+ Filled 9510 voxels in small holes; now have 225525 voxels
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+ Filled 6456 voxels in large holes; now have 231981 voxels
+ Clustering voxels ...
+ Largest cluster has 231978 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 670420 voxels
+ Mask now has 232209 voxels
++ 232209 voxels in the mask [out of 902629: 25.73%]
++ first 9 x-planes are zero [from L]
++ last 8 x-planes are zero [from R]
++ first 10 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 140 146
num TRs per run (censored): 6 0
fraction censored per run : 0.0410959 0
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 41.396595 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_xbFSwitdEj3
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.815663, 892.073425]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_xbFSwitdEj3/tmp_ulay.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 286
degrees of freedom used : 20
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
degrees of freedom left : 266
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_xbFSwitdEj3/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 101.023346
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
TRs censored : 6
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_xbFSwitdEj3/ulay_box_0.nii
censor fraction : 0.020548
num regs of interest : 2
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_xbFSwitdEj3/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_29fF1-3TsSDii9DOVJVt_A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_29fF1-3TsSDii9DOVJVt_A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :683
[1] 2469846
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
num TRs per stim (orig) : 120 115
num TRs censored per stim : 3 0
fraction TRs censored : 0.025 0.000
ave mot per sresp (orig) : 0.064565 0.056228
ave mot per sresp (cens) : 0.058830 0.056228
TSNR average : 143.405
global correlation (GCOR) : 0.0697554
anat/EPI mask Dice coef : 0.917844
anat/templ mask Dice coef : 0.956878
maximum F-stat (masked) : 54.1967
blur estimates (ACF) : 0.84362 3.35238 12.3604
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_F78SW5oqxEW++ Writing palette image to QC_sub-02/media/qc_00_vorig_EPI.pbar.jpg
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 37 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:6:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-02/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 881.000000 -com SET_FUNC_RANGE 881.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 4.5 15.9578 -19.73494 -com SAVE_JPEG sagittalimage QC_sub-02/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-02/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-02/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-02/media/__tmp_chauf_F78SW5oqxEW
+* Removing temporary image directory 'QC_sub-02/media/__tmp_chauf_F78SW5oqxEW'.
[1] Done Xvfb :305 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-02/media/qc_00_vorig_EPI*
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__workdir_EAC_rEICjqm728c/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__workdir_EAC_rEICjqm728c/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__workdir_EAC_rEICjqm728c/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-02/media/__workdir_EAC_rEICjqm728c/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__workdir_EAC_rEICjqm728c/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 117.500801
+ Used gradual clip level = 106.002403 .. 129.026794
+ Number voxels above clip level = 225638
+ Clustering voxels ...
+ Largest cluster has 225284 voxels
+ Clustering voxels ...
+ Largest cluster has 219424 voxels
+ Filled 8050 voxels in small holes; now have 227474 voxels
+ Filled 6863 voxels in large holes; now have 234337 voxels
+ Clustering voxels ...
+ Largest cluster has 234330 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 668065 voxels
+ Mask now has 234564 voxels
++ 234564 voxels in the mask [out of 902629: 25.99%]
++ first 9 x-planes are zero [from L]
++ last 7 x-planes are zero [from R]
++ first 10 y-planes are zero [from P]
++ last 8 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 853.000000 -func_range 853.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-01/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-01/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
++ Output dataset ./eac_5_mfilt.nii
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_xbFSwitdEj3++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
[0, 853.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.815663 892.073425 -com SET_FUNC_RANGE 101.023346 -com SET_THRESHNEW 41.396595 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_xbFSwitdEj3
+* Removing temporary image directory '../__tmp_chauf_xbFSwitdEj3'.
[1] Done Xvfb :683 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-03/media/__workdir_EAC_d1KlWnkhvgr*'
++ DONE! Image output:
QC_sub-03/media/qc_01_ve2a_epi2anat
++ Output dataset ./QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG/tmp_olay.nii
++ User-entered function range value value (853.000000)
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 34.605473 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ Using blowup factor: 2
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_gRi88pR0qSs
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=8..56 y=1..60 z=0..34
++ 3dAutobox: output dataset = QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG/ulay_box_0.nii
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.402436, 911.744141]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_gRi88pR0qSs/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_gRi88pR0qSs/tmp_olay.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_BzNWo3TQGXEJMlruPBjLMw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ For olay, the 33%ile value leads to
--> upper range value: 94.521233
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_BzNWo3TQGXEJMlruPBjLMw+orig.BRIK
++ Output dataset ./QC_sub-03/media/__workdir_EAC_nrr0CME6ooo/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 102900 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ 3dAutobox: output dataset = ../__tmp_chauf_gRi88pR0qSs/ulay_box_0.nii
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 2.222893 0.162651 -10
++ Will have the ref box central gapord: 7 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :982
[1] 2472877
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Output dataset ./QC_sub-03/media/__workdir_EAC_nrr0CME6ooo/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-03/media/__workdir_EAC_nrr0CME6ooo/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_gRi88pR0qSs/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Zd82SSjTlflDaw_s7ZBjnw argv[10]=-verbose
+ Clustering voxels ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
++ Computing sub-brick 0
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Zd82SSjTlflDaw_s7ZBjnw+tlrc.BRIK
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :529
[1] 2473155
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_G89f8zF2c5c
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 485.345262]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_G89f8zF2c5c/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_G89f8zF2c5c/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_G89f8zF2c5c/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_G89f8zF2c5c/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_-wNymRkT6zZlGawHxY3cWg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_-wNymRkT6zZlGawHxY3cWg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :842
[1] 2473508
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG++ Writing palette image to QC_sub-01/media/qc_00_vorig_EPI.pbar.jpg
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-01/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 853.000000 -com SET_FUNC_RANGE 853.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 2.222893 0.162651 -10 -com SAVE_JPEG sagittalimage QC_sub-01/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-01/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-01/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG
+* Removing temporary image directory 'QC_sub-01/media/__tmp_chauf_2BhSd2lGYAG'.
[1] Done Xvfb :982 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-01/media/qc_00_vorig_EPI*
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_gRi88pR0qSs++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.402436 911.744141 -com SET_FUNC_RANGE 94.521233 -com SET_THRESHNEW 34.605473 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_gRi88pR0qSs
+* Removing temporary image directory '../__tmp_chauf_gRi88pR0qSs'.
[1] Done Xvfb :529 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-02/media/__workdir_EAC_rEICjqm728c*'
++ DONE! Image output:
QC_sub-02/media/qc_01_ve2a_epi2anat
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 120.036797
+ Used gradual clip level = 114.503998 .. 127.510201
+ Number voxels above clip level = 220844
+ Clustering voxels ...
+ Largest cluster has 220704 voxels
+ Clustering voxels ...
+ Largest cluster has 216180 voxels
+ Filled 11507 voxels in small holes; now have 227687 voxels
+ Filled 8446 voxels in large holes; now have 236133 voxels
+ Clustering voxels ...
++ Output dataset ./QC_sub-02/media/__workdir_EAC_RdE9vzsFBeS/REFBOX.nii
+ Largest cluster has 236128 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Largest cluster has 665503 voxels
+ Mask now has 237126 voxels
++ 237126 voxels in the mask [out of 902629: 26.27%]
++ first 10 x-planes are zero [from L]
++ last 7 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 2 z-planes are zero [from I]
++ last 11 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./QC_sub-02/media/__workdir_EAC_RdE9vzsFBeS/eac_0_cp.nii
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-02/media/__workdir_EAC_RdE9vzsFBeS/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
++ olay_alpha has known value: No
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 39.569855 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_wI0eVhhqLjJ
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_G89f8zF2c5c++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 485.345262 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_G89f8zF2c5c
+* Removing temporary image directory '../__tmp_chauf_G89f8zF2c5c'.
[1] Done Xvfb :842 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-03/media/__workdir_EAC_nrr0CME6ooo*'
++ DONE! Image output:
QC_sub-03/media/qc_02_va2t_anat2temp
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.904045, 878.626160]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ olay_alpha has known value: No
++ Output dataset ./../__tmp_chauf_wI0eVhhqLjJ/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_g4hCnO6aZUG
++ Output dataset ./../__tmp_chauf_wI0eVhhqLjJ/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 106.050217
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_wI0eVhhqLjJ/ulay_box_0.nii
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 460.565648]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_g4hCnO6aZUG/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_g4hCnO6aZUG/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_wI0eVhhqLjJ/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_pC2kR5SrbWiXBp9c1JdvGA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Computing sub-brick 0
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_g4hCnO6aZUG/ulay_box_0.nii
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_pC2kR5SrbWiXBp9c1JdvGA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :999
[1] 2475424
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_g4hCnO6aZUG/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_x8PDf0XKoeOr-xvikN1KpA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_x8PDf0XKoeOr-xvikN1KpA+tlrc.BRIK
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :514
[1] 2475668
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-03_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 8.931915 -thr_olay 3.272364 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-03/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-03/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-03_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-03/media/__tmp_chauf_TreW1c1ptEv
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_TreW1c1ptEv/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_TreW1c1ptEv/tmp_olay.nii
++ User-entered function range value value (8.931915)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-03/media/__tmp_chauf_TreW1c1ptEv/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-03/media/__tmp_chauf_TreW1c1ptEv/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_V0H11ew0yfae2cHqgA5-oA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_V0H11ew0yfae2cHqgA5-oA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :548
[1] 2475998
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__tmp_chauf_wI0eVhhqLjJ++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.904045 878.626160 -com SET_FUNC_RANGE 106.050217 -com SET_THRESHNEW 39.569855 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_wI0eVhhqLjJ
+* Removing temporary image directory '../__tmp_chauf_wI0eVhhqLjJ'.
[1] Done Xvfb :999 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-01/media/__workdir_EAC_mTqxEiaVTSV*'
++ DONE! Image output:
QC_sub-01/media/qc_01_ve2a_epi2anat
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_g4hCnO6aZUG++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 460.565648 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_g4hCnO6aZUG
+* Removing temporary image directory '../__tmp_chauf_g4hCnO6aZUG'.
[1] Done Xvfb :514 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-02/media/__workdir_EAC_RdE9vzsFBeS*'
++ DONE! Image output:
QC_sub-02/media/qc_02_va2t_anat2temp
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__workdir_EAC_AqwNeHk31Cs/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__workdir_EAC_AqwNeHk31Cs/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-01/media/__workdir_EAC_AqwNeHk31Cs/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_vKLJasUj5R1
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_TreW1c1ptEv++ Writing palette image to QC_sub-03/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 476.934997]
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-02_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 14.947501 -thr_olay 5.344243 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-02/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-02/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-02_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-02/media/__tmp_chauf_59BbiDFYCiw
++ Output dataset ./../__tmp_chauf_vKLJasUj5R1/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Output dataset ./../__tmp_chauf_vKLJasUj5R1/tmp_olay.nii
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-03/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 8.931915 -com SET_THRESHNEW 3.272364 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-03/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-03/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-03/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-03/media/__tmp_chauf_TreW1c1ptEv
+* Removing temporary image directory 'QC_sub-03/media/__tmp_chauf_TreW1c1ptEv'.
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
[1] Done Xvfb :548 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-03/media/qc_03_vstat_Full_Fstat*
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_vKLJasUj5R1/ulay_box_0.nii
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__tmp_chauf_59BbiDFYCiw/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_vKLJasUj5R1/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Qt8k7nOKJYlUJv71aR3dPQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Qt8k7nOKJYlUJv71aR3dPQ+tlrc.BRIK
++ Output dataset ./QC_sub-02/media/__tmp_chauf_59BbiDFYCiw/tmp_olay.nii
++ User-entered function range value value (14.947501)
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :487
[1] 2477683
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-02/media/__tmp_chauf_59BbiDFYCiw/ulay_box_0.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-02/media/__tmp_chauf_59BbiDFYCiw/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_pqZpCgB_EMQsfG-TFhky3Q argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_pqZpCgB_EMQsfG-TFhky3Q+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :573
[1] 2478146
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-03_REML+tlrc.HEAD
+ Number of voxels in mask = 68545
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__tmp_chauf_vKLJasUj5R1++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 476.934997 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_vKLJasUj5R1
+* Removing temporary image directory '../__tmp_chauf_vKLJasUj5R1'.
[1] Done Xvfb :487 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-01/media/__workdir_EAC_AqwNeHk31Cs*'
++ DONE! Image output:
QC_sub-01/media/qc_02_va2t_anat2temp
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_59BbiDFYCiw++ Writing palette image to QC_sub-02/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-02/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 14.947501 -com SET_THRESHNEW 5.344243 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-02/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-02/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-02/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-02/media/__tmp_chauf_59BbiDFYCiw
+* Removing temporary image directory 'QC_sub-02/media/__tmp_chauf_59BbiDFYCiw'.
[1] Done Xvfb :573 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-02/media/qc_03_vstat_Full_Fstat*
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-01_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 13.157135 -thr_olay 4.919535 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-01/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-01/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-01_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-01/media/__tmp_chauf_NjbipMnhoU1
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__tmp_chauf_NjbipMnhoU1/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Output dataset ./QC_sub-01/media/__tmp_chauf_NjbipMnhoU1/tmp_olay.nii
++ User-entered function range value value (13.157135)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-01/media/__tmp_chauf_NjbipMnhoU1/ulay_box_0.nii
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-02_REML+tlrc.HEAD
+ Number of voxels in mask = 69370
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-01/media/__tmp_chauf_NjbipMnhoU1/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_fJaxsVSMgjNAk74gf09nhA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_fJaxsVSMgjNAk74gf09nhA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :853
[1] 2479632
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+ 3dGrayplot: Elapsed = 8.8 s
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-03+tlrc.HEAD -olay full_mask.sub-03+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-03/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-03+tlrc.HEAD
++ Found input file: full_mask.sub-03+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_rOkg72lU3zf
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_rOkg72lU3zf/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_rOkg72lU3zf/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_rOkg72lU3zf/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_rOkg72lU3zf/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_2fAKos0_lpu7X7G9Baam5w argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_2fAKos0_lpu7X7G9Baam5w+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :818
[1] 2480008
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__tmp_chauf_NjbipMnhoU1++ Writing palette image to QC_sub-01/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-01/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 13.157135 -com SET_THRESHNEW 4.919535 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-01/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-01/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-01/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-01/media/__tmp_chauf_NjbipMnhoU1
+* Removing temporary image directory 'QC_sub-01/media/__tmp_chauf_NjbipMnhoU1'.
[1] Done Xvfb :853 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-01/media/qc_03_vstat_Full_Fstat*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-01_REML+tlrc.HEAD
+ Number of voxels in mask = 69731
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/__tmp_chauf_rOkg72lU3zf++ Writing palette image to QC_sub-03/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-03/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_rOkg72lU3zf
+* Removing temporary image directory './__tmp_chauf_rOkg72lU3zf'.
[1] Done Xvfb :818 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
+ 3dGrayplot: Elapsed = 8.9 s
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-03/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-02+tlrc.HEAD -olay full_mask.sub-02+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-02/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-02+tlrc.HEAD
++ Found input file: full_mask.sub-02+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_33D69u071eR
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Output dataset ././__tmp_chauf_33D69u071eR/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_33D69u071eR/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ 3dAutobox: output dataset = ./__tmp_chauf_33D69u071eR/ulay_box_0.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_33D69u071eR/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_7UuV9B6m0lS6AP59osvm5Q argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_7UuV9B6m0lS6AP59osvm5Q+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :997
[1] 2481509
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-03/media/qc_09_regr_df.dat
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-03/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-03/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-03/media/__tmp_chauf_glK4FL9KO6e
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_glK4FL9KO6e/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_glK4FL9KO6e/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-03/media/__tmp_chauf_glK4FL9KO6e/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-03/media/__tmp_chauf_glK4FL9KO6e/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_hK3Vm3CTlhsuCD3aT9ZBKg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_hK3Vm3CTlhsuCD3aT9ZBKg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :97
[1] 2482035
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/__tmp_chauf_33D69u071eR++ Writing palette image to QC_sub-02/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-02/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_33D69u071eR
+* Removing temporary image directory './__tmp_chauf_33D69u071eR'.
[1] Done Xvfb :997 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-02/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
+ 3dGrayplot: Elapsed = 9.0 s
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-01+tlrc.HEAD -olay full_mask.sub-01+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-01/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-01+tlrc.HEAD
++ Found input file: full_mask.sub-01+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_L9VZqsID8He
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Output dataset ././__tmp_chauf_L9VZqsID8He/tmp_ulay.nii
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_glK4FL9KO6e++ Writing palette image to QC_sub-03/media/qc_10_regr_corr_errts.pbar.jpg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_L9VZqsID8He/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_10_regr_corr_errts.axi.jpg'
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
AFNI QUITTs!
++ 3dAutobox: output dataset = ./__tmp_chauf_L9VZqsID8He/ulay_box_0.nii
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-03/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-03/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-03/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-03/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-03/media/__tmp_chauf_glK4FL9KO6e
+* Removing temporary image directory 'QC_sub-03/media/__tmp_chauf_glK4FL9KO6e'.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
[1] Done Xvfb :97 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-03/media/qc_10_regr_corr_errts*
++ Check summary of degrees of freedom in: QC_sub-02/media/qc_09_regr_df.dat
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_L9VZqsID8He/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_4jsBAjPcMA0dME4J3I2tlw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_4jsBAjPcMA0dME4J3I2tlw+tlrc.BRIK
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-02/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-02/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
-- trying to start Xvfb :709
[1] 2483456
++ Found input file: corr_brain+tlrc.HEAD
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-02/media/__tmp_chauf_Qn77tlHidt9
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ Output dataset ./QC_sub-02/media/__tmp_chauf_Qn77tlHidt9/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-02/media/__tmp_chauf_Qn77tlHidt9/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ olay_alpha has known value: No
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-03+tlrc.HEAD -ulay_range 0% 120% -func_range 176 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 176=red 155=oran-red 134=orange 113=oran-yell 92=yell-oran 71=yellow 51=lt-blue2 29=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-03/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 51 - 176 -prefix QC_sub-03/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-03+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-03/media/__tmp_chauf_6nGDZqpMg66
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-02/media/__tmp_chauf_Qn77tlHidt9/ulay_box_0.nii
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-02/media/__tmp_chauf_Qn77tlHidt9/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_9nDKohvwEU43E5FgaiaW8A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_9nDKohvwEU43E5FgaiaW8A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
++ Output dataset ./QC_sub-03/media/__tmp_chauf_6nGDZqpMg66/tmp_ulay.nii
-- trying to start Xvfb :12
[1] 2484133
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-03/media/__tmp_chauf_6nGDZqpMg66/tmp_olay.nii
++ User-entered function range value value (176)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-03/media/__tmp_chauf_6nGDZqpMg66/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-03/media/__tmp_chauf_6nGDZqpMg66/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_4L0rK24bS7vKw8SzcSzopw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_4L0rK24bS7vKw8SzcSzopw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :488
[1] 2484361
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/__tmp_chauf_L9VZqsID8He++ Writing palette image to QC_sub-01/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-01/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_L9VZqsID8He
+* Removing temporary image directory './__tmp_chauf_L9VZqsID8He'.
[1] Done Xvfb :709 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-01/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_Qn77tlHidt9++ Writing palette image to QC_sub-02/media/qc_10_regr_corr_errts.pbar.jpg
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-02/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-02/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-02/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-02/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-02/media/__tmp_chauf_Qn77tlHidt9
+* Removing temporary image directory 'QC_sub-02/media/__tmp_chauf_Qn77tlHidt9'.
[1] Done Xvfb :12 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-02/media/qc_10_regr_corr_errts*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-01/media/qc_09_regr_df.dat
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/QC_sub-03/media/__tmp_chauf_6nGDZqpMg66++ Writing palette image to QC_sub-03/media/qc_11_regr_tsnr_fin.pbar.jpg
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-01/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-01/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-03/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 176=red 155=oran-red 134=orange 113=oran-yell 92=yell-oran 71=yellow 51=lt-blue2 29=blue 0=none -com PBAR_SAVEIM QC_sub-03/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 176 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-03/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-03/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-03/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-03/media/__tmp_chauf_6nGDZqpMg66
+* Removing temporary image directory 'QC_sub-03/media/__tmp_chauf_6nGDZqpMg66'.
[1] Done Xvfb :488 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-03/media/qc_11_regr_tsnr_fin*
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-02+tlrc.HEAD -ulay_range 0% 120% -func_range 153 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 153=red 138=oran-red 124=orange 110=oran-yell 96=yell-oran 82=yellow 68=lt-blue2 38=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-02/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 68 - 153 -prefix QC_sub-02/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-02+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw
++ Output dataset ./QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ 3dAutobox: output dataset = QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ/ulay_box_0.nii
++ Check for corr matrix warnings in: QC_sub-03/media/qc_12_warns_xmat.dat
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Check for censor fraction warnings (general): QC_sub-03/media/qc_13_warns_cen_total.dat
++ Output dataset ./QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Check for censor fraction warnings (per stim): QC_sub-03/media/qc_14_warns_cen_stim.dat
++ Output dataset ./QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw/tmp_olay.nii
++ User-entered function range value value (153)
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_S6KxDhB711qqEktG2FfnDQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset /tmp/3dcalc_XYZ_S6KxDhB711qqEktG2FfnDQ+tlrc.BRIK
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw/ulay_box_0.nii
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :610
[1] 2486555
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Check basic summary quants from proc in: QC_sub-03/media/qc_16_qsumm_ssrev.dat
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-03
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-03+tlrc.HEAD
final stats dset : stats.sub-03_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 8
average motion (per TR) : 0.0976639
average censored motion : 0.0819588
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.81088
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.79726
average outlier frac (TR) : 0.00255359
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 139 139
num TRs per run (censored): 7 7
fraction censored per run : 0.0479452 0.0479452
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 278
degrees of freedom used : 20
degrees of freedom left : 258
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 14
censor fraction : 0.047945
num regs of interest : 2
num TRs per stim (orig) : 120 115
num TRs censored per stim : 7 5
fraction TRs censored : 0.058 0.043
ave mot per sresp (orig) : 0.104433 0.088395
ave mot per sresp (cens) : 0.082356 0.078794
TSNR average : 122.576
global correlation (GCOR) : 0.137945
anat/EPI mask Dice coef : 0.917524
anat/templ mask Dice coef : 0.949783
maximum F-stat (masked) : 42.0812
blur estimates (ACF) : 0.824937 3.35061 13.1355
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-03
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_j49dKonCxk0Fs-T7e96b_Q argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_j49dKonCxk0Fs-T7e96b_Q+tlrc.BRIK
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-03/index.html
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-03.results/QC_sub-03/index.html\n
++ Will have the ref box central gapord: 10 13 10
consider running:
------------------- end of optionizing -------------------
afni_open -b ./afni_pro_glm/sub-03.results/QC_sub-03/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:04:04 UTC 2025
-- trying to start Xvfb :951
[1] 2487004
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
echo auto-generated by afni_proc.py, Tue Nov 4 05:04:05 2025
auto-generated by afni_proc.py, Tue Nov 4 05:04:05 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 05:04:05 UTC 2025
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-04_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-04.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-04.tcsh |& tee ./afni_pro_glm/output.proc.sub-04.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-04.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-04.tcsh
set subj = sub-04
endif
set output_dir = ./afni_pro_glm/sub-04.results
if ( -d ./afni_pro_glm/sub-04.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_pro_glm/sub-04.results
mkdir ./afni_pro_glm/sub-04.results/stimuli
cp ./ds000102/sub-04/func/congruent.1D ./ds000102/sub-04/func/incongruent.1D ./afni_pro_glm/sub-04.results/stimuli
3dcopy ds000102/sub-04/anat/sub-04_T1w.nii.gz ./afni_pro_glm/sub-04.results/sub-04_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_pro_glm/sub-04.results/pb00.sub-04.r01.tcat ds000102/sub-04/func/sub-04_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
3dTcat -prefix ./afni_pro_glm/sub-04.results/pb00.sub-04.r02.tcat ds000102/sub-04/func/sub-04_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.8 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-04.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-04.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ++ Writing palette image to QC_sub-01/media/qc_10_regr_corr_errts.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-01/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-01/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-01/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-01/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-01/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ
+* Removing temporary image directory 'QC_sub-01/media/__tmp_chauf_pUx4Rc8sXcJ'.
[1] Done Xvfb :610 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-01/media/qc_10_regr_corr_errts*
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw++ Writing palette image to QC_sub-02/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-02/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 153=red 138=oran-red 124=orange 110=oran-yell 96=yell-oran 82=yellow 68=lt-blue2 38=blue 0=none -com PBAR_SAVEIM QC_sub-02/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 153 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-02/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-02/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-02/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw
+* Removing temporary image directory 'QC_sub-02/media/__tmp_chauf_ebA8q1JHnFw'.
[1] Done Xvfb :951 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-02/media/qc_11_regr_tsnr_fin*
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ 34848 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-04.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-01+tlrc.HEAD -ulay_range 0% 120% -func_range 188 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 188=red 168=oran-red 148=orange 129=oran-yell 109=yell-oran 89=yellow 70=lt-blue2 43=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-01/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 70 - 188 -prefix QC_sub-01/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-01+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-01/media/__tmp_chauf_tqiuSp42liI
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Copy ulay to visualize (volumetric) within user's range:
++ Check for corr matrix warnings in: QC_sub-02/media/qc_12_warns_xmat.dat
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Check for censor fraction warnings (general): QC_sub-02/media/qc_13_warns_cen_total.dat
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Check for censor fraction warnings (per stim): QC_sub-02/media/qc_14_warns_cen_stim.dat
++ Output dataset ./QC_sub-01/media/__tmp_chauf_tqiuSp42liI/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-01/media/__tmp_chauf_tqiuSp42liI/tmp_olay.nii
++ User-entered function range value value (188)
++ Check basic summary quants from proc in: QC_sub-02/media/qc_16_qsumm_ssrev.dat
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-01/media/__tmp_chauf_tqiuSp42liI/ulay_box_0.nii
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-02
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-02+tlrc.HEAD
#Script is running (command trimmed):
3dinfo ./__tt_sub-04_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/__tt_sub-04_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 330.981995
+ Used gradual clip level = 329.589508 .. 344.044403
+ Number voxels above clip level = 36802
+ Clustering voxels ...
+ Largest cluster has 34732 voxels
+ Clustering voxels ...
+ Largest cluster has 34509 voxels
+ Filled 246 voxels in small holes; now have 34755 voxels
+ Clustering voxels ...
+ Largest cluster has 34754 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 129086 voxels
+ Mask now has 34754 voxels
++ 34754 voxels in the mask [out of 163840: 21.21%]
++ first 11 x-planes are zero [from L]
++ last 10 x-planes are zero [from R]
++ first 3 y-planes are zero [from P]
++ last 6 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 5 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 815.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/815.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-04_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-04_T1w_ns+orig -prefix ./sub-04_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-04_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-04_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 1362585 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=5 ytop=2
++ Zero-pad: zbot=8 ztop=3
++ 34754 voxels [15.0%] in weight mask
++ Output dataset ./__tt_sub-04_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 34754
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.524 32.434 27.547 (index)
+ source center of mass = 89.867 113.672 123.954 (index)
+ source-target CM = 85.601 -1.260 -3.733 (xyz)
+ estimated center of mass shifts = 85.601 -1.260 -3.733
++ shift param auto-range: 24.9..146.3 -68.7..66.1 -67.9..60.5
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 75.600967 .. 95.600967 center = 85.600967
+ Range param#2 [y-shift] = -11.260391 .. 8.739609 center = -1.260391
+ Range param#3 [z-shift] = -13.733498 .. 6.266502 center = -3.733498
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 45.600967 .. 125.600967 center = 85.600967
+ Range param#2 [y-shift] = -41.260391 .. 38.739609 center = -1.260391
+ Range param#3 [z-shift] = -43.733498 .. 36.266502 center = -3.733498
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000091
++ Final parameter search ranges:
+ x-shift = 45.601 .. 125.601
+ y-shift = -41.260 .. 38.740
+ z-shift = -43.733 .. 36.267
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1362585 [out of 11534336] voxels
+ base mask has 46916 [out of 231744] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=40 usiz=154.5
+ - copying weight image
+ - using 34754 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 28400 total points stored in 55 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
+ - Test (64+191)*64 params [top5=*o+-.]:#oo+..+.-.+o-o.o.+.ooo-+o$-+oo + - best 88 costs found:
o= 0 v=-0.057173: 85.60 -1.26 -3.73 0.00 0.00 0.00 [grid]
o= 1 v=-0.031811: 83.31 -8.44 -35.43 -10.97 18.37 5.04 [rand]
o= 2 v=-0.030876: 87.90 -8.44 -35.43 -10.97 18.37 5.04 [rand]
o= 3 v=-0.026428: 78.93 -27.93 -30.40 -30.00 30.00 -7.50 [grid]
o= 4 v=-0.025526: 118.09 -28.25 28.16 9.69 -21.75 -40.77 [rand]
o= 5 v=-0.024647: 83.31 -8.44 -35.43 -10.97 18.37 -5.04 [rand]
o= 6 v=-0.024520: 92.27 5.41 -30.40 30.00 7.50 -7.50 [grid]
o= 7 v=-0.024304: 114.62 -28.40 -37.35 7.44 27.42 -25.63 [rand]
o= 8 v=-0.024124: 79.19 32.68 -40.31 -19.28 27.13 15.13 [rand]
o= 9 v=-0.023976: 78.93 -27.93 -30.40 -7.50 30.00 -7.50 [grid]
o=10 v=-0.023535: 102.40 -19.72 -36.09 -10.10 16.32 39.31 [rand]
o=11 v=-0.022785: 83.32 -19.42 -32.25 -37.05 26.99 -2.67 [rand]
o=12 v=-0.022748: 48.96 -18.49 18.52 -5.26 -41.39 40.03 [rand]
o=13 v=-0.022659: 74.11 15.05 -37.49 19.17 -13.96 2.56 [rand]
o=14 v=-0.022605: 92.27 5.41 -30.40 30.00 7.50 7.50 [grid]
o=15 v=-0.022559: 60.08 -32.60 30.55 42.46 -41.07 35.63 [rand]
o=16 v=-0.022241: 87.90 5.92 -35.43 10.97 18.37 5.04 [rand]
o=17 v=-0.022108: 56.59 25.88 -37.35 -7.44 -27.42 -25.63 [rand]
o=18 v=-0.022033: 112.27 -27.93 -30.40 7.50 30.00 -30.00 [grid]
o=19 v=-0.021949: 118.09 -28.25 28.16 -9.69 -21.75 -40.77 [rand]
o=20 v=-0.021623: 110.57 -30.31 -39.18 17.69 14.58 -34.92 [rand]
o=21 v=-0.021512: 92.27 -7.93 -30.40 -7.50 30.00 7.50 [grid]
o=22 v=-0.021219: 92.27 5.41 -30.40 30.00 30.00 -7.50 [grid]
o=23 v=-0.021171: 83.32 -19.42 -32.25 -37.05 26.99 2.67 [rand]
o=24 v=-0.020908: 94.80 -13.03 -25.47 -6.82 34.51 24.39 [rand]
o=25 v=-0.020618: 74.11 15.05 -37.49 19.17 -13.96 -2.56 [rand]
o=26 v=-0.020561: 112.53 26.88 34.21 -15.05 -4.71 -12.78 [rand]
o=27 v=-0.020199: 53.39 27.79 32.41 40.41 19.09 40.75 [rand]
o=28 v=-0.020134: 78.93 -7.93 -30.40 -7.50 7.50 -7.50 [grid]
o=29 v=-0.020111: 87.89 -19.42 -32.25 -37.05 26.99 -2.67 [rand]
o=30 v=-0.019873: 78.93 -27.93 -30.40 -30.00 30.00 7.50 [grid]
o=31 v=-0.019844: 92.27 -7.93 -30.40 -30.00 30.00 30.00 [grid]
o=32 v=-0.019767: 60.23 25.99 33.77 41.23 -14.90 41.72 [rand]
o=33 v=-0.019743: 78.93 -7.93 -30.40 -30.00 30.00 7.50 [grid]
o=34 v=-0.019725: 92.27 -7.93 -30.40 7.50 7.50 7.50 [grid]
o=35 v=-0.019649: 92.27 -7.93 -30.40 -7.50 7.50 7.50 [grid]
o=36 v=-0.019564: 79.19 -35.20 32.84 19.28 -27.13 15.13 [rand]
o=37 v=-0.019558: 53.11 25.73 -35.62 -9.69 -21.75 -40.77 [rand]
o=38 v=-0.019556: 48.96 -18.49 18.52 5.26 -41.39 40.03 [rand]
o=39 v=-0.019507: 78.93 -7.93 -30.40 -30.00 7.50 -7.50 [grid]
o=40 v=-0.019490: 87.90 -8.44 -35.43 10.97 18.37 5.04 [rand]
o=41 v=-0.019330: 60.63 27.79 31.71 17.69 -14.58 34.92 [rand]
o=42 v=-0.019318: 49.01 26.04 27.50 25.97 -37.53 40.21 [rand]
o=43 v=-0.019284: 60.63 27.79 -39.18 17.69 -14.58 -34.92 [rand]
o=44 v=-0.019178: 53.17 -15.60 -9.33 -8.22 -9.03 36.72 [rand]
o=45 v=-0.019158: 92.27 -7.93 2.93 -7.50 -7.50 -7.50 [grid]
o=46 v=-0.019108: 87.89 -19.42 -32.25 -37.05 26.99 2.67 [rand]
o=47 v=-0.018873: 71.66 -25.13 -21.91 -5.58 38.34 -17.97 [rand]
o=48 v=-0.018833: 83.27 -33.63 -30.44 -4.84 28.19 -20.57 [rand]
o=49 v=-0.018679: 111.02 6.16 -30.85 5.61 -11.01 -21.77 [rand]
o=50 v=-0.018650: 92.27 5.41 -30.40 -7.50 7.50 -7.50 [grid]
o=51 v=-0.018624: 104.72 -24.68 -37.03 -37.88 13.88 18.42 [rand]
o=52 v=-0.018547: 92.27 5.41 2.93 -7.50 7.50 -7.50 [grid]
o=53 v=-0.018521: 66.49 22.15 -37.03 37.88 13.88 -18.42 [rand]
o=54 v=-0.018422: 96.90 -35.39 -9.71 -4.11 32.66 25.74 [rand]
o=55 v=-0.018300: 92.27 5.41 -30.40 30.00 30.00 7.50 [grid]
o=56 v=-0.018268: 87.90 -8.44 -35.43 -10.97 18.37 -5.04 [rand]
o=57 v=-0.018152: 92.27 5.41 -30.40 30.00 -7.50 7.50 [grid]
o=58 v=-0.018124: 95.66 -24.16 -25.48 23.24 8.32 39.75 [rand]
o=59 v=-0.018114: 66.49 22.15 -37.03 37.88 13.88 18.42 [rand]
o=60 v=-0.018096: 114.62 -28.40 29.88 -7.44 -27.42 -25.63 [rand]
o=61 v=-0.018075: 60.23 -28.51 33.77 41.23 -14.90 41.72 [rand]
o=62 v=-0.017918: 100.62 24.95 3.17 30.75 -34.00 35.37 [rand]
o=63 v=-0.017913: 55.98 33.35 33.08 -24.31 -11.89 -2.84 [rand]
o=64 v=-0.017890: 92.02 -35.20 32.84 19.28 -27.13 -15.13 [rand]
o=65 v=-0.017782: 53.39 27.79 32.41 40.41 -19.09 40.75 [rand]
o=66 v=-0.017717: 76.94 -25.42 -40.31 -27.41 42.10 -24.35 [rand]
o=67 v=-0.017560: 92.27 -7.93 -30.40 7.50 7.50 -7.50 [grid]
o=68 v=-0.017556: 58.93 25.41 22.93 7.50 -7.50 30.00 [grid]
o=69 v=-0.017550: 99.31 -3.95 -38.03 -41.38 19.47 -31.64 [rand]
o=70 v=-0.017523: 92.27 25.41 -30.40 -7.50 -7.50 -7.50 [grid]
o=71 v=-0.017493: 58.67 26.88 34.21 15.05 -4.71 12.78 [rand]
o=72 v=-0.017464: 68.81 -19.72 -36.09 -10.10 16.32 -39.31 [rand]
o=73 v=-0.017438: 57.58 2.86 28.18 18.18 -25.37 33.48 [rand]
o=74 v=-0.017374: 92.27 -7.93 -30.40 -7.50 7.50 -7.50 [grid]
o=75 v=-0.017343: 75.35 7.58 -19.37 7.51 21.00 -38.82 [rand]
o=76 v=-0.017323: 120.40 -15.04 -37.75 10.62 7.10 -10.42 [rand]
o=77 v=-0.017233: 49.01 -28.56 27.50 25.97 -37.53 40.21 [rand]
o=78 v=-0.017229: 65.35 26.02 -38.22 37.90 -5.03 3.08 [rand]
o=79 v=-0.017227: 113.42 -24.48 -30.27 3.35 33.01 -26.09 [rand]
o=80 v=-0.017126: 76.40 -13.03 -25.47 -6.82 34.51 -24.39 [rand]
o=81 v=-0.017022: 114.62 -28.40 29.88 7.44 -27.42 -25.63 [rand]
o=82 v=-0.016995: 99.31 1.43 -38.03 -41.38 19.47 -31.64 [rand]
o=83 v=-0.016974: 51.27 -17.85 -12.34 -16.34 -8.64 40.54 [rand]
o=84 v=-0.016970: 111.12 -32.60 30.55 -42.46 -41.07 -35.63 [rand]
o=85 v=-0.016944: 94.21 -22.89 -32.44 -14.86 11.51 26.00 [rand]
o=86 v=-0.016944: 102.65 21.43 -27.98 12.15 -12.44 39.75 [rand]
o=87 v=-0.016864: 53.11 25.73 -35.62 9.69 -21.75 -40.77 [rand]
#Script is running (command trimmed):
3dinfo ./__tt_sub-05_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/__tt_sub-05_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - A little optimization:*[#16324=-0.242186] *[#16333=-0.244621] *[#16338=-0.24742] *[#16339=-0.250965] *[#16345=-0.252477] *[#16348=-0.252626] *[#16350=-0.252908] *[#16351=-0.253194] *[#16352=-0.253219] *[#16354=-0.253413] *[#16357=-0.253437] *[#16359=-0.253456] *[#16360=-0.253484] *[#16363=-0.253485] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.253485: 86.04 -1.07 0.23 -1.05 -0.42 0.75 [grid] [f=43]
o= 1 v=-0.034663: 87.22 -8.63 -35.19 -11.40 18.78 5.31 [rand] [f=34]
o= 2 v=-0.033728: 85.17 -8.30 -34.31 -10.39 17.05 5.99 [rand] [f=40]
o= 3 v=-0.031684: 79.77 -24.85 -31.46 -31.05 30.52 -7.06 [grid] [f=56]
o= 4 v=-0.030601: 118.50 -27.77 27.75 9.33 -22.36 -36.07 [rand] [f=48]
o= 5 v=-0.036415: 83.68 -8.62 -35.10 -11.84 15.10 0.86 [rand] [f=64]
o= 6 v=-0.036301: 91.43 4.87 -33.92 27.79 7.49 -0.90 [grid] [f=51]
o= 7 v=-0.026917: 110.23 -27.93 -37.46 7.46 27.32 -25.82 [rand] [f=47]
o= 8 v=-0.027718: 79.08 32.21 -39.74 -14.73 26.33 15.61 [rand] [f=45]
o= 9 v=-0.026469: 78.37 -26.97 -30.22 -7.81 29.92 -6.41 [grid] [f=55]
o=10 v=-0.027382: 101.34 -19.60 -35.37 -9.83 16.65 39.30 [rand] [f=60]
o=11 v=-0.027308: 81.56 -19.58 -32.08 -31.53 25.98 -3.67 [rand] [f=49]
o=12 v=-0.027088: 48.93 -20.17 18.84 -6.67 -44.04 39.30 [rand] [f=38]
o=13 v=-0.038798: 77.78 20.41 -38.24 18.90 -7.58 2.00 [rand] [f=83]
o=14 v=-0.040608: 91.04 1.66 -36.41 25.63 16.76 1.26 [grid] [f=66]
o=15 v=-0.040391: 55.31 -33.58 35.63 40.59 -43.00 37.55 [rand] [f=55]
o=16 v=-0.041994: 91.87 1.51 -35.86 19.26 18.12 2.58 [rand] [f=74]
o=17 v=-0.025003: 56.65 25.75 -37.07 -8.04 -28.81 -26.69 [rand] [f=45]
o=18 v=-0.024459: 112.80 -27.71 -30.48 7.78 29.87 -25.77 [grid] [f=37]
o=19 v=-0.026129: 117.60 -28.09 28.62 -9.43 -16.27 -40.55 [rand] [f=47]
o=20 v=-0.026307: 110.84 -26.29 -39.33 17.14 15.09 -34.97 [rand] [f=38]
o=21 v=-0.027581: 86.79 -8.28 -34.57 -7.22 29.18 6.71 [grid] [f=51]
o=22 v=-0.037283: 93.34 2.86 -35.07 26.59 27.82 -2.79 [grid] [f=45]
o=23 v=-0.026669: 83.49 -26.95 -31.41 -34.67 26.63 2.46 [rand] [f=53]
o=24 v=-0.030974: 96.62 -12.06 -27.14 -2.69 33.04 26.28 [rand] [f=83]
o=25 v=-0.040564: 78.31 20.13 -37.41 15.51 -7.19 5.44 [rand] [f=77]
o=26 v=-0.027584: 110.85 28.32 35.60 -6.25 -3.36 -16.85 [rand] [f=49]
o=27 v=-0.029853: 49.25 30.95 31.64 40.14 18.13 42.35 [rand] [f=39]
o=28 v=-0.030851: 80.29 -7.56 -33.94 -9.27 12.75 -6.59 [grid] [f=49]
o=29 v=-0.027366: 83.99 -19.52 -33.29 -36.77 27.60 -0.97 [rand] [f=47]
o=30 v=-0.022590: 79.61 -28.12 -30.01 -30.11 34.02 7.40 [grid] [f=45]
o=31 v=-0.028459: 96.70 -8.14 -30.90 -32.24 34.10 30.39 [grid] [f=38]
o=32 v=-0.031030: 55.81 26.02 36.17 38.65 -15.19 39.89 [rand] [f=35]
o=33 v=-0.030595: 77.69 -2.57 -33.01 -32.34 31.91 6.07 [grid] [f=52]
o=34 v=-0.034006: 95.78 -6.16 -35.12 10.78 6.99 7.13 [grid] [f=60]
o=35 v=-0.032570: 91.12 -14.08 -34.09 -7.60 7.36 8.63 [grid] [f=53]
o=36 v=-0.026929: 79.93 -36.19 35.44 18.32 -27.81 15.52 [rand] [f=66]
o=37 v=-0.036532: 55.79 26.10 -35.27 -9.73 -31.33 -44.71 [rand] [f=83]
o=38 v=-0.024983: 48.78 -18.40 19.68 4.38 -40.52 42.14 [rand] [f=42]
o=39 v=-0.028658: 79.92 -3.44 -30.21 -28.80 7.60 -8.10 [grid] [f=61]
o=40 v=-0.037945: 92.54 0.62 -38.64 16.43 13.59 4.79 [rand] [f=64]
o=41 v=-0.041558: 54.24 22.81 30.88 21.16 -7.87 37.02 [rand] [f=66]
o=42 v=-0.024399: 49.60 21.90 26.47 25.97 -37.55 39.06 [rand] [f=51]
o=43 v=-0.030976: 59.92 33.19 -39.18 16.56 -21.10 -33.41 [rand] [f=50]
o=44 v=-0.027816: 52.51 -13.36 -9.74 -11.68 -12.86 34.20 [rand] [f=36]
o=45 v=-0.253401: 86.03 -1.09 0.16 -1.10 -0.53 0.91 [grid] [f=81]
o=46 v=-0.026839: 82.87 -18.31 -34.26 -35.50 27.34 -4.59 [rand] [f=83]
o=47 v=-0.026081: 73.37 -22.35 -23.06 -6.80 39.34 -20.63 [rand] [f=76]
o=48 v=-0.021237: 82.31 -33.40 -30.15 -5.94 28.07 -19.94 [rand] [f=48]
o=49 v=-0.024798: 109.81 7.32 -31.42 3.09 -15.95 -21.96 [rand] [f=36]
o=50 v=-0.022814: 92.52 5.27 -32.07 -7.66 6.43 -6.63 [grid] [f=39]
o=51 v=-0.020789: 104.91 -24.21 -36.53 -38.17 14.14 22.34 [rand] [f=41]
o=52 v=-0.253416: 86.02 -1.08 0.17 -1.08 -0.56 0.91 [grid] [f=75]
o=53 v=-0.032465: 64.59 21.50 -31.91 35.52 20.84 -16.51 [rand] [f=75]
o=54 v=-0.021177: 97.62 -35.61 -9.21 -4.60 32.39 26.19 [rand] [f=32]
o=55 v=-0.038013: 91.23 2.11 -35.16 28.56 30.55 6.14 [grid] [f=53]
o=56 v=-0.042857: 82.34 -5.29 -35.15 -16.01 10.92 0.26 [rand] [f=83]
o=57 v=-0.025160: 96.26 3.32 -29.76 29.53 -7.32 7.04 [grid] [f=37]
o=58 v=-0.020675: 96.14 -23.74 -25.31 22.90 9.13 39.18 [rand] [f=44]
o=59 v=-0.029224: 70.50 21.37 -38.12 38.64 14.12 14.95 [rand] [f=44]
o=60 v=-0.022978: 115.09 -28.64 33.93 -7.79 -26.82 -25.97 [rand] [f=36]
o=61 v=-0.025912: 60.61 -30.85 36.20 41.65 -18.74 43.73 [rand] [f=64]
o=62 v=-0.025461: 99.45 24.77 5.15 26.63 -36.31 41.46 [rand] [f=52]
o=63 v=-0.029830: 52.48 32.70 33.47 -25.26 -6.15 1.93 [rand] [f=54]
o=64 v=-0.027404: 92.51 -39.15 36.26 19.26 -30.37 -14.13 [rand] [f=68]
o=65 v=-0.031599: 55.84 30.93 36.26 38.58 -18.58 40.89 [rand] [f=50]
o=66 v=-0.021848: 73.69 -25.29 -39.96 -28.61 41.18 -24.34 [rand] [f=47]
o=67 v=-0.023416: 91.39 -4.64 -31.11 5.49 7.45 -7.43 [grid] [f=40]
o=68 v=-0.045084: 54.48 27.90 30.46 12.20 -3.12 30.25 [grid] [f=60]
o=69 v=-0.028540: 95.53 -8.27 -37.95 -42.29 22.56 -31.61 [rand] [f=55]
o=70 v=-0.024842: 86.85 25.08 -32.07 -10.38 0.08 -8.90 [grid] [f=65]
o=71 v=-0.038695: 58.65 27.63 35.13 12.77 -2.37 21.42 [rand] [f=83]
o=72 v=-0.025694: 72.11 -19.13 -35.50 -9.51 17.51 -39.07 [rand] [f=55]
o=73 v=-0.039314: 56.59 1.31 35.89 18.55 -23.68 29.66 [rand] [f=56]
o=74 v=-0.032174: 90.43 -13.89 -30.50 -6.01 8.40 -5.44 [grid] [f=54]
o=75 v=-0.024419: 71.24 7.52 -20.41 7.69 20.99 -39.36 [rand] [f=49]
o=76 v=-0.026622: 119.13 -16.67 -36.03 9.70 5.92 -10.00 [rand] [f=48]
o=77 v=-0.026246: 56.63 -29.00 27.80 25.75 -38.16 41.36 [rand] [f=42]
o=78 v=-0.026011: 65.85 22.82 -39.28 37.06 -5.28 3.05 [rand] [f=48]
o=79 v=-0.022744: 112.45 -29.88 -33.65 4.92 32.72 -25.79 [rand] [f=37]
o=80 v=-0.022894: 75.73 -13.06 -25.57 -6.89 34.35 -19.53 [rand] [f=48]
o=81 v=-0.023827: 114.66 -29.18 33.84 7.59 -27.56 -25.58 [rand] [f=42]
o=82 v=-0.025322: 95.46 -0.21 -38.66 -40.89 19.48 -31.45 [rand] [f=53]
o=83 v=-0.025980: 52.61 -19.29 -11.91 -17.24 -8.29 35.25 [rand] [f=49]
o=84 v=-0.033877: 113.31 -38.87 34.34 -40.11 -40.90 -31.27 [rand] [f=48]
#++ removing skull or area outside brain
o=85 v=-0.023807: 99.98 -22.85 -31.33 -14.29 11.11 26.30 [rand] [f=41]
#Script is running (command trimmed):
o=86 v=-0.033325: 106.78 23.27 -32.06 12.63 -8.19 42.82 [rand] [f=79]
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
o=87 v=-0.035975: 60.06 30.75 -37.17 6.05 -24.52 -40.91 [rand] [f=64]
+ - saving # 0 for use with twobest
+ - skip #52 for twobest: too close to set # 0
+ - skip #45 for twobest: too close to set # 0
+ - saving #68 for use with twobest
+ - saving #56 for use with twobest
+ - saving #16 for use with twobest
+ - saving #41 for use with twobest
+ - saving #14 for use with twobest
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - saving #25 for use with twobest
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
+ - saving #15 for use with twobest
+ - saving #73 for use with twobest
+ - skip #13 for twobest: too close to set #25
+ - saving #71 for use with twobest
+ - saving #55 for use with twobest
+ - saving #40 for use with twobest
+ - saving #22 for use with twobest
+ - saving #37 for use with twobest
+ - skip # 5 for twobest: too close to set #56
+ - saving # 6 for use with twobest
+ - saving #87 for use with twobest
+ - skip # 1 for twobest: too close to set # 5
+ - saving #34 for use with twobest
+ - saving #84 for use with twobest
+ - skip # 2 for twobest: too close to set # 1
+ - saving #86 for use with twobest
+ - saving #35 for use with twobest
+ - saving #53 for use with twobest
+ - saving #74 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #65 for use with twobest
+ - saving #32 for use with twobest
+ - saving #43 for use with twobest
+ - saving #24 for use with twobest
+ - saving #28 for use with twobest
+ - saving # 4 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
++ Forming automask
+ Fixed clip level = 358.511993
+ Used gradual clip level = 346.521515 .. 381.547607
+ Number voxels above clip level = 39458
+ Clustering voxels ...
+ Largest cluster has 38691 voxels
+ Clustering voxels ...
+ Largest cluster has 38271 voxels
+ Filled 290 voxels in small holes; now have 38561 voxels
+ Clustering voxels ...
+ Largest cluster has 38559 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 125281 voxels
+ Mask now has 38559 voxels
++ 38559 voxels in the mask [out of 163840: 23.53%]
++ first 10 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 1 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 4 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
+ !source mask fill: ubot=40 usiz=154.5
#++ Applying threshold of 855.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/855.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-05_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-05_T1w_ns+orig -prefix ./sub-05_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-05_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
+ - retaining old weight image
+ - using 34754 points from base image [use_all=2]
+ * Exit alignment setup routine
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-05_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
+ 28518 total points stored in 57 'TOHD(17.3748)' bloks (0 duplicates)
++ Loading datasets into memory
*[#21113=-0.311188] *[#21121=-0.313303] *[#21133=-0.313477] *[#21135=-0.314695] *[#21136=-0.315877] *[#21137=-0.315915] *[#21139=-0.318374] *[#21142=-0.319829] *[#21143=-0.321291] *[#21146=-0.327842] *[#21148=-0.328051] *[#21149=-0.330311] *[#21150=-0.332388] *[#21151=-0.336105] *[#21152=-0.337194] *[#21153=-0.337717] *[#21156=-0.33781] *[#21159=-0.340286] *[#21161=-0.340357] *[#21164=-0.342145] *[#21165=-0.34222] *[#21167=-0.34351] *[#21168=-0.343666] *[#21169=-0.344317] *[#21174=-0.344549] *[#21180=-0.345821] *[#21188=-0.34592] *[#21189=-0.345947] *[#21191=-0.345961] *[#21192=-0.346051] *[#21193=-0.346109] *[#21194=-0.346198] *[#21196=-0.346206] *[#21197=-0.346277] *[#21198=-0.346309] *[#21199=-0.346479] *[#21200=-0.346515] *[#21201=-0.346622] *[#21202=-0.346664] *[#21203=-0.346837] *[#21204=-0.346982] *[#21205=-0.34726] *[#21206=-0.347267] *[#21207=-0.347327] *[#21208=-0.347422]
+ - param set #1 has cost=-0.347422 [o=0 t=0]
+ -- Parameters = 85.9643 -1.7949 0.3122 -0.4703 -0.7726 0.8519 0.9816 1.0358 0.9658 0.0035 0.0057 -0.0065
+ - param set #2 has cost=-0.039158 [o=68 t=1]
+ -- Parameters = 54.0290 27.7201 30.4721 12.1561 -3.4631 30.1582 0.9903 1.0035 1.0417 -0.0029 -0.0015 -0.0020
+ - param set #3 has cost=-0.035005 [o=56 t=2]
+ -- Parameters = 82.3732 -5.0851 -34.8720 -15.8591 11.1959 -0.1241 1.0067 0.9996 0.9995 -0.0005 0.0001 0.0001
+ - param set #4 has cost=-0.032134 [o=16 t=3]
+ -- Parameters = 91.8575 1.2588 -35.6472 19.1750 17.9715 2.4605 1.0049 0.9974 1.0068 0.0009 -0.0005 0.0004
+ - param set #5 has cost=-0.036884 [o=41 t=4]
+ -- Parameters = 53.8409 22.5922 31.1065 20.2181 -9.0379 33.3659 0.9559 0.9974 0.9984 0.0016 -0.0021 -0.0010
+ - param set #6 has cost=-0.029590 [o=14 t=5]
+ -- Parameters = 91.0032 1.6218 -36.2291 25.5183 16.5996 1.1633 1.0022 1.0107 1.0046 -0.0012 -0.0029 0.0021
+ - param set #7 has cost=-0.033721 [o=25 t=6]
+ -- Parameters = 78.4297 20.1750 -37.4369 15.8513 -7.2865 5.1719 0.9990 1.0020 0.9995 -0.0002 -0.0003 0.0127
+ - param set #8 has cost=-0.031759 [o=15 t=7]
+ -- Parameters = 55.4370 -33.4423 35.7918 41.1451 -41.8368 36.7889 0.9939 1.0359 1.0001 0.0006 0.0002 0.0014
+ - param set #9 has cost=-0.035902 [o=73 t=8]
+ -- Parameters = 56.6838 1.1316 36.0091 18.7391 -23.0193 29.9342 0.9616 0.9937 1.0023 -0.0026 0.0017 0.0018
+ - param set #10 has cost=-0.034729 [o=71 t=9]
+ -- Parameters = 58.7633 27.8458 34.9625 13.0946 -1.6964 21.4780 0.9933 0.9936 0.9994 0.0019 -0.0014 -0.0002
+ - param set #11 has cost=-0.026924 [o=55 t=10]
+ -- Parameters = 90.4013 1.9897 -34.9308 28.0351 30.4796 6.7218 1.0068 1.0168 1.0408 0.0007 0.0007 0.0014
+ - param set #12 has cost=-0.030518 [o=40 t=11]
+ -- Parameters = 92.1294 0.0687 -34.2256 16.0356 14.0022 5.3822 1.0071 1.0007 0.9952 0.0007 0.0012 0.0008
+ - param set #13 has cost=-0.025845 [o=22 t=12]
+ -- Parameters = 93.1288 2.6503 -35.2692 26.5644 27.4675 -2.4092 1.0059 1.0099 1.0017 0.0009 -0.0012 0.0020
+ - param set #14 has cost=-0.032309 [o=37 t=13]
+ -- Parameters = 55.8893 25.9920 -35.3750 -9.9108 -31.4140 -44.8763 0.9981 1.0052 0.9928 -0.0029 0.0005 0.0012
+ - param set #15 has cost=-0.028886 [o=6 t=14]
+ -- Parameters = 91.1831 4.2142 -33.9879 22.6386 7.0358 -1.7092 0.9936 0.9993 1.0007 -0.0000 -0.0010 -0.0009
+ - param set #16 has cost=-0.029800 [o=87 t=15]
+ -- Parameters = 60.1060 30.5791 -37.5140 5.9448 -24.7952 -41.1523 1.0007 0.9932 0.9926 0.0006 0.0005 0.0118
+ - param set #17 has cost=-0.032892 [o=34 t=16]
+ -- Parameters = 95.1499 -5.6258 -34.4535 9.8702 5.9750 4.3450 1.0119 0.9555 0.9921 0.0055 -0.0028 -0.0057
+ - param set #18 has cost=-0.028610 [o=84 t=17]
+ -- Parameters = 113.9468 -38.8993 33.8385 -34.7749 -41.5473 -31.6373 0.9975 1.0006 0.9922 0.0008 0.0008 0.0003
+ - param set #19 has cost=-0.029598 [o=86 t=18]
+ -- Parameters = 107.5828 23.3591 -32.3760 12.3311 -8.3093 44.7174 0.9544 1.0137 1.0752 -0.0000 0.0131 -0.0024
+ - param set #20 has cost=-0.027739 [o=35 t=19]
+ -- Parameters = 91.0398 -14.1420 -34.0999 -7.5936 7.3886 8.6531 1.0053 1.0085 1.0011 0.0000 0.0002 -0.0002
+ - param set #21 has cost=-0.024607 [o=53 t=20]
+ -- Parameters = 64.5502 21.5474 -31.9704 35.4574 20.8796 -16.3494 1.0005 0.9987 0.9996 -0.0004 -0.0002 -0.0001
+ - param set #22 has cost=-0.027085 [o=74 t=21]
+ -- Parameters = 90.2406 -14.1206 -30.4472 -5.9380 8.1771 -5.3494 1.0036 1.0038 1.0021 0.0002 0.0007 -0.0000
+ - param set #23 has cost=-0.024005 [o=3 t=22]
+ -- Parameters = 79.8391 -24.8211 -31.8010 -31.1880 30.3270 -6.7864 0.9993 0.9989 0.9952 0.0004 0.0003 0.0013
+ - param set #24 has cost=-0.027502 [o=65 t=23]
+ -- Parameters = 55.7818 31.8091 36.2633 37.7605 -22.4381 38.6090 0.9748 0.9799 1.0077 0.0030 0.0069 0.0015
+ - param set #25 has cost=-0.025462 [o=32 t=24]
+ -- Parameters = 56.0696 26.0749 36.2661 38.2948 -17.5320 37.7678 0.9768 0.9967 0.9916 -0.0025 0.0001 -0.0021
+ - param set #26 has cost=-0.027644 [o=43 t=25]
+ -- Parameters = 60.8473 32.9166 -39.6221 15.1801 -21.3614 -33.4714 0.9904 0.9944 0.9914 0.0002 0.0102 -0.0020
+ - param set #27 has cost=-0.027666 [o=24 t=26]
+ -- Parameters = 96.4862 -11.4494 -26.9797 -2.7932 33.7695 26.3981 0.9968 0.9947 0.9970 -0.0027 -0.0010 0.0014
+ - param set #28 has cost=-0.032047 [o=28 t=27]
+ -- Parameters = 80.5500 -6.6915 -34.1502 -9.1686 9.3476 -0.2770 1.0102 0.9869 0.9792 -0.0265 0.0134 0.0106
+ - param set #29 has cost=-0.024754 [o=4 t=28]
+ -- Parameters = 118.3152 -27.5828 27.9556 9.1787 -22.4434 -36.2781 1.0094 0.9984 0.9965 -0.0001 0.0014 0.0126
+ - param set #30 has cost=-0.341670 [o=-1 t=-1]
+ -- Parameters = 86.2265 -1.8670 0.2280 -0.6524 -0.4414 0.7763 0.9721 1.0332 0.9781 -0.0047 0.0029 0.0006
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0371
+ --- dist(#3,#1) = 0.399
+ --- dist(#4,#1) = 0.402
+ --- dist(#5,#1) = 0.446
+ --- dist(#6,#1) = 0.44
+ --- dist(#7,#1) = 0.433
+ --- dist(#8,#1) = 0.472
+ --- dist(#9,#1) = 0.435
+ --- dist(#10,#1) = 0.508
+ --- dist(#11,#1) = 0.449
+ --- dist(#12,#1) = 0.431
+ --- dist(#13,#1) = 0.462
+ --- dist(#14,#1) = 0.432
+ --- dist(#15,#1) = 0.473
+ --- dist(#16,#1) = 0.487
+ --- dist(#17,#1) = 0.457
+ --- dist(#18,#1) = 0.429
+ --- dist(#19,#1) = 0.464
+ --- dist(#20,#1) = 0.43
+ --- dist(#21,#1) = 0.384
+ --- dist(#22,#1) = 0.499
+ --- dist(#23,#1) = 0.449
+ --- dist(#24,#1) = 0.384
+ --- dist(#25,#1) = 0.441
+ --- dist(#26,#1) = 0.445
+ --- dist(#27,#1) = 0.449
+ --- dist(#28,#1) = 0.413
+ --- dist(#29,#1) = 0.404
+ --- dist(#30,#1) = 0.401
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=40 usiz=154.5
+ - retaining old weight image
+ - using 34754 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 28602 total points stored in 58 'TOHD(17.2644)' bloks (0 duplicates)
*[#24035=-0.387379] *[#24055=-0.387807] *[#24056=-0.388207] *[#24057=-0.388861] *[#24059=-0.389542] *[#24060=-0.390113] *[#24061=-0.390566] *[#24062=-0.391177] *[#24063=-0.391771] *[#24064=-0.391909] *[#24065=-0.392164] *[#24068=-0.392668] *[#24079=-0.393428] *[#24081=-0.39349] *[#24082=-0.393757] *[#24083=-0.393847] *[#24085=-0.393913] *[#24090=-0.394086] *[#24093=-0.39474] *[#24098=-0.394915] *[#24100=-0.395116] *[#24103=-0.395481] *[#24104=-0.395701] *[#24107=-0.396044] *[#24108=-0.396119] *[#24111=-0.396233] *[#24112=-0.396386] *[#24115=-0.396504] *[#24121=-0.396889] *[#24122=-0.396915] *[#24125=-0.397064] *[#24126=-0.397391] *[#24131=-0.397539] *[#24132=-0.397629] *[#24135=-0.397878] *[#24136=-0.397882] *[#24142=-0.397956] *[#24144=-0.398334]
+ - param set #1 has cost=-0.398334 [o=0 t=0]
+ -- Parameters = 85.9948 -1.6757 0.2024 -0.5807 -1.2674 0.5224 0.9906 1.0333 0.9703 -0.0113 0.0067 -0.0162
+ - param set #2 has cost=-0.391270 [o=-1 t=-1]
+ -- Parameters = 86.0647 -1.6291 0.1202 -0.6119 -0.7255 0.8135 0.9893 1.0303 0.9794 -0.0093 0.0062 -0.0047
+ - param set #3 has cost=-0.032957 [o=68 t=1]
+ -- Parameters = 53.9330 27.7445 30.4762 12.1192 -3.5773 30.1660 0.9833 1.0002 1.0424 -0.0025 -0.0022 -0.0009
+ - param set #4 has cost=-0.033183 [o=41 t=4]
+ -- Parameters = 54.8349 23.2596 30.9279 20.6890 -9.7344 32.5777 0.9367 0.9653 1.0283 0.0030 -0.0075 -0.0007
+ - param set #5 has cost=-0.029469 [o=73 t=8]
+ -- Parameters = 56.7591 1.1107 36.0937 18.9681 -22.7969 29.8249 0.9607 0.9837 1.0039 -0.0002 0.0019 0.0014
+ - param set #6 has cost=-0.028166 [o=56 t=2]
+ -- Parameters = 82.4666 -5.0089 -34.8337 -15.6526 11.3355 -0.3319 1.0118 0.9999 1.0001 0.0015 -0.0007 -0.0005
+ - param set #7 has cost=-0.029421 [o=71 t=9]
+ -- Parameters = 58.7550 27.8345 34.9971 13.0924 -1.6760 21.4497 0.9932 0.9933 1.0011 0.0027 -0.0016 -0.0001
+ - param set #8 has cost=-0.030243 [o=25 t=6]
+ -- Parameters = 78.4041 20.4264 -37.3512 15.8945 -7.1336 5.1484 0.9982 0.9993 1.0144 -0.0021 -0.0008 0.0133
+ - param set #9 has cost=-0.025613 [o=34 t=16]
+ -- Parameters = 95.2990 -5.6234 -34.1612 9.9732 5.9040 4.0870 1.0120 0.9533 0.9999 0.0036 -0.0027 -0.0061
+ - param set #10 has cost=-0.029568 [o=37 t=13]
+ -- Parameters = 55.6254 26.0592 -35.8745 -10.2819 -31.3088 -44.7663 1.0020 1.0328 0.9643 -0.0036 -0.0066 0.0052
+ - param set #11 has cost=-0.027494 [o=16 t=3]
+ -- Parameters = 91.6399 0.7526 -35.7373 19.0313 18.4502 2.7874 1.0067 0.9854 1.0052 0.0029 -0.0047 0.0011
+ - param set #12 has cost=-0.025810 [o=28 t=27]
+ -- Parameters = 80.6062 -6.6092 -34.0348 -9.3436 9.2935 -0.2429 1.0065 0.9886 0.9791 -0.0276 0.0133 0.0106
+ - param set #13 has cost=-0.025071 [o=15 t=7]
+ -- Parameters = 55.4693 -33.4767 35.8285 41.2135 -42.1278 36.8028 0.9953 1.0364 0.9993 0.0007 0.0011 0.0013
+ - param set #14 has cost=-0.029626 [o=40 t=11]
+ -- Parameters = 91.9248 0.1631 -34.9643 19.0825 15.4470 5.3994 0.9894 0.9710 0.9916 -0.0044 -0.0020 -0.0077
+ - param set #15 has cost=-0.027539 [o=87 t=15]
+ -- Parameters = 60.7066 30.8152 -37.4908 6.0785 -25.2633 -40.7765 0.9788 0.9699 0.9838 0.0092 -0.0007 0.0112
+ - param set #16 has cost=-0.023713 [o=86 t=18]
+ -- Parameters = 107.2420 23.3292 -32.4170 12.5935 -8.1852 44.7417 0.9594 1.0145 1.0733 0.0082 0.0140 -0.0028
+ - param set #17 has cost=-0.023208 [o=14 t=5]
+ -- Parameters = 90.9572 1.5239 -36.2139 25.5564 16.4695 1.0689 1.0030 1.0105 1.0048 -0.0015 -0.0028 0.0016
+ - param set #18 has cost=-0.024146 [o=6 t=14]
+ -- Parameters = 91.0558 4.1600 -33.9114 22.6141 6.9413 -1.5941 0.9917 0.9961 1.0114 0.0032 -0.0026 0.0005
+ - param set #19 has cost=-0.024226 [o=84 t=17]
+ -- Parameters = 113.9607 -38.9018 33.8273 -34.8139 -41.5599 -31.6493 0.9971 1.0007 0.9922 0.0007 0.0009 0.0003
+ - param set #20 has cost=-0.023411 [o=35 t=19]
+ -- Parameters = 91.1202 -14.1911 -33.9851 -7.5718 7.4552 8.5570 1.0056 1.0068 1.0018 0.0011 -0.0008 0.0003
+ - param set #21 has cost=-0.025745 [o=24 t=26]
+ -- Parameters = 96.7929 -11.4799 -27.3389 -2.8087 34.1713 26.8517 0.9939 0.9912 0.9991 -0.0044 -0.0013 0.0028
+ - param set #22 has cost=-0.032102 [o=43 t=25]
+ -- Parameters = 59.7482 35.6211 -41.7882 13.1726 -24.4694 -34.2436 0.9536 0.9776 0.9574 -0.0051 0.0119 -0.0045
+ - param set #23 has cost=-0.022848 [o=65 t=23]
+ -- Parameters = 55.6020 31.7058 36.2633 37.9380 -21.9343 38.7087 0.9612 0.9793 1.0078 0.0009 0.0062 -0.0006
+ - param set #24 has cost=-0.022057 [o=74 t=21]
+ -- Parameters = 90.2561 -14.1427 -30.4496 -5.9594 8.1515 -5.4085 1.0046 1.0048 1.0024 0.0000 0.0012 0.0000
+ - param set #25 has cost=-0.021020 [o=55 t=10]
+ -- Parameters = 90.3337 1.9814 -34.7795 28.0055 30.5132 6.6220 1.0094 1.0187 1.0404 0.0001 0.0007 0.0012
+ - param set #26 has cost=-0.020982 [o=22 t=12]
+ -- Parameters = 92.2350 2.4060 -35.6815 26.3583 27.5284 -1.5271 1.0109 1.0103 1.0049 0.0020 -0.0024 0.0002
+ - param set #27 has cost=-0.022513 [o=32 t=24]
+ -- Parameters = 56.3170 22.0671 36.2659 37.8346 -17.3304 38.3624 0.9802 0.9941 0.9893 -0.0032 -0.0007 -0.0031
+ - param set #28 has cost=-0.021362 [o=4 t=28]
+ -- Parameters = 118.3676 -27.2696 27.9475 8.9470 -22.5320 -36.4482 1.0088 0.9991 0.9968 -0.0001 0.0014 0.0207
+ - param set #29 has cost=-0.020002 [o=53 t=20]
+ -- Parameters = 64.5883 21.3793 -32.0766 35.1403 21.1180 -15.7527 0.9940 0.9957 0.9987 0.0006 -0.0017 0.0011
+ - param set #30 has cost=-0.020025 [o=3 t=22]
+ -- Parameters = 79.9313 -24.8004 -31.8045 -31.2924 30.4304 -6.7779 1.0002 0.9985 0.9919 -0.0019 0.0008 0.0005
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0519
+ --- dist(#3,#1) = 0.389
+ --- dist(#4,#1) = 0.401
+ --- dist(#5,#1) = 0.525
+ --- dist(#6,#1) = 0.469
+ --- dist(#7,#1) = 0.44
+ --- dist(#8,#1) = 0.503
+ --- dist(#9,#1) = 0.449
+ --- dist(#10,#1) = 0.435
+ --- dist(#11,#1) = 0.438
+ --- dist(#12,#1) = 0.471
+ --- dist(#13,#1) = 0.449
+ --- dist(#14,#1) = 0.428
+ --- dist(#15,#1) = 0.394
+ --- dist(#16,#1) = 0.43
+ --- dist(#17,#1) = 0.464
+ --- dist(#18,#1) = 0.465
+ --- dist(#19,#1) = 0.426
+ --- dist(#20,#1) = 0.491
+ --- dist(#21,#1) = 0.427
+ --- dist(#22,#1) = 0.455
+ --- dist(#23,#1) = 0.451
+ --- dist(#24,#1) = 0.451
+ --- dist(#25,#1) = 0.383
+ --- dist(#26,#1) = 0.411
+ --- dist(#27,#1) = 0.437
+ --- dist(#28,#1) = 0.449
+ --- dist(#29,#1) = 0.4
+ --- dist(#30,#1) = 0.403
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=40 usiz=154.5
+ - retaining old weight image
+ - using 34754 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 28464 total points stored in 58 'TOHD(17.1973)' bloks (0 duplicates)
*[#27190=-0.41914] *[#27211=-0.419351] *[#27212=-0.420232] *[#27214=-0.42065] *[#27215=-0.42118] *[#27216=-0.42179] *[#27217=-0.422054] *[#27218=-0.422077] *[#27220=-0.422188] *[#27223=-0.422202] *[#27226=-0.422292] *[#27233=-0.422596] *[#27234=-0.422672] *[#27235=-0.422784] *[#27236=-0.422897] *[#27238=-0.423] *[#27241=-0.423203] *[#27242=-0.423269] *[#27245=-0.423359] *[#27248=-0.423489] *[#27249=-0.423491] *[#27251=-0.423524] *[#27253=-0.423663] *[#27260=-0.423674] *[#27262=-0.423717] *[#27265=-0.423738] *[#27267=-0.42383] *[#27268=-0.423838] *[#27272=-0.423863] *[#27277=-0.423913] *[#27280=-0.423983] *[#27281=-0.423996] *[#27284=-0.424007] *[#27285=-0.424013] *[#27287=-0.424019] *[#27289=-0.424061] *[#27290=-0.4241] *[#27291=-0.424125] *[#27292=-0.424131] *[#27295=-0.424131] *[#27300=-0.424137] *[#27302=-0.424163] *[#27303=-0.424195] *[#27304=-0.424222] *[#27305=-0.424238] *[#27307=-0.424248] *[#27308=-0.424273] *[#27309=-0.424302] *[#27310=-0.424319]
+ - param set #1 has cost=-0.424319 [o=0 t=0]
+ -- Parameters = 85.9667 -1.4696 0.2530 -0.7177 -1.5839 0.5770 0.9961 1.0277 0.9735 -0.0147 0.0041 -0.0230
+ - param set #2 has cost=-0.422691 [o=-1 t=-1]
+ -- Parameters = 86.0262 -1.5482 0.2838 -0.8058 -1.3721 0.5924 0.9938 1.0296 0.9721 -0.0178 0.0031 -0.0195
+ - param set #3 has cost=-0.029682 [o=41 t=4]
+ -- Parameters = 54.6741 23.6412 30.9118 21.3776 -9.4409 32.6205 0.9384 0.9605 1.0335 0.0034 -0.0088 -0.0002
+ - param set #4 has cost=-0.028577 [o=68 t=1]
+ -- Parameters = 53.9136 27.7617 30.4155 12.1164 -3.6337 30.1770 0.9626 1.0003 1.0439 -0.0033 -0.0016 -0.0004
+ - param set #5 has cost=-0.028295 [o=43 t=25]
+ -- Parameters = 59.8625 35.6027 -41.6959 13.3501 -24.1515 -34.0146 0.9517 0.9626 0.9583 -0.0058 0.0123 -0.0056
+ - param set #6 has cost=-0.025969 [o=25 t=6]
+ -- Parameters = 78.3137 20.4528 -37.2483 15.8740 -7.1573 5.2136 1.0009 0.9984 1.0202 -0.0021 -0.0016 0.0125
+ - param set #7 has cost=-0.025060 [o=40 t=11]
+ -- Parameters = 91.8896 0.2242 -34.6980 18.9386 15.4311 5.4156 0.9925 0.9721 1.0015 -0.0073 -0.0008 -0.0065
+ - param set #8 has cost=-0.025081 [o=37 t=13]
+ -- Parameters = 55.5225 26.0681 -35.9785 -10.1538 -31.3126 -44.3648 1.0054 1.0310 0.9625 -0.0032 -0.0083 0.0045
+ - param set #9 has cost=-0.028956 [o=73 t=8]
+ -- Parameters = 56.0728 1.7040 35.8976 17.8097 -22.3322 30.8839 0.8913 0.9396 0.9997 0.0002 -0.0144 -0.0291
+ - param set #10 has cost=-0.025084 [o=71 t=9]
+ -- Parameters = 58.8017 27.8376 34.9828 13.3577 -1.5013 21.5499 0.9918 0.9943 1.0016 0.0098 -0.0033 -0.0002
+ - param set #11 has cost=-0.023383 [o=56 t=2]
+ -- Parameters = 82.4633 -5.0061 -34.8144 -15.5620 11.3903 -0.3813 1.0115 1.0020 1.0040 0.0023 -0.0007 -0.0000
+ - param set #12 has cost=-0.024304 [o=87 t=15]
+ -- Parameters = 60.3847 30.9327 -37.6401 6.0453 -25.5512 -41.1651 0.9788 0.9698 0.9817 0.0095 -0.0012 0.0112
+ - param set #13 has cost=-0.023983 [o=16 t=3]
+ -- Parameters = 90.9910 0.4101 -35.1943 18.9435 18.1207 2.3513 1.0304 0.9806 1.0062 0.0053 -0.0157 0.0014
+ - param set #14 has cost=-0.021126 [o=28 t=27]
+ -- Parameters = 80.5442 -6.7002 -34.0353 -9.3537 9.2080 -0.1625 1.0048 0.9890 0.9803 -0.0280 0.0136 0.0109
+ - param set #15 has cost=-0.022578 [o=24 t=26]
+ -- Parameters = 96.6607 -11.5556 -27.4103 -2.7363 34.4311 26.8934 0.9933 0.9947 1.0004 -0.0037 -0.0015 0.0031
+ - param set #16 has cost=-0.020681 [o=34 t=16]
+ -- Parameters = 95.2552 -5.5862 -34.1743 9.9962 5.9110 4.1380 1.0118 0.9530 1.0007 0.0036 -0.0027 -0.0061
+ - param set #17 has cost=-0.021274 [o=15 t=7]
+ -- Parameters = 55.3733 -33.5093 35.7853 41.2473 -42.2740 36.8229 0.9952 1.0379 0.9993 0.0005 0.0020 0.0001
+ - param set #18 has cost=-0.021141 [o=84 t=17]
+ -- Parameters = 113.9945 -38.8945 33.8606 -34.9284 -41.6897 -31.7283 0.9944 1.0009 0.9915 0.0012 0.0067 -0.0002
+ - param set #19 has cost=-0.021270 [o=6 t=14]
+ -- Parameters = 90.7151 3.8936 -33.9877 22.4410 6.7870 -1.1853 0.9927 0.9971 1.0205 0.0030 -0.0039 0.0004
+ - param set #20 has cost=-0.020280 [o=86 t=18]
+ -- Parameters = 106.5885 23.4699 -32.0372 12.7314 -7.5688 44.2667 0.9620 1.0099 1.0748 0.0097 0.0141 -0.0026
+ - param set #21 has cost=-0.021134 [o=35 t=19]
+ -- Parameters = 91.0372 -14.2350 -33.7480 -7.4184 7.8336 8.2678 1.0087 1.0090 1.0034 0.0096 -0.0022 0.0014
+ - param set #22 has cost=-0.018173 [o=14 t=5]
+ -- Parameters = 90.9530 1.5118 -36.2530 25.5732 16.4518 1.0717 1.0026 1.0116 1.0045 -0.0014 -0.0031 0.0018
+ - param set #23 has cost=-0.018660 [o=65 t=23]
+ -- Parameters = 55.3498 31.6240 36.0806 38.1167 -22.0353 38.3394 0.9449 0.9823 1.0063 0.0005 0.0055 -0.0005
+ - param set #24 has cost=-0.020135 [o=32 t=24]
+ -- Parameters = 56.6301 22.1420 36.2611 38.1986 -17.2012 38.7358 0.9715 0.9727 0.9973 -0.0034 0.0004 0.0006
+ - param set #25 has cost=-0.018921 [o=74 t=21]
+ -- Parameters = 90.2756 -14.1447 -30.4249 -5.9190 8.1138 -5.3645 1.0048 1.0046 1.0037 -0.0003 0.0013 0.0003
+ - param set #26 has cost=-0.020450 [o=4 t=28]
+ -- Parameters = 118.3205 -27.1630 28.0824 8.9770 -22.7657 -36.5031 1.0083 0.9998 0.9944 -0.0001 0.0013 0.0208
+ - param set #27 has cost=-0.018101 [o=55 t=10]
+ -- Parameters = 90.6406 2.1000 -34.6104 27.8560 30.6729 6.4731 1.0037 1.0193 1.0419 0.0001 0.0007 0.0018
+ - param set #28 has cost=-0.017690 [o=22 t=12]
+ -- Parameters = 92.1339 2.4421 -35.7201 26.3731 27.4962 -1.5480 1.0171 1.0107 1.0051 0.0015 -0.0031 0.0011
+ - param set #29 has cost=-0.017084 [o=3 t=22]
+ -- Parameters = 79.9683 -24.8274 -31.9786 -31.3220 30.3711 -6.7567 1.0004 0.9992 0.9916 -0.0021 -0.0014 0.0008
+ - param set #30 has cost=-0.017092 [o=53 t=20]
+ -- Parameters = 64.7865 20.9741 -31.7376 34.5643 21.4058 -15.3077 0.9970 1.0019 0.9833 -0.0028 0.0007 -0.0001
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0158 XXX
+ --- dist(#3,#1) = 0.446
+ --- dist(#4,#1) = 0.401
+ --- dist(#5,#1) = 0.524
+ --- dist(#6,#1) = 0.469
+ --- dist(#7,#1) = 0.434
+ --- dist(#8,#1) = 0.499
+ --- dist(#9,#1) = 0.437
+ --- dist(#10,#1) = 0.474
+ --- dist(#11,#1) = 0.443
+ --- dist(#12,#1) = 0.438
+ --- dist(#13,#1) = 0.4
+ --- dist(#14,#1) = 0.466
+ --- dist(#15,#1) = 0.428
+ --- dist(#16,#1) = 0.468
+ --- dist(#17,#1) = 0.425
+ --- dist(#18,#1) = 0.429
+ --- dist(#19,#1) = 0.43
+ --- dist(#20,#1) = 0.412
+ --- dist(#21,#1) = 0.485
+ --- dist(#22,#1) = 0.45
+ --- dist(#23,#1) = 0.383
+ --- dist(#24,#1) = 0.448
+ --- dist(#25,#1) = 0.456
+ --- dist(#26,#1) = 0.436
+ --- dist(#27,#1) = 0.45
+ --- dist(#28,#1) = 0.4
+ --- dist(#29,#1) = 0.403
+ - cast out 1 parameter set for being too close to best set
+ - Total coarse refinement net CPU time = 0.0 s; 9500 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=40 usiz=154.5
++ 1504356 voxels in -source_automask+2
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 30 cases
++ largeness ==> set -twobest 29
+ 27965 total points stored in 57 'TOHD(17.1233)' bloks (0 duplicates)
++ Zero-pad: ybot=7 ytop=1
++ Zero-pad: zbot=8 ztop=4
++ 38558 voxels [16.1%] in weight mask
++ Output dataset ./__tt_sub-05_T1w_ns_al_junk_wtal+orig.BRIK
*[#30703=-0.429271] + - cost(#1)=-0.429271 * [o=0 t=0]
++ Number of points for matching = 38558
+ -- Parameters = 85.9667 -1.4696 0.2530 -0.7177 -1.5839 0.5770 0.9961 1.0277 0.9735 -0.0147 0.0041 -0.0230
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - cost(#2)=-0.028134 [o=41 t=4]
+ -- Parameters = 54.6741 23.6412 30.9118 21.3776 -9.4409 32.6205 0.9384 0.9605 1.0335 0.0034 -0.0088 -0.0002
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - cost(#3)=-0.027469 [o=73 t=8]
+ - But it is always important to check the alignment visually to be sure.
+ -- Parameters = 56.0728 1.7040 35.8976 17.8097 -22.3322 30.8839 0.8913 0.9396 0.9997 0.0002 -0.0144 -0.0291
+ - cost(#4)=-0.027607 [o=68 t=1]
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 30.748 32.931 28.178 (index)
+ -- Parameters = 53.9136 27.7617 30.4155 12.1164 -3.6337 30.1770 0.9626 1.0003 1.0439 -0.0033 -0.0016 -0.0004
+ - cost(#5)=-0.025490 [o=43 t=25]
+ -- Parameters = 59.8625 35.6027 -41.6959 13.3501 -24.1515 -34.0146 0.9517 0.9626 0.9583 -0.0058 0.0123 -0.0056
+ - cost(#6)=-0.022790 [o=25 t=6]
+ -- Parameters = 78.3137 20.4528 -37.2483 15.8740 -7.1573 5.2136 1.0009 0.9984 1.0202 -0.0021 -0.0016 0.0125
+ - cost(#7)=-0.024403 [o=71 t=9]
+ -- Parameters = 58.8017 27.8376 34.9828 13.3577 -1.5013 21.5499 0.9918 0.9943 1.0016 0.0098 -0.0033 -0.0002
+ - cost(#8)=-0.022843 [o=37 t=13]
+ -- Parameters = 55.5225 26.0681 -35.9785 -10.1538 -31.3126 -44.3648 1.0054 1.0310 0.9625 -0.0032 -0.0083 0.0045
+ - cost(#9)=-0.021327 [o=40 t=11]
+ -- Parameters = 91.8896 0.2242 -34.6980 18.9386 15.4311 5.4156 0.9925 0.9721 1.0015 -0.0073 -0.0008 -0.0065
+ - cost(#10)=-0.021988 [o=87 t=15]
+ -- Parameters = 60.3847 30.9327 -37.6401 6.0453 -25.5512 -41.1651 0.9788 0.9698 0.9817 0.0095 -0.0012 0.0112
+ - cost(#11)=-0.020153 [o=16 t=3]
+ -- Parameters = 90.9910 0.4101 -35.1943 18.9435 18.1207 2.3513 1.0304 0.9806 1.0062 0.0053 -0.0157 0.0014
+ - cost(#12)=-0.020314 [o=56 t=2]
+ -- Parameters = 82.4633 -5.0061 -34.8144 -15.5620 11.3903 -0.3813 1.0115 1.0020 1.0040 0.0023 -0.0007 -0.0000
+ - cost(#13)=-0.018779 [o=24 t=26]
+ -- Parameters = 96.6607 -11.5556 -27.4103 -2.7363 34.4311 26.8934 0.9933 0.9947 1.0004 -0.0037 -0.0015 0.0031
+ - cost(#14)=-0.019332 [o=15 t=7]
+ -- Parameters = 55.3733 -33.5093 35.7853 41.2473 -42.2740 36.8229 0.9952 1.0379 0.9993 0.0005 0.0020 0.0001
+ - cost(#15)=-0.019253 [o=6 t=14]
+ -- Parameters = 90.7151 3.8936 -33.9877 22.4410 6.7870 -1.1853 0.9927 0.9971 1.0205 0.0030 -0.0039 0.0004
+ - cost(#16)=-0.020009 [o=84 t=17]
+ -- Parameters = 113.9945 -38.8945 33.8606 -34.9284 -41.6897 -31.7283 0.9944 1.0009 0.9915 0.0012 0.0067 -0.0002
+ - cost(#17)=-0.018500 [o=35 t=19]
+ -- Parameters = 91.0372 -14.2350 -33.7480 -7.4184 7.8336 8.2678 1.0087 1.0090 1.0034 0.0096 -0.0022 0.0014
+ - cost(#18)=-0.019523 [o=28 t=27]
+ -- Parameters = 80.5442 -6.7002 -34.0353 -9.3537 9.2080 -0.1625 1.0048 0.9890 0.9803 -0.0280 0.0136 0.0109
+ - cost(#19)=-0.018090 [o=34 t=16]
+ -- Parameters = 95.2552 -5.5862 -34.1743 9.9962 5.9110 4.1380 1.0118 0.9530 1.0007 0.0036 -0.0027 -0.0061
+ - cost(#20)=-0.019163 [o=4 t=28]
+ -- Parameters = 118.3205 -27.1630 28.0824 8.9770 -22.7657 -36.5031 1.0083 0.9998 0.9944 -0.0001 0.0013 0.0208
+ - cost(#21)=-0.017317 [o=86 t=18]
+ -- Parameters = 106.5885 23.4699 -32.0372 12.7314 -7.5688 44.2667 0.9620 1.0099 1.0748 0.0097 0.0141 -0.0026
+ - cost(#22)=-0.018773 [o=32 t=24]
+ -- Parameters = 56.6301 22.1420 36.2611 38.1986 -17.2012 38.7358 0.9715 0.9727 0.9973 -0.0034 0.0004 0.0006
+ - cost(#23)=-0.017287 [o=74 t=21]
+ -- Parameters = 90.2756 -14.1447 -30.4249 -5.9190 8.1138 -5.3645 1.0048 1.0046 1.0037 -0.0003 0.0013 0.0003
+ - cost(#24)=-0.017106 [o=65 t=23]
+ -- Parameters = 55.3498 31.6240 36.0806 38.1167 -22.0353 38.3394 0.9449 0.9823 1.0063 0.0005 0.0055 -0.0005
+ - cost(#25)=-0.015174 [o=14 t=5]
+ -- Parameters = 90.9530 1.5118 -36.2530 25.5732 16.4518 1.0717 1.0026 1.0116 1.0045 -0.0014 -0.0031 0.0018
+ - cost(#26)=-0.015719 [o=55 t=10]
+ -- Parameters = 90.6406 2.1000 -34.6104 27.8560 30.6729 6.4731 1.0037 1.0193 1.0419 0.0001 0.0007 0.0018
+ - cost(#27)=-0.015535 [o=22 t=12]
+ -- Parameters = 92.1339 2.4421 -35.7201 26.3731 27.4962 -1.5480 1.0171 1.0107 1.0051 0.0015 -0.0031 0.0011
+ source center of mass = 90.531 101.706 114.658 (index)
+ - cost(#28)=-0.014225 [o=53 t=20]
+ source-target CM = 86.108 -4.649 -2.541 (xyz)
+ estimated center of mass shifts = 86.108 -4.649 -2.541
++ shift param auto-range: 25.4..146.8 -73.0..63.7 -68.0..62.9
+ -- Parameters = 64.7865 20.9741 -31.7376 34.5643 21.4058 -15.3077 0.9970 1.0019 0.9833 -0.0028 0.0007 -0.0001
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ - cost(#29)=-0.015936 [o=3 t=22]
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ -- Parameters = 79.9683 -24.8274 -31.9786 -31.3220 30.3711 -6.7567 1.0004 0.9992 0.9916 -0.0021 -0.0014 0.0008
+ - cost(#30)=-0.088224 [o=-2 t=-2]
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ -- Parameters = 85.6010 -1.2604 -3.7335 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 30 cases
+ Range param#1 [x-shift] = 76.107513 .. 96.107513 center = 86.107513
*[#30770=-0.429348] *[#30771=-0.429551] *[#30783=-0.429666] *[#30800=-0.429704] *[#30801=-0.429856] *[#30807=-0.429881] *[#30808=-0.429916] *[#30809=-0.429962] *[#30810=-0.430034] *[#30811=-0.430056] *[#30812=-0.430091] *[#30815=-0.430105] *[#30818=-0.430129] *[#30821=-0.43013] *[#30823=-0.430141] *[#30828=-0.430146] *[#30831=-0.430149]
+ - cost(#1)=-0.430149 * [o=0 t=0]
+ -- Parameters = 85.9607 -1.3934 0.2466 -0.6693 -1.6296 0.5782 0.9960 1.0218 0.9743 -0.0148 0.0032 -0.0233
+ Range param#2 [y-shift] = -14.649185 .. 5.350815 center = -4.649185
+ Range param#3 [z-shift] = -12.541183 .. 7.458817 center = -2.541183
+ - cost(#2)=-0.028486 [o=41 t=4]
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ -- Parameters = 54.6044 23.6193 30.8180 21.5258 -9.3082 32.4110 0.9374 0.9601 1.0365 0.0091 -0.0084 0.0007
+ - cost(#3)=-0.029051 [o=73 t=8]
+ -- Parameters = 55.9456 1.7821 35.5120 17.5176 -22.3301 30.9390 0.8914 0.9039 1.0021 -0.0003 -0.0146 -0.0295
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.107513 .. 126.107513 center = 86.107513
+ Range param#2 [y-shift] = -44.649185 .. 35.350815 center = -4.649185
+ Range param#3 [z-shift] = -42.541183 .. 37.458817 center = -2.541183
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000089
+ - cost(#4)=-0.028419 [o=68 t=1]
+ -- Parameters = 54.1551 27.8815 30.2462 12.0759 -3.1454 30.1718 0.9629 1.0033 1.0446 -0.0032 -0.0022 -0.0061
++ Final parameter search ranges:
+ x-shift = 46.108 .. 126.108
+ y-shift = -44.649 .. 35.351
+ z-shift = -42.541 .. 37.459
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1504356 [out of 11534336] voxels
+ - cost(#5)=-0.025994 [o=43 t=25]
+ base mask has 52016 [out of 239616] voxels
+ -- Parameters = 59.6695 35.8149 -41.7336 13.1393 -24.0445 -34.2011 0.9492 0.9663 0.9562 -0.0076 0.0130 -0.0050
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
+ - cost(#6)=-0.022851 [o=25 t=6]
+ -- Parameters = 78.2638 20.4058 -37.2300 15.8427 -7.1562 5.2811 1.0010 0.9994 1.0204 -0.0024 -0.0022 0.0123
+ - cost(#7)=-0.024807 [o=71 t=9]
+ -- Parameters = 58.8138 27.9217 34.8916 13.3030 -1.5897 21.4949 0.9928 0.9985 1.0066 0.0095 -0.0024 -0.0077
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - cost(#8)=-0.022862 [o=37 t=13]
+ -- Parameters = 55.5295 26.0430 -35.9889 -10.1778 -31.3033 -44.3984 1.0052 1.0315 0.9629 -0.0033 -0.0084 0.0053
+ - cost(#9)=-0.021391 [o=40 t=11]
+ -- Parameters = 91.8834 0.2227 -34.6768 18.9290 15.4197 5.4098 0.9936 0.9718 1.0015 -0.0066 -0.0017 -0.0054
+ - cost(#10)=-0.022081 [o=87 t=15]
+ -- Parameters = 60.4035 30.9708 -37.6736 6.1280 -25.5166 -41.2092 0.9786 0.9705 0.9828 0.0097 0.0058 0.0111
+ - cost(#11)=-0.020514 [o=16 t=3]
+ -- Parameters = 91.0448 0.2316 -35.1882 18.8965 18.2647 2.3267 1.0384 0.9846 1.0036 0.0054 -0.0160 0.0010
+ - cost(#12)=-0.020716 [o=56 t=2]
+ -- Parameters = 82.4689 -5.2902 -34.8050 -15.8919 11.3659 -0.2331 1.0085 1.0077 1.0044 0.0040 -0.0008 0.0011
+ - cost(#13)=-0.020284 [o=24 t=26]
+ -- Parameters = 96.6392 -11.1700 -27.0952 -1.7988 34.5260 29.1374 0.9931 0.9995 1.0042 -0.0059 0.0020 0.0045
+ - cost(#14)=-0.019428 [o=15 t=7]
+ -- Parameters = 55.3311 -33.4960 35.7614 41.2766 -42.3738 36.7771 0.9943 1.0386 1.0019 0.0009 0.0001 0.0003
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ - cost(#15)=-0.019521 [o=6 t=14]
+ -- Parameters = 90.4037 3.9104 -34.0087 22.6799 6.6714 -1.0515 0.9934 0.9947 1.0231 0.0053 -0.0060 0.0003
+ - cost(#16)=-0.020110 [o=84 t=17]
+ -- Parameters = 114.0290 -38.8908 33.8417 -34.8871 -41.7556 -31.6798 0.9934 1.0023 0.9913 0.0084 0.0068 -0.0002
+ - cost(#17)=-0.018857 [o=35 t=19]
+ -- Parameters = 90.8830 -14.2864 -33.8525 -7.3527 7.6722 8.3444 1.0122 1.0055 1.0064 0.0101 -0.0031 0.0084
+ - cost(#18)=-0.019768 [o=28 t=27]
+ -- Parameters = 80.5799 -6.7661 -34.0352 -9.3938 9.1395 -0.2118 1.0049 0.9890 0.9817 -0.0287 0.0206 0.0107
+ - cost(#19)=-0.018103 [o=34 t=16]
+ -- Parameters = 95.2571 -5.5983 -34.1847 9.9958 5.8965 4.1410 1.0118 0.9529 1.0005 0.0036 -0.0027 -0.0069
+ - cost(#20)=-0.019210 [o=4 t=28]
+ -- Parameters = 118.3248 -27.1443 28.0940 9.0053 -22.7830 -36.4849 1.0086 0.9997 0.9940 -0.0001 0.0021 0.0208
+ - cost(#21)=-0.017583 [o=86 t=18]
+ -- Parameters = 106.5466 23.7121 -32.1022 12.7656 -7.7097 44.3291 0.9626 1.0037 1.0755 0.0128 0.0135 -0.0035
+ !source mask fill: ubot=32 usiz=159.5
+ - cost(#22)=-0.018841 [o=32 t=24]
+ -- Parameters = 56.6396 22.1347 36.1852 38.1732 -17.1814 38.7621 0.9711 0.9740 0.9994 -0.0037 0.0008 0.0013
+ - cost(#23)=-0.020786 [o=74 t=21]
+ -- Parameters = 86.9450 -14.6949 -30.5789 -6.4642 7.7437 -4.1465 1.0037 1.0047 1.0035 0.0062 0.0009 -0.0007
+ - cost(#24)=-0.019042 [o=65 t=23]
+ -- Parameters = 52.3837 28.9634 36.1377 38.8351 -21.3466 38.3499 0.9451 0.9805 1.0045 -0.0000 0.0056 0.0003
+ - cost(#25)=-0.018163 [o=14 t=5]
+ -- Parameters = 91.1204 3.3952 -33.0953 25.0904 15.1428 0.9175 1.0029 1.0081 1.0031 0.0014 -0.0044 0.0026
+ - cost(#26)=-0.015970 [o=55 t=10]
+ -- Parameters = 90.6806 2.1301 -34.5359 27.7506 30.8155 6.5583 1.0023 1.0176 1.0419 -0.0005 0.0007 0.0092
+ - copying weight image
+ - using 38558 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ - cost(#27)=-0.015880 [o=22 t=12]
+ -- Parameters = 92.1517 2.3663 -35.8051 26.3704 27.2222 -1.6678 1.0216 1.0110 1.0059 0.0018 -0.0032 0.0007
+ 31408 total points stored in 61 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
+ - cost(#28)=-0.015045 [o=53 t=20]
+ -- Parameters = 65.1391 20.4747 -31.7443 33.4650 20.7445 -15.1351 0.9944 1.0068 0.9836 -0.0032 0.0002 -0.0045
+ - cost(#29)=-0.016026 [o=3 t=22]
+ -- Parameters = 79.8935 -24.8377 -32.0224 -31.3134 30.3895 -6.7103 0.9999 0.9999 0.9908 -0.0029 -0.0024 -0.0001
+ - cost(#30)=-0.422963 [o=-2 t=-2]
+ -- Parameters = 85.8052 -1.6324 0.1445 -0.2365 -1.3612 0.8297 0.9960 1.0293 0.9809 0.0001 -0.0034 -0.0157
+ - case #1 [o=0 t=0] is now the best
+ - Initial cost = -0.430149
+ - Initial fine Parameters = 85.9607 -1.3934 0.2466 -0.6693 -1.6296 0.5782 0.9960 1.0218 0.9743 -0.0148 0.0032 -0.0233
*[#33874=-0.430154] *[#33888=-0.430189] *[#33889=-0.430199] *[#33892=-0.430211] *[#33897=-0.430238] *[#33898=-0.430245] *[#33899=-0.430246] *[#33900=-0.430256] *[#33901=-0.430277] *[#33902=-0.430302] *[#33903=-0.430303] *[#33905=-0.430304] *[#33908=-0.430336] *[#33909=-0.430371] *[#33910=-0.430382] *[#33911=-0.430382] *[#33912=-0.430387] *[#33913=-0.430403] *[#33914=-0.430423] *[#33915=-0.430429] *[#33916=-0.430431] *[#33918=-0.430456] *[#33919=-0.430471] *[#33922=-0.430494] *[#33923=-0.430511] *[#33924=-0.430539] *[#33925=-0.430542] *[#33927=-0.430547] *[#33928=-0.430583] *[#33931=-0.430603] *[#33932=-0.430617] *[#33933=-0.430644] *[#33934=-0.430672] *[#33935=-0.430708] *[#33936=-0.430812] *[#33941=-0.430876] *[#33942=-0.430965] *[#33943=-0.431059] *[#33944=-0.431174] *[#33945=-0.431207] *[#33946=-0.43129] *[#33949=-0.431454] *[#33950=-0.431537] *[#33955=-0.43159] *[#33956=-0.431635] *[#33961=-0.431668] *[#33962=-0.431685] *[#33963=-0.431716] *[#33964=-0.431761] *[#33967=-0.431814] *[#33968=-0.431867] *[#33971=-0.431943] *[#33974=-0.43198] *[#33976=-0.432046] *[#33979=-0.432088] *[#33980=-0.432104] *[#33983=-0.432173] *[#33984=-0.432203] *[#33985=-0.432215] *[#33986=-0.432219] *[#33987=-0.432229] *[#33990=-0.432263] *[#33991=-0.43228] *[#33992=-0.432291] *[#33993=-0.432293] *[#33996=-0.432307] *[#33997=-0.432314] *[#34000=-0.432331] *[#34001=-0.432343] *[#34002=-0.432357] *[#34003=-0.432363] *[#34008=-0.432366] *[#34009=-0.432367] *[#34010=-0.432408] *[#34011=-0.432428] *[#34012=-0.43244] *[#34013=-0.432446] *[#34014=-0.432455] *[#34015=-0.432462] *[#34016=-0.432465] *[#34017=-0.432496] *[#34018=-0.432505] *[#34019=-0.43251] *[#34020=-0.432524] *[#34025=-0.432536] *[#34030=-0.432558] *[#34033=-0.432561] *[#34038=-0.432566] *[#34039=-0.43257] *[#34040=-0.43257] *[#34042=-0.432578] *[#34043=-0.432581] *[#34044=-0.432586] *[#34045=-0.432598] *[#34048=-0.432606] *[#34051=-0.432621] *[#34052=-0.432631] *[#34055=-0.432634] *[#34057=-0.43264] *[#34060=-0.432655] *[#34061=-0.432673] *[#34062=-0.432686] *[#34064=-0.432687] *[#34067=-0.432689] *[#34071=-0.432713] *[#34078=-0.43272] *[#34081=-0.432725] *[#34087=-0.432727] *[#34088=-0.432738] *[#34089=-0.432742] *[#34090=-0.432747] *[#34091=-0.432752] *[#34092=-0.432753] *[#34096=-0.43277] *[#34097=-0.432773] *[#34098=-0.432774] *[#34100=-0.432776] *[#34101=-0.432784] *[#34102=-0.432787] *[#34103=-0.432797] *[#34106=-0.432799] *[#34113=-0.432801] *[#34118=-0.432809] *[#34121=-0.43281] *[#34127=-0.432812] *[#34130=-0.432813] *[#34133=-0.432815] *[#34136=-0.43282] *[#34141=-0.43282] *[#34145=-0.432823] *[#34146=-0.432825] *[#34149=-0.432825] *[#34150=-0.432826] *[#34151=-0.432827] *[#34152=-0.432827] *[#34153=-0.432829] *[#34156=-0.432829] *[#34157=-0.432832] *[#34158=-0.432833] *[#34159=-0.432834] *[#34160=-0.432836] *[#34163=-0.432837] *[#34166=-0.432838] *[#34167=-0.432839] *[#34171=-0.432839] *[#34172=-0.432839] *[#34173=-0.43284] *[#34174=-0.432841] *[#34177=-0.432841] *[#34188=-0.432841] *[#34190=-0.432841] *[#34191=-0.432841] *[#34199=-0.432842] *[#34206=-0.432842] *[#34210=-0.432842] *[#34211=-0.432842] *[#34225=-0.432842] *[#34231=-0.432842] *[#34253=-0.432842]
+ - Finalish cost = -0.432842 ; 427 funcs
+ - ini Finalish Parameters = 85.8572 -1.2949 0.3524 -0.4250 -2.2043 0.5585 0.9968 1.0223 0.9752 -0.0070 0.0035 -0.0388
*[#34455=-0.432843] *[#34488=-0.432843] *[#34501=-0.432843] *[#34524=-0.432843]
+ - Final cost = -0.432843 ; 279 funcs
+ Final fine fit Parameters:
x-shift= 85.8559 y-shift= -1.2943 z-shift= 0.3525 ... enorm= 85.8663 mm
z-angle= -0.4220 x-angle= -2.2064 y-angle= 0.5566 ... total= 2.3163 deg
x-scale= 0.9968 y-scale= 1.0224 z-scale= 0.9752 ... vol3D= 0.9938 = base bigger than source
y/x-shear= -0.0070 z/x-shear= 0.0036 z/y-shear= -0.0388
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 85.8559 -1.2943 0.3525 -0.4220 -2.2064 0.5566 0.9968 1.0224 0.9752 -0.0070 0.0036 -0.0388
++ Unloading unneeded data
++ Output dataset ./sub-04_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-04_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 16.7
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/__tt_sub-04_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/sub-04_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-04_T1w_ns+orig sub-04_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-04_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-04_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-04_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-04_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
Performing center alignment with @Align_Centers
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-04_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-04_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-04_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-04_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-04_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-04_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-04_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Center distance of 0.000000 mm
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o.-o-.-+.+o.-++.+.+o+-++oo-+--+o..+.$+.+.o--+-- + - best 88 costs found:
o= 0 v=-0.060670: 86.11 -4.65 -2.54 0.00 0.00 0.00 [grid]
o= 1 v=-0.026718: 94.62 -33.66 28.83 -12.98 -30.57 -5.92 [rand]
o= 2 v=-0.023077: 58.09 -0.53 29.37 18.18 -25.37 33.48 [rand]
o= 3 v=-0.021513: 81.97 -25.22 27.51 19.78 -42.00 3.76 [rand]
o= 4 v=-0.021368: 58.09 -8.77 29.37 18.18 -25.37 33.48 [rand]
o= 5 v=-0.021254: 59.44 -11.32 -29.21 30.00 30.00 7.50 [grid]
o= 6 v=-0.021248: 92.52 -38.59 34.03 -19.28 -27.13 -15.13 [rand]
o= 7 v=-0.020676: 90.24 -25.22 27.51 -19.78 -42.00 -3.76 [rand]
o= 8 v=-0.020155: 65.86 22.63 -37.03 37.90 5.03 3.08 [rand]
o= 9 v=-0.019960: 59.33 -19.27 28.81 16.09 -31.75 34.71 [rand]
o=10 v=-0.019670: 57.09 22.49 31.07 7.44 -27.42 25.63 [rand]
o=11 v=-0.019615: 77.59 -33.66 28.83 12.98 -30.57 5.92 [rand]
o=12 v=-0.019278: 59.44 -31.32 24.13 7.50 -30.00 30.00 [grid]
o=13 v=-0.019217: 99.44 -28.90 28.25 -6.84 -32.76 -20.90 [rand]
o=14 v=-0.018971: 71.17 -28.74 27.38 19.13 -33.53 22.48 [rand]
o=15 v=-0.018653: 112.88 -19.27 -33.89 -16.09 31.75 -34.71 [rand]
o=16 v=-0.018563: 60.74 22.60 34.97 41.23 -14.90 41.72 [rand]
o=17 v=-0.018085: 53.08 0.37 34.27 11.98 -4.28 19.20 [rand]
o=18 v=-0.017828: 94.77 -28.81 34.03 -27.41 -42.10 -24.35 [rand]
o=19 v=-0.017390: 118.32 24.40 33.60 -40.41 -19.09 -40.75 [rand]
o=20 v=-0.017263: 79.44 -11.32 -29.21 -7.50 7.50 -7.50 [grid]
o=21 v=-0.017108: 90.24 -25.22 27.51 -19.78 -42.00 3.76 [rand]
o=22 v=-0.017049: 59.18 -32.79 35.40 15.05 -4.71 12.78 [rand]
o=23 v=-0.016979: 83.81 -11.83 29.15 -10.97 -18.37 5.04 [rand]
o=24 v=-0.016905: 115.74 -36.49 -26.15 4.69 15.37 -11.33 [rand]
o=25 v=-0.016644: 111.48 22.60 34.97 -41.23 -14.90 -41.72 [rand]
o=26 v=-0.016333: 59.33 9.98 28.81 -16.09 -31.75 -34.71 [rand]
o=27 v=-0.016166: 69.31 -23.11 29.82 -10.10 -16.32 -39.31 [rand]
o=28 v=-0.016154: 114.13 -0.53 29.37 -18.18 -25.37 -33.48 [rand]
o=29 v=-0.016098: 102.32 9.46 32.59 -15.74 -34.09 -7.78 [rand]
o=30 v=-0.016056: 72.77 -28.90 28.25 6.84 -32.76 20.90 [rand]
o=31 v=-0.015977: 66.99 18.77 -35.84 37.88 13.88 18.42 [rand]
o=32 v=-0.015904: 99.27 33.24 34.65 -14.03 -8.39 -25.20 [rand]
o=33 v=-0.015768: 101.05 -28.74 27.38 -19.13 -33.53 -22.48 [rand]
o=34 v=-0.015315: 65.86 22.63 31.94 -37.90 -5.03 -3.08 [rand]
o=35 v=-0.015278: 83.66 -17.56 -10.24 14.12 31.85 -31.94 [rand]
o=36 v=-0.015246: 108.57 -33.80 -32.57 27.43 -11.56 -19.13 [rand]
o=37 v=-0.015205: 92.77 -11.32 24.13 -30.00 -30.00 30.00 [grid]
o=38 v=-0.015197: 115.73 29.96 34.27 24.31 -11.89 -2.84 [rand]
o=39 v=-0.015113: 59.44 -31.32 24.13 30.00 -30.00 30.00 [grid]
o=40 v=-0.015025: 59.33 -19.27 -33.89 16.09 31.75 34.71 [rand]
o=41 v=-0.014986: 79.69 -38.59 34.03 19.28 -27.13 15.13 [rand]
o=42 v=-0.014871: 58.98 14.22 25.20 5.55 -17.35 23.42 [rand]
o=43 v=-0.014690: 81.97 -25.22 27.51 19.78 -42.00 -3.76 [rand]
o=44 v=-0.014684: 92.77 -31.32 -29.21 -30.00 30.00 -30.00 [grid]
o=45 v=-0.014658: 112.88 -19.27 28.81 16.09 -31.75 -34.71 [rand]
o=46 v=-0.014524: 100.11 -20.18 33.14 -14.61 -39.00 -28.71 [rand]
o=47 v=-0.014406: 92.77 -11.32 24.13 -30.00 -30.00 -7.50 [grid]
o=48 v=-0.014396: 102.90 13.81 29.82 -10.10 -16.32 39.31 [rand]
o=49 v=-0.014379: 71.11 -41.60 -20.79 34.16 13.44 -39.30 [rand]
o=50 v=-0.014230: 69.89 -18.76 32.59 15.74 -34.09 7.78 [rand]
o=51 v=-0.014042: 92.77 22.02 24.13 7.50 -7.50 30.00 [grid]
o=52 v=-0.014015: 59.44 -31.32 24.13 7.50 -7.50 30.00 [grid]
o=53 v=-0.013997: 65.13 -33.03 29.71 3.65 -28.33 14.78 [rand]
o=54 v=-0.013863: 115.12 22.49 31.07 -7.44 -27.42 25.63 [rand]
o=55 v=-0.013735: 101.40 -25.54 27.85 -14.56 -29.40 -15.92 [rand]
o=56 v=-0.013712: 111.48 22.60 -40.05 -41.23 14.90 -41.72 [rand]
o=57 v=-0.013686: 57.12 -19.91 -23.82 32.35 9.77 6.77 [rand]
o=58 v=-0.013440: 115.09 -19.91 -23.82 -32.35 9.77 -6.77 [rand]
o=59 v=-0.013409: 57.09 22.49 31.07 -7.44 -27.42 -25.63 [rand]
o=60 v=-0.013385: 114.13 -8.77 29.37 -18.18 -25.37 -33.48 [rand]
o=61 v=-0.013383: 105.22 18.77 -35.84 -37.88 13.88 -18.42 [rand]
o=62 v=-0.013087: 112.88 9.98 28.81 -16.09 -31.75 -34.71 [rand]
o=63 v=-0.012987: 88.40 -11.83 -34.23 -10.97 18.37 -5.04 [rand]
o=64 v=-0.012901: 92.77 22.02 24.13 -30.00 -30.00 7.50 [grid]
o=65 v=-0.012760: 112.77 -31.32 -29.21 30.00 -7.50 -30.00 [grid]
o=66 v=-0.012645: 77.59 -33.66 28.83 -12.98 -30.57 5.92 [rand]
o=67 v=-0.012533: 55.08 -24.24 19.06 24.25 -27.21 33.81 [rand]
o=68 v=-0.012466: 61.14 24.40 32.91 17.69 -14.58 34.92 [rand]
o=69 v=-0.012457: 79.69 -38.59 34.03 -19.28 -27.13 15.13 [rand]
o=70 v=-0.012446: 59.44 2.02 24.13 -7.50 -7.50 30.00 [grid]
o=71 v=-0.012373: 79.44 -11.32 24.13 -7.50 -30.00 30.00 [grid]
o=72 v=-0.012326: 59.44 2.02 24.13 7.50 -30.00 30.00 [grid]
o=73 v=-0.012300: 105.22 -28.06 -35.84 37.88 -13.88 -18.42 [rand]
o=74 v=-0.012296: 120.96 -40.33 11.71 40.22 24.26 41.52 [rand]
o=75 v=-0.012210: 65.86 22.63 31.94 -37.90 -5.03 3.08 [rand]
o=76 v=-0.012150: 59.30 -14.86 27.29 31.57 -38.60 41.85 [rand]
o=77 v=-0.012087: 54.13 -34.84 -25.49 14.28 39.88 18.16 [rand]
o=78 v=-0.012058: 59.44 -11.32 -29.21 7.50 7.50 7.50 [grid]
o=79 v=-0.011957: 69.31 13.81 29.82 10.10 -16.32 -39.31 [rand]
o=80 v=-0.011927: 119.14 -9.67 -39.36 -11.98 -4.28 -19.20 [rand]
o=81 v=-0.011908: 64.11 -34.08 -24.80 28.45 9.40 -40.53 [rand]
o=82 v=-0.011892: 60.69 -12.07 24.57 5.61 -11.01 21.77 [rand]
o=83 v=-0.011852: 112.77 -31.32 -29.21 -7.50 7.50 -30.00 [grid]
o=84 v=-0.011850: 92.77 -11.32 -29.21 -30.00 7.50 -7.50 [grid]
o=85 v=-0.011846: 112.77 -11.32 -9.21 30.00 -30.00 -30.00 [grid]
o=86 v=-0.011813: 83.81 -11.83 -34.23 -10.97 18.37 -5.04 [rand]
o=87 v=-0.011778: 83.99 -11.16 27.56 6.85 -38.25 36.13 [rand]
Resampling ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
+ - A little optimization:*[#16323=-0.125016] *[#16324=-0.131339] *[#16333=-0.15837] *[#16335=-0.166736] *[#16336=-0.176502] *[#16338=-0.190424] *[#16341=-0.191658] *[#16342=-0.193783] *[#16343=-0.193831] *[#16345=-0.200328] *[#16348=-0.201383] *[#16349=-0.202721] *[#16353=-0.202919] *[#16354=-0.203104] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.203104: 86.34 -2.35 0.18 -0.88 0.60 -0.18 [grid] [f=41]
o= 1 v=-0.033073: 92.46 -37.63 28.19 -8.34 -37.02 -7.31 [rand] [f=61]
o= 2 v=-0.030296: 56.22 0.66 28.77 19.35 -21.14 33.59 [rand] [f=61]
o= 3 v=-0.025514: 82.08 -25.67 28.56 19.34 -42.49 3.02 [rand] [f=69]
o= 4 v=-0.024501: 58.24 -3.56 31.21 18.24 -24.80 33.25 [rand] [f=34]
o= 5 v=-0.034916: 53.16 -11.26 -28.24 32.92 31.81 8.04 [grid] [f=52]
o= 6 v=-0.024052: 92.85 -38.28 32.69 -18.39 -28.47 -13.52 [rand] [f=48]
o= 7 v=-0.028171: 91.76 -24.65 29.96 -20.46 -44.41 -2.56 [rand] [f=79]
o= 8 v=-0.030284: 67.39 24.19 -35.81 44.59 9.16 7.06 [rand] [f=56]
o= 9 v=-0.027550: 58.79 -19.30 27.00 17.84 -24.36 33.63 [rand] [f=49]
o=10 v=-0.029677: 56.28 21.90 27.83 9.69 -20.06 23.12 [rand] [f=46]
o=11 v=-0.026884: 79.21 -33.44 34.86 13.31 -30.01 4.23 [rand] [f=67]
o=12 v=-0.020925: 60.31 -31.72 23.99 7.35 -30.42 30.03 [grid] [f=44]
o=13 v=-0.028393: 98.55 -31.03 31.94 -8.25 -31.30 -19.97 [rand] [f=50]
o=14 v=-0.024557: 71.33 -32.50 28.22 19.37 -32.09 21.34 [rand] [f=64]
o=15 v=-0.020768: 116.90 -19.04 -33.41 -16.12 31.59 -34.73 [rand] [f=34]
o=16 v=-0.027824: 67.35 23.81 36.18 40.85 -19.20 41.65 [rand] [f=64]
o=17 v=-0.024526: 53.28 -0.98 36.35 10.51 -3.89 19.66 [rand] [f=64]
o=18 v=-0.023677: 94.80 -29.34 36.77 -27.27 -41.87 -24.14 [rand] [f=32]
o=19 v=-0.023842: 115.89 23.29 34.46 -41.48 -21.02 -42.87 [rand] [f=59]
o=20 v=-0.024819: 77.69 -10.92 -30.56 -9.44 4.63 -8.10 [grid] [f=75]
o=21 v=-0.026518: 89.74 -23.90 31.03 -16.10 -44.71 3.97 [rand] [f=47]
o=22 v=-0.025053: 50.98 -33.03 36.73 17.95 -1.77 13.79 [rand] [f=65]
o=23 v=-0.037105: 83.54 -16.07 37.30 -14.47 -15.55 7.56 [rand] [f=58]
o=24 v=-0.018093: 115.74 -36.55 -25.53 4.78 15.39 -11.68 [rand] [f=35]
o=25 v=-0.024320: 112.82 22.50 36.38 -42.61 -21.77 -43.41 [rand] [f=75]
o=26 v=-0.025164: 62.28 9.02 30.47 -17.94 -35.19 -33.58 [rand] [f=56]
o=27 v=-0.020572: 70.17 -18.32 27.47 -12.76 -18.34 -39.87 [rand] [f=56]
o=28 v=-0.028007: 116.16 0.70 36.77 -18.78 -25.03 -38.74 [rand] [f=72]
o=29 v=-0.024620: 97.84 8.79 32.63 -16.08 -34.24 -8.36 [rand] [f=44]
o=30 v=-0.028219: 76.97 -29.66 29.70 7.93 -31.54 20.73 [rand] [f=57]
o=31 v=-0.025972: 66.67 23.15 -39.34 37.93 12.68 16.00 [rand] [f=57]
o=32 v=-0.026795: 99.98 30.36 33.24 -18.08 -11.17 -13.96 [rand] [f=54]
o=33 v=-0.025714: 101.43 -28.19 32.48 -17.86 -30.69 -22.23 [rand] [f=48]
o=34 v=-0.027036: 65.34 25.97 27.90 -40.82 -7.90 -4.54 [rand] [f=48]
o=35 v=-0.026465: 80.50 -18.96 -10.49 15.04 31.08 -25.51 [rand] [f=61]
o=36 v=-0.019541: 104.63 -33.76 -32.47 27.72 -11.38 -19.25 [rand] [f=41]
o=37 v=-0.025550: 93.08 -18.21 19.13 -26.58 -31.76 30.59 [grid] [f=63]
o=38 v=-0.017509: 115.39 26.84 34.82 25.84 -11.78 -2.90 [rand] [f=41]
o=39 v=-0.018477: 59.87 -35.50 24.54 29.85 -29.02 30.27 [grid] [f=51]
o=40 v=-0.017353: 60.71 -18.86 -34.34 15.69 31.62 33.94 [rand] [f=47]
o=41 v=-0.035021: 82.40 -35.67 30.47 12.88 -34.86 6.71 [rand] [f=57]
o=42 v=-0.025034: 55.01 14.95 25.72 5.14 -18.10 24.82 [rand] [f=39]
o=43 v=-0.022304: 82.28 -24.06 31.84 19.12 -41.86 -2.97 [rand] [f=52]
o=44 v=-0.017218: 93.92 -31.63 -29.41 -29.85 29.89 -30.35 [grid] [f=49]
o=45 v=-0.020587: 117.23 -19.62 27.93 15.66 -30.52 -34.64 [rand] [f=44]
o=46 v=-0.028545: 101.87 -20.44 37.24 -14.53 -39.36 -23.89 [rand] [f=57]
o=47 v=-0.034070: 93.61 -15.68 34.16 -31.52 -24.23 -10.88 [grid] [f=74]
o=48 v=-0.021859: 103.37 13.38 27.28 -10.64 -18.96 44.58 [rand] [f=53]
o=49 v=-0.020068: 69.18 -42.12 -16.94 30.56 25.76 -42.60 [rand] [f=44]
o=50 v=-0.032635: 82.55 -22.48 35.84 19.85 -26.86 4.41 [rand] [f=56]
o=51 v=-0.017117: 92.73 20.70 25.13 6.11 -6.78 30.24 [grid] [f=39]
o=52 v=-0.021487: 54.97 -31.57 25.54 7.93 -6.57 31.44 [grid] [f=63]
o=53 v=-0.018796: 64.51 -33.47 29.65 3.38 -27.60 20.09 [rand] [f=47]
o=54 v=-0.018544: 115.07 23.47 30.42 -6.47 -29.33 29.86 [rand] [f=51]
o=55 v=-0.023688: 99.92 -26.80 31.99 -12.95 -29.39 -15.84 [rand] [f=69]
o=56 v=-0.024072: 115.45 21.67 -41.71 -40.13 14.23 -43.04 [rand] [f=56]
o=57 v=-0.022200: 59.24 -18.22 -26.98 31.84 11.41 9.14 [rand] [f=55]
o=58 v=-0.024556: 113.28 -21.66 -24.34 -31.21 15.46 -4.85 [rand] [f=42]
o=59 v=-0.021205: 56.30 17.60 31.68 -5.63 -27.16 -25.01 [rand] [f=50]
o=60 v=-0.026916: 113.26 -6.34 36.36 -15.70 -27.48 -37.26 [rand] [f=60]
o=61 v=-0.028783: 105.81 18.40 -33.78 -44.15 7.61 -8.72 [rand] [f=48]
o=62 v=-0.018460: 114.23 10.51 29.46 -16.27 -31.87 -29.31 [rand] [f=51]
o=63 v=-0.017111: 88.18 -12.46 -30.71 -12.01 18.23 -5.76 [rand] [f=48]
o=64 v=-0.018821: 92.63 20.89 24.67 -31.17 -29.72 10.91 [grid] [f=72]
o=65 v=-0.017884: 113.18 -32.64 -28.55 30.78 -6.51 -24.27 [grid] [f=50]
o=66 v=-0.028975: 86.06 -38.53 30.93 -10.17 -29.79 7.34 [rand] [f=59]
o=67 v=-0.020730: 56.32 -24.32 23.57 23.49 -27.26 34.56 [rand] [f=68]
o=68 v=-0.031381: 54.86 21.49 29.23 12.11 -22.68 28.58 [rand] [f=82]
o=69 v=-0.024127: 83.60 -39.37 34.04 -18.42 -27.41 14.66 [rand] [f=52]
o=70 v=-0.032829: 58.63 -0.27 30.52 -3.17 -6.71 27.29 [grid] [f=60]
o=71 v=-0.023056: 83.02 -10.10 22.00 -9.82 -31.15 27.54 [grid] [f=62]
o=72 v=-0.031459: 57.93 6.49 35.13 12.48 -26.35 28.56 [grid] [f=68]
o=73 v=-0.016145: 105.60 -27.81 -34.69 37.76 -14.07 -14.56 [rand] [f=83]
o=74 v=-0.026345: 122.63 -43.27 6.99 41.20 27.19 40.93 [rand] [f=53]
o=75 v=-0.026289: 65.69 22.99 27.99 -41.86 -8.79 -3.19 [rand] [f=56]
o=76 v=-0.027993: 56.50 -14.76 34.29 25.64 -39.30 41.77 [rand] [f=66]
o=77 v=-0.017703: 53.88 -35.04 -27.58 13.50 41.59 22.98 [rand] [f=45]
o=78 v=-0.016557: 55.61 -10.79 -28.40 6.79 7.40 7.89 [grid] [f=44]
o=79 v=-0.022606: 68.64 13.72 34.97 8.67 -16.73 -40.38 [rand] [f=59]
o=80 v=-0.016017: 118.58 -8.86 -37.80 -6.57 -2.24 -17.85 [rand] [f=50]
o=81 v=-0.018811: 64.69 -35.16 -25.17 27.23 12.57 -42.30 [rand] [f=42]
o=82 v=-0.031736: 59.31 -8.60 35.56 -0.51 -11.47 20.49 [rand] [f=67]
o=83 v=-0.020152: 111.07 -25.99 -33.09 -8.30 7.66 -29.73 [grid] [f=42]
o=84 v=-0.024157: 92.04 -11.72 -33.28 -26.41 5.95 -5.99 [grid] [f=76]
o=85 v=-0.018456: 113.12 -13.78 -9.80 26.48 -30.15 -32.94 [grid] [f=67]
o=86 v=-0.021813: 84.52 -7.30 -34.16 -11.65 10.17 -7.86 [rand] [f=76]
o=87 v=-0.018415: 83.51 -11.26 25.40 4.08 -41.12 32.16 [rand] [f=78]
+ - saving # 0 for use with twobest
+ - saving #23 for use with twobest
+ - saving #41 for use with twobest
+ - saving # 5 for use with twobest
+ - saving #47 for use with twobest
+ - saving # 1 for use with twobest
+ - saving #70 for use with twobest
+ - saving #50 for use with twobest
+ - saving #82 for use with twobest
+ - saving #72 for use with twobest
+ - saving #68 for use with twobest
+ - saving # 2 for use with twobest
+ - saving # 8 for use with twobest
+ - saving #10 for use with twobest
+ - saving #66 for use with twobest
+ - saving #61 for use with twobest
+ - saving #46 for use with twobest
+ - saving #13 for use with twobest
+ - saving #30 for use with twobest
+ - saving # 7 for use with twobest
+ - saving #28 for use with twobest
+ - saving #76 for use with twobest
+ - saving #16 for use with twobest
+ - saving # 9 for use with twobest
+ - saving #34 for use with twobest
+ - saving #60 for use with twobest
+ - saving #11 for use with twobest
+ - saving #32 for use with twobest
+ - saving #21 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ !source mask fill: ubot=32 usiz=159.5
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
+ - retaining old weight image
+ - using 38558 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31422 total points stored in 63 'TOHD(17.3748)' bloks (0 duplicates)
*[#21315=-0.231518] *[#21346=-0.231572] *[#21349=-0.233306] *[#21353=-0.23479] *[#21357=-0.23558] *[#21358=-0.235959] *[#21361=-0.236266] *[#21367=-0.238035] *[#21369=-0.23899] *[#21372=-0.24024] *[#21379=-0.240835] *[#21382=-0.241235] *[#21386=-0.24193] *[#21389=-0.242199] *[#21390=-0.243043] *[#21393=-0.243383] *[#21394=-0.243739] *[#21395=-0.245077] *[#21396=-0.245915] *[#21397=-0.246264] *[#21400=-0.246279] *[#21402=-0.247398] *[#21410=-0.247695] *[#21413=-0.248044]
+ - param set #1 has cost=-0.248044 [o=0 t=0]
+ -- Parameters = 85.8391 -2.8840 -0.3705 0.0359 -0.1677 0.1105 0.9766 1.0096 0.9787 0.0033 0.0010 0.0017
+ - param set #2 has cost=-0.034536 [o=23 t=1]
+ -- Parameters = 83.2613 -16.1186 37.1174 -14.2433 -15.8829 7.1507 0.9908 1.0008 1.0000 -0.0000 -0.0002 0.0003
+ - param set #3 has cost=-0.030768 [o=41 t=2]
+ -- Parameters = 82.4645 -35.6527 30.4712 12.9120 -34.7659 6.7165 0.9969 0.9952 1.0050 0.0003 0.0023 0.0001
Clipping -0.000100 1683.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - param set #4 has cost=-0.029411 [o=5 t=3]
+ -- Parameters = 52.8967 -11.1629 -28.1962 33.0525 31.8463 8.1248 1.0001 1.0003 1.0004 0.0003 -0.0003 0.0005
+ - param set #5 has cost=-0.031270 [o=47 t=4]
+ -- Parameters = 93.7022 -16.4058 34.2499 -31.1535 -23.4445 -10.2592 1.0061 1.0001 1.0061 0.0038 0.0162 0.0015
+ - param set #6 has cost=-0.032329 [o=1 t=5]
+ -- Parameters = 92.8017 -37.9462 28.4960 -8.6936 -35.8831 -6.8396 0.9979 1.0118 1.0520 0.0010 0.0017 0.0009
+ - param set #7 has cost=-0.027999 [o=70 t=6]
+ -- Parameters = 57.9600 -0.4272 30.4620 -2.8299 -6.9850 26.5374 0.9906 0.9931 1.0357 -0.0006 -0.0018 0.0012
+ - param set #8 has cost=-0.032304 [o=50 t=7]
+ -- Parameters = 82.6376 -26.6869 36.0745 18.6984 -27.3640 4.4822 0.9989 1.0042 0.9937 -0.0051 -0.0018 0.0004
+ - param set #9 has cost=-0.030114 [o=82 t=8]
+ -- Parameters = 59.2537 -8.6928 35.6841 -0.4484 -11.3348 20.3385 0.9906 0.9975 0.9997 0.0002 0.0004 -0.0002
+ - param set #10 has cost=-0.023271 [o=72 t=9]
+ -- Parameters = 56.3740 6.7476 36.2658 13.3572 -29.3716 26.0317 1.0063 0.9947 0.9971 -0.0017 -0.0006 -0.0022
+ - param set #11 has cost=-0.022143 [o=68 t=10]
+ -- Parameters = 54.7773 21.3644 29.1654 12.3221 -22.7292 28.4258 0.9992 1.0029 1.0047 0.0129 -0.0001 -0.0008
+ - param set #12 has cost=-0.023408 [o=2 t=11]
+ -- Parameters = 56.0809 0.5809 28.7073 19.5563 -21.5058 33.0976 0.9994 1.0074 0.9986 -0.0019 -0.0008 -0.0001
+ - param set #13 has cost=-0.029657 [o=8 t=12]
+ -- Parameters = 67.1214 25.3271 -36.1979 44.5546 10.2042 7.3711 0.9910 0.9904 0.9923 -0.0031 0.0099 -0.0013
++ Output dataset ./__ats_tmp___rs_pre.sub-04_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-04_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
+ - param set #14 has cost=-0.026963 [o=10 t=13]
++ 3drefit processed 1 datasets
+ -- Parameters = 54.9859 22.3098 30.0685 8.0498 -18.9186 26.7173 0.9527 0.9566 1.0323 0.0103 0.0038 0.0057
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ - param set #15 has cost=-0.026186 [o=66 t=14]
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
+ -- Parameters = 85.9630 -38.6729 31.5617 -15.6482 -29.0135 6.6453 0.9952 1.0013 1.0034 0.0000 -0.0014 -0.0012
+ - param set #16 has cost=-0.021923 [o=61 t=15]
+ -- Parameters = 106.1639 18.8012 -34.5088 -39.8640 7.2301 -8.6502 1.0040 0.9890 0.9896 0.0026 -0.0003 0.0009
+ - param set #17 has cost=-0.022054 [o=46 t=16]
+ -- Parameters = 101.8248 -20.0526 37.4516 -14.4989 -34.6427 -23.9928 0.9983 1.0029 0.9997 -0.0015 -0.0008 0.0000
+ - param set #18 has cost=-0.026127 [o=13 t=17]
+ -- Parameters = 98.5812 -31.1132 32.4595 -8.3686 -30.6121 -19.7642 1.0044 0.9959 0.9997 0.0006 0.0164 -0.0004
Registration (cubic final interpolation) ...
+ - param set #19 has cost=-0.026353 [o=30 t=18]
+ -- Parameters = 76.8435 -29.7031 29.8120 7.8811 -31.4235 20.5153 1.0018 1.0045 0.9996 -0.0008 -0.0000 -0.0003
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ - param set #20 has cost=-0.024600 [o=7 t=19]
+ -- Parameters = 92.0566 -24.2807 30.2045 -15.0765 -44.8444 -3.0335 1.0043 1.0126 0.9991 -0.0005 -0.0002 0.0023
++ Initial scale factor set to 0.30/112.00=3.8e+02
+ - param set #21 has cost=-0.020287 [o=28 t=20]
+ -- Parameters = 115.4880 0.4603 36.7319 -18.6372 -25.3592 -38.3159 0.9933 0.9947 0.9977 -0.0006 -0.0000 0.0001
+ - param set #22 has cost=-0.020465 [o=76 t=21]
+ -- Parameters = 56.2535 -14.7031 34.4005 25.7212 -39.4552 41.7565 1.0016 0.9985 0.9986 -0.0000 0.0007 -0.0008
+ - param set #23 has cost=-0.019234 [o=16 t=22]
+ -- Parameters = 67.2304 23.7907 36.3596 40.8676 -19.2146 41.5663 1.0034 0.9979 1.0000 0.0001 -0.0001 0.0002
+ - param set #24 has cost=-0.026490 [o=9 t=23]
+ -- Parameters = 59.4812 -18.9257 27.3501 17.6650 -23.7659 32.6820 0.9974 0.9994 1.0471 0.0002 0.0013 0.0004
+ - param set #25 has cost=-0.019673 [o=34 t=24]
+ -- Parameters = 65.3920 25.9598 27.9515 -40.8716 -7.8825 -4.5674 1.0003 0.9998 1.0026 -0.0007 0.0124 -0.0003
+ - param set #26 has cost=-0.020875 [o=60 t=25]
+ -- Parameters = 113.2836 -6.4181 36.6128 -15.5927 -26.5816 -36.9760 1.0010 0.9905 0.9917 0.0011 0.0018 0.0035
+ - param set #27 has cost=-0.032912 [o=11 t=26]
+ -- Parameters = 81.8630 -30.8406 35.9458 18.4741 -22.8703 0.2417 1.0494 0.9705 0.9647 0.0343 -0.0038 -0.0058
+ - param set #28 has cost=-0.023880 [o=32 t=27]
+ -- Parameters = 99.7513 30.0814 33.1231 -18.0710 -11.6322 -9.1746 1.0023 1.0015 1.0009 0.0011 0.0013 0.0016
+ - param set #29 has cost=-0.024096 [o=21 t=28]
+ -- Parameters = 89.5632 -24.1154 30.9296 -10.3431 -44.3548 3.5820 1.0014 0.9999 1.0020 0.0006 0.0012 -0.0001
*[#24172=-0.249814] *[#24174=-0.25036] *[#24175=-0.2508] *[#24176=-0.251951] *[#24177=-0.252731] *[#24178=-0.253175] *[#24182=-0.253386] *[#24183=-0.253801]
+ - param set #30 has cost=-0.253801 [o=-1 t=-1]
+ -- Parameters = 85.9927 -3.8236 0.0987 0.0717 -0.0998 -0.4927 0.9752 1.0299 0.9833 -0.0008 0.0082 -0.0086
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.046
+ --- dist(#3,#1) = 0.463
+ --- dist(#4,#1) = 0.448
+ --- dist(#5,#1) = 0.427
+ --- dist(#6,#1) = 0.45
+ --- dist(#7,#1) = 0.427
+ --- dist(#8,#1) = 0.398
+ --- dist(#9,#1) = 0.445
+ --- dist(#10,#1) = 0.494
+ --- dist(#11,#1) = 0.414
+ --- dist(#12,#1) = 0.38
+ --- dist(#13,#1) = 0.388
+ --- dist(#14,#1) = 0.369
+ --- dist(#15,#1) = 0.371
+ --- dist(#16,#1) = 0.436
+ --- dist(#17,#1) = 0.405
+ --- dist(#18,#1) = 0.497
+ --- dist(#19,#1) = 0.492
+ --- dist(#20,#1) = 0.424
+ --- dist(#21,#1) = 0.374
+ --- dist(#22,#1) = 0.452
+ --- dist(#23,#1) = 0.39
+ --- dist(#24,#1) = 0.467
+ --- dist(#25,#1) = 0.444
+ --- dist(#26,#1) = 0.456
+ --- dist(#27,#1) = 0.469
+ --- dist(#28,#1) = 0.458
+ --- dist(#29,#1) = 0.455
+ --- dist(#30,#1) = 0.467
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=32 usiz=159.5
+ - retaining old weight image
+ - using 38558 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30897 total points stored in 62 'TOHD(17.2644)' bloks (0 duplicates)
*[#24185=-0.286138] *[#24195=-0.286989] *[#24214=-0.287422] *[#24215=-0.287455] *[#24216=-0.287959] *[#24218=-0.28804] *[#24221=-0.288214] *[#24228=-0.288324] *[#24238=-0.288634] *[#24240=-0.288648] *[#24245=-0.288675] *[#24246=-0.28875] *[#24247=-0.288817] *[#24250=-0.288842] *[#24251=-0.288897] *[#24252=-0.288957] *[#24255=-0.28899] *[#24257=-0.289032] *[#24260=-0.289042] *[#24263=-0.289055] *[#24266=-0.289065] *[#24267=-0.289066] *[#24268=-0.289125] *[#24269=-0.289217] *[#24270=-0.289327] *[#24271=-0.289331] *[#24272=-0.289354] *[#24274=-0.28941] *[#24275=-0.289444] *[#24276=-0.289476] *[#24277=-0.289528] *[#24278=-0.289562] *[#24280=-0.289616] *[#24281=-0.289677] *[#24284=-0.289722] *[#24285=-0.289737] *[#24287=-0.289766] *[#24288=-0.289802] *[#24289=-0.28987] *[#24290=-0.289984] *[#24293=-0.290039] *[#24294=-0.290074]
+ - param set #1 has cost=-0.290074 [o=-1 t=-1]
+ -- Parameters = 85.9758 -3.8214 0.1923 0.0378 -0.1004 -0.2164 0.9778 1.0295 0.9794 0.0078 0.0066 -0.0115
+ - param set #2 has cost=-0.287927 [o=0 t=0]
+ -- Parameters = 85.8539 -3.8289 0.3059 0.1350 0.4703 0.1500 0.9748 1.0306 0.9683 0.0154 -0.0037 -0.0035
+ - param set #3 has cost=-0.028055 [o=23 t=1]
+ -- Parameters = 83.1932 -16.1545 37.2036 -14.2404 -15.8543 7.0742 0.9907 0.9990 0.9995 0.0006 -0.0008 0.0090
+ - param set #4 has cost=-0.027540 [o=11 t=26]
+ -- Parameters = 81.7576 -30.7913 36.0675 18.5476 -22.8072 -0.0657 1.0534 0.9653 0.9600 0.0346 -0.0026 -0.0051
+ - param set #5 has cost=-0.028256 [o=1 t=5]
+ -- Parameters = 92.6838 -38.0921 28.6974 -8.1478 -35.9527 -6.7006 1.0018 1.0117 1.0501 0.0070 0.0019 -0.0039
+ - param set #6 has cost=-0.030557 [o=50 t=7]
+ -- Parameters = 83.1046 -26.6738 35.8761 18.8792 -27.1847 5.0154 0.9824 0.9952 0.9852 -0.0046 0.0074 -0.0013
+ - param set #7 has cost=-0.027449 [o=47 t=4]
+ -- Parameters = 93.6044 -16.5165 34.2212 -31.1671 -23.3602 -10.7409 1.0083 1.0025 1.0111 0.0032 0.0252 0.0021
+ - param set #8 has cost=-0.027870 [o=41 t=2]
+ -- Parameters = 82.6376 -35.5992 30.4717 12.9384 -34.6156 6.7072 0.9926 0.9905 1.0071 0.0008 0.0041 0.0004
+ - param set #9 has cost=-0.024669 [o=82 t=8]
+ -- Parameters = 59.3853 -8.7471 35.6115 -0.4837 -11.0354 20.6566 0.9859 0.9967 1.0079 -0.0000 0.0045 0.0005
+ - param set #10 has cost=-0.025225 [o=8 t=12]
+ -- Parameters = 67.1156 25.3325 -36.2050 44.5502 10.2525 7.3511 0.9909 0.9913 0.9916 -0.0030 0.0099 -0.0012
+ - param set #11 has cost=-0.030161 [o=5 t=3]
+ -- Parameters = 51.9480 -10.6129 -28.5880 32.6869 31.3979 8.3407 0.9834 0.9890 0.9836 0.0040 0.0016 0.0123
+ - param set #12 has cost=-0.025181 [o=70 t=6]
+ -- Parameters = 57.5653 -0.2467 31.4941 -2.8804 -6.8651 26.7755 0.9555 0.9513 1.0383 -0.0025 -0.0026 0.0017
+ - param set #13 has cost=-0.021371 [o=10 t=13]
+ -- Parameters = 54.8790 22.1145 29.9904 8.1478 -18.8987 26.8215 0.9523 0.9600 1.0386 0.0100 0.0041 0.0045
+ - param set #14 has cost=-0.022621 [o=9 t=23]
+ -- Parameters = 59.4707 -18.9198 27.3703 17.6579 -23.7515 32.4791 0.9970 0.9983 1.0534 0.0014 0.0012 0.0005
+ - param set #15 has cost=-0.022840 [o=30 t=18]
+ -- Parameters = 76.7508 -29.6402 29.9215 7.8342 -31.5189 20.3524 1.0030 1.0119 0.9897 -0.0012 0.0003 -0.0005
+ - param set #16 has cost=-0.022496 [o=66 t=14]
+ -- Parameters = 86.1478 -38.6887 31.5846 -15.5735 -29.1022 6.9992 0.9930 1.0119 1.0102 -0.0002 -0.0042 -0.0009
+ - param set #17 has cost=-0.020479 [o=13 t=17]
+ -- Parameters = 98.6075 -31.1060 32.5190 -8.3389 -30.6755 -19.8035 1.0035 0.9965 0.9957 0.0017 0.0168 -0.0005
+ - param set #18 has cost=-0.019883 [o=7 t=19]
+ -- Parameters = 92.3045 -23.8203 30.5040 -14.9487 -44.9941 -3.1108 1.0055 1.0365 1.0027 -0.0002 -0.0008 0.0013
+ - param set #19 has cost=-0.019388 [o=21 t=28]
+ -- Parameters = 89.5036 -24.0454 30.9838 -10.3314 -44.4868 3.5983 0.9974 1.0019 1.0006 0.0021 0.0012 -0.0013
+ - param set #20 has cost=-0.021253 [o=32 t=27]
+ -- Parameters = 99.7168 31.4649 33.0909 -18.3619 -12.0227 -8.8334 1.0081 0.9842 0.9964 0.0030 -0.0141 -0.0088
+ - param set #21 has cost=-0.019367 [o=2 t=11]
+ -- Parameters = 56.0765 0.3968 28.5631 19.5790 -21.5340 33.0952 0.9985 1.0112 1.0004 -0.0007 -0.0005 -0.0004
+ - param set #22 has cost=-0.018226 [o=72 t=9]
+ -- Parameters = 56.3088 6.6685 36.3295 13.3673 -29.1932 25.9669 1.0053 0.9925 0.9969 0.0073 -0.0000 -0.0011
+ - param set #23 has cost=-0.016555 [o=68 t=10]
+ -- Parameters = 54.8317 21.5076 29.0896 12.5451 -22.8274 28.4102 0.9939 1.0029 1.0086 0.0127 0.0004 -0.0013
+ - param set #24 has cost=-0.019182 [o=46 t=16]
+ -- Parameters = 102.2024 -18.0483 37.3725 -14.5448 -34.8577 -23.3682 0.9939 1.0063 0.9937 -0.0017 -0.0017 -0.0016
+ - param set #25 has cost=-0.020977 [o=61 t=15]
+ -- Parameters = 106.1544 18.5915 -34.7671 -39.9194 7.0612 -8.6723 1.0050 0.9903 0.9881 0.0028 -0.0011 0.0008
+ - param set #26 has cost=-0.016697 [o=60 t=25]
+ -- Parameters = 113.1413 -6.4776 36.4662 -15.6665 -26.5311 -36.8912 1.0011 0.9901 0.9921 0.0007 0.0022 0.0027
+ - param set #27 has cost=-0.016898 [o=76 t=21]
+ -- Parameters = 56.2070 -14.9242 34.3935 25.7645 -39.3312 41.1948 1.0063 0.9922 0.9997 -0.0025 0.0054 -0.0035
+ - param set #28 has cost=-0.015678 [o=28 t=20]
+ -- Parameters = 115.4606 0.3702 36.7868 -18.4447 -25.3428 -38.4100 0.9881 0.9935 0.9951 -0.0013 -0.0002 -0.0001
+ - param set #29 has cost=-0.014625 [o=34 t=24]
+ -- Parameters = 65.3385 25.9339 28.0278 -40.7996 -7.6755 -4.6696 1.0005 0.9991 1.0039 -0.0005 0.0122 -0.0006
+ - param set #30 has cost=-0.015239 [o=16 t=22]
+ -- Parameters = 67.0939 23.9251 36.4312 41.0873 -18.9181 41.7880 1.0115 0.9978 0.9970 -0.0001 -0.0027 0.0006
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0465
+ --- dist(#3,#1) = 0.446
+ --- dist(#4,#1) = 0.425
+ --- dist(#5,#1) = 0.428
+ --- dist(#6,#1) = 0.463
+ --- dist(#7,#1) = 0.397
+ --- dist(#8,#1) = 0.448
+ --- dist(#9,#1) = 0.425
+ --- dist(#10,#1) = 0.495
+ --- dist(#11,#1) = 0.391
+ --- dist(#12,#1) = 0.443
+ --- dist(#13,#1) = 0.372
+ --- dist(#14,#1) = 0.363
+ --- dist(#15,#1) = 0.436
+ --- dist(#16,#1) = 0.389
+ --- dist(#17,#1) = 0.441
+ --- dist(#18,#1) = 0.444
+ --- dist(#19,#1) = 0.404
+ --- dist(#20,#1) = 0.499
+ --- dist(#21,#1) = 0.493
+ --- dist(#22,#1) = 0.374
+ --- dist(#23,#1) = 0.465
+ --- dist(#24,#1) = 0.452
+ --- dist(#25,#1) = 0.46
+ --- dist(#26,#1) = 0.453
+ --- dist(#27,#1) = 0.389
+ --- dist(#28,#1) = 0.457
+ --- dist(#29,#1) = 0.467
+ --- dist(#30,#1) = 0.454
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=32 usiz=159.5
+ - retaining old weight image
+ - using 38558 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31493 total points stored in 65 'TOHD(17.1973)' bloks (0 duplicates)
*[#27452=-0.300431] *[#27473=-0.301851] *[#27479=-0.301978] *[#27480=-0.30204] *[#27483=-0.30229] *[#27484=-0.302532] *[#27485=-0.302611] *[#27488=-0.303107] *[#27489=-0.303671] *[#27490=-0.303794] *[#27491=-0.303993] *[#27492=-0.304029] *[#27498=-0.305142] *[#27499=-0.305378] *[#27500=-0.305878] *[#27505=-0.306123] *[#27506=-0.306147] *[#27508=-0.306312] *[#27513=-0.30638] *[#27514=-0.30641] *[#27515=-0.306556] *[#27516=-0.306642] *[#27535=-0.306831] *[#27538=-0.306859] *[#27542=-0.306877] *[#27543=-0.306896] *[#27546=-0.306948] *[#27547=-0.306955] *[#27550=-0.306972] *[#27551=-0.306991] *[#27552=-0.306996] *[#27553=-0.307008] *[#27554=-0.307013] *[#27557=-0.307062] *[#27558=-0.307098] *[#27560=-0.307103] *[#27561=-0.30712] *[#27562=-0.307135] *[#27563=-0.307166] *[#27564=-0.307195] *[#27569=-0.307214] *[#27571=-0.307224]
+ - param set #1 has cost=-0.307224 [o=-1 t=-1]
+ -- Parameters = 85.9054 -3.6902 0.4121 0.0332 -0.6549 0.0045 0.9824 1.0272 0.9787 0.0081 -0.0001 -0.0257
+ - param set #2 has cost=-0.307209 [o=0 t=0]
+ -- Parameters = 86.0652 -3.8990 0.3792 -0.1428 -0.5782 0.0538 0.9833 1.0314 0.9781 0.0030 -0.0029 -0.0246
+ - param set #3 has cost=-0.027203 [o=50 t=7]
+ -- Parameters = 83.2382 -26.7059 36.1136 19.0448 -27.2880 5.1066 0.9791 1.0027 0.9763 -0.0033 0.0053 -0.0014
+ - param set #4 has cost=-0.025894 [o=5 t=3]
+ -- Parameters = 51.9010 -10.7103 -28.5408 32.7364 31.3042 8.2606 0.9841 0.9882 0.9885 0.0055 0.0012 0.0120
+ - param set #5 has cost=-0.024525 [o=1 t=5]
+ -- Parameters = 92.6066 -38.2336 28.7261 -8.1241 -35.8233 -6.9938 1.0039 1.0098 1.0518 0.0063 0.0076 -0.0040
+ - param set #6 has cost=-0.023625 [o=23 t=1]
+ -- Parameters = 83.0606 -16.2042 37.4548 -14.2738 -15.5294 6.4461 0.9935 0.9993 0.9933 0.0010 0.0002 0.0100
+ - param set #7 has cost=-0.023581 [o=41 t=2]
+ -- Parameters = 82.6620 -35.7617 30.5613 12.9840 -34.6601 6.5954 0.9922 0.9890 1.0105 0.0048 0.0042 0.0004
+ - param set #8 has cost=-0.024048 [o=11 t=26]
+ -- Parameters = 81.7295 -30.6915 36.0088 18.5638 -22.3283 -0.0392 1.0532 0.9601 0.9594 0.0329 -0.0025 -0.0045
+ - param set #9 has cost=-0.021897 [o=47 t=4]
+ -- Parameters = 93.8947 -16.8226 35.2226 -31.5273 -22.8180 -10.7062 1.0354 1.0034 1.0034 0.0014 0.0303 -0.0042
+ - param set #10 has cost=-0.021430 [o=8 t=12]
+ -- Parameters = 67.0466 25.4498 -36.2010 44.5865 10.3420 7.3508 0.9920 0.9925 0.9907 -0.0030 0.0097 -0.0013
+ - param set #11 has cost=-0.023053 [o=70 t=6]
+ -- Parameters = 57.5182 -1.2167 31.3834 -3.1385 -6.4630 27.0882 0.9509 0.9600 1.0545 -0.0046 -0.0050 0.0018
+ - param set #12 has cost=-0.021762 [o=82 t=8]
+ -- Parameters = 59.3527 -8.7942 35.5393 -0.4460 -10.9955 20.7763 0.9824 0.9968 1.0118 -0.0001 0.0040 0.0003
+ - param set #13 has cost=-0.020294 [o=30 t=18]
+ -- Parameters = 76.8296 -29.5672 29.8179 7.7623 -31.6844 20.2664 1.0082 1.0127 0.9864 -0.0014 0.0005 0.0007
+ - param set #14 has cost=-0.019753 [o=9 t=23]
+ -- Parameters = 59.2112 -18.8959 27.3255 17.5466 -23.8663 32.5254 1.0045 0.9977 1.0572 0.0021 0.0030 -0.0004
+ - param set #15 has cost=-0.019866 [o=66 t=14]
+ -- Parameters = 86.0281 -38.7103 31.4910 -15.5881 -29.0130 6.7000 0.9929 1.0130 1.0213 0.0001 -0.0051 -0.0003
+ - param set #16 has cost=-0.018705 [o=10 t=13]
+ -- Parameters = 54.6265 21.9881 29.5423 7.9969 -18.9127 26.4641 0.9450 0.9612 1.0510 0.0170 0.0039 0.0054
+ - param set #17 has cost=-0.018423 [o=32 t=27]
+ -- Parameters = 99.9600 31.6076 32.7940 -18.4650 -12.2970 -8.8602 1.0083 0.9838 0.9964 0.0083 -0.0141 -0.0089
+ - param set #18 has cost=-0.017817 [o=61 t=15]
+ -- Parameters = 106.4878 18.6940 -34.9053 -39.9529 7.0115 -8.8179 1.0029 0.9899 0.9854 0.0023 -0.0012 0.0008
+ - param set #19 has cost=-0.017879 [o=13 t=17]
+ -- Parameters = 98.5322 -31.1941 32.5155 -8.2373 -30.6176 -19.7030 1.0034 0.9975 0.9975 0.0078 0.0168 -0.0003
+ - param set #20 has cost=-0.015836 [o=7 t=19]
+ -- Parameters = 92.2734 -23.6804 30.5074 -14.8764 -44.9932 -2.9733 1.0087 1.0352 1.0012 -0.0006 -0.0016 0.0027
+ - param set #21 has cost=-0.014812 [o=21 t=28]
+ -- Parameters = 89.4450 -24.1551 31.0190 -10.1488 -44.4515 3.7800 0.9949 1.0045 0.9993 0.0022 0.0015 -0.0014
+ - param set #22 has cost=-0.014649 [o=2 t=11]
+ -- Parameters = 56.0463 0.5252 28.6334 19.6722 -21.4917 33.0869 0.9954 1.0098 0.9985 -0.0000 0.0001 -0.0004
+ - param set #23 has cost=-0.016393 [o=46 t=16]
+ -- Parameters = 102.1209 -17.4818 36.9526 -14.6687 -35.3503 -23.0428 0.9932 1.0040 0.9933 -0.0030 -0.0016 -0.0039
+ - param set #24 has cost=-0.014796 [o=72 t=9]
+ -- Parameters = 56.4200 6.6527 36.2981 13.3174 -29.2528 26.0149 1.0047 0.9951 0.9983 0.0133 0.0007 -0.0013
+ - param set #25 has cost=-0.013697 [o=76 t=21]
+ -- Parameters = 56.1829 -14.9541 34.3799 25.7761 -39.2599 41.1881 1.0091 0.9923 0.9997 -0.0032 0.0057 -0.0034
+ - param set #26 has cost=-0.012947 [o=60 t=25]
+ -- Parameters = 113.1542 -6.2886 36.6353 -15.4288 -26.3551 -36.7076 0.9998 0.9898 1.0012 -0.0005 0.0017 0.0021
+ - param set #27 has cost=-0.014937 [o=68 t=10]
+ -- Parameters = 55.1230 21.9602 28.9433 11.4549 -22.5690 28.3615 0.9643 0.9963 1.0085 0.0125 -0.0032 -0.0007
+ - param set #28 has cost=-0.012520 [o=28 t=20]
+ -- Parameters = 115.3924 0.2828 36.8483 -18.3169 -25.4448 -38.4442 0.9889 0.9951 0.9915 -0.0013 0.0001 -0.0002
+ - param set #29 has cost=-0.012719 [o=16 t=22]
+ -- Parameters = 67.1621 23.8101 36.5434 40.9681 -18.8296 41.9114 1.0107 0.9951 0.9915 0.0002 -0.0025 0.0057
+ - param set #30 has cost=-0.012230 [o=34 t=24]
+ -- Parameters = 65.3401 25.8786 28.1359 -40.8710 -7.6475 -4.7579 1.0014 1.0007 1.0049 -0.0004 0.0183 -0.0004
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0229
+ --- dist(#3,#1) = 0.446
+ --- dist(#4,#1) = 0.425
+ --- dist(#5,#1) = 0.432
+ --- dist(#6,#1) = 0.445
+ --- dist(#7,#1) = 0.463
+ --- dist(#8,#1) = 0.401
+ --- dist(#9,#1) = 0.387
+ --- dist(#10,#1) = 0.435
+ --- dist(#11,#1) = 0.439
+ --- dist(#12,#1) = 0.495
+ --- dist(#13,#1) = 0.368
+ --- dist(#14,#1) = 0.438
+ --- dist(#15,#1) = 0.361
+ --- dist(#16,#1) = 0.391
+ --- dist(#17,#1) = 0.441
+ --- dist(#18,#1) = 0.401
+ --- dist(#19,#1) = 0.444
+ --- dist(#20,#1) = 0.457
+ --- dist(#21,#1) = 0.493
+ --- dist(#22,#1) = 0.385
+ --- dist(#23,#1) = 0.487
+ --- dist(#24,#1) = 0.449
+ --- dist(#25,#1) = 0.373
+ --- dist(#26,#1) = 0.458
+ --- dist(#27,#1) = 0.453
+ --- dist(#28,#1) = 0.466
+ --- dist(#29,#1) = 0.455
+ --- dist(#30,#1) = 0.454
+ - Total coarse refinement net CPU time = 0.0 s; 9670 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=32 usiz=159.5
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 31325 total points stored in 65 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.302740 * [o=-1 t=-1]
+ -- Parameters = 85.9054 -3.6902 0.4121 0.0332 -0.6549 0.0045 0.9824 1.0272 0.9787 0.0081 -0.0001 -0.0257
+ - cost(#2)=-0.303091 * [o=0 t=0]
+ -- Parameters = 86.0652 -3.8990 0.3792 -0.1428 -0.5782 0.0538 0.9833 1.0314 0.9781 0.0030 -0.0029 -0.0246
+ - cost(#3)=-0.026371 [o=50 t=7]
+ -- Parameters = 83.2382 -26.7059 36.1136 19.0448 -27.2880 5.1066 0.9791 1.0027 0.9763 -0.0033 0.0053 -0.0014
+ - cost(#4)=-0.023682 [o=5 t=3]
+ -- Parameters = 51.9010 -10.7103 -28.5408 32.7364 31.3042 8.2606 0.9841 0.9882 0.9885 0.0055 0.0012 0.0120
+ - cost(#5)=-0.023162 [o=1 t=5]
+ -- Parameters = 92.6066 -38.2336 28.7261 -8.1241 -35.8233 -6.9938 1.0039 1.0098 1.0518 0.0063 0.0076 -0.0040
+ - cost(#6)=-0.022707 [o=11 t=26]
+ -- Parameters = 81.7295 -30.6915 36.0088 18.5638 -22.3283 -0.0392 1.0532 0.9601 0.9594 0.0329 -0.0025 -0.0045
+ - cost(#7)=-0.022342 [o=23 t=1]
+ -- Parameters = 83.0606 -16.2042 37.4548 -14.2738 -15.5294 6.4461 0.9935 0.9993 0.9933 0.0010 0.0002 0.0100
+ - cost(#8)=-0.022427 [o=41 t=2]
+ -- Parameters = 82.6620 -35.7617 30.5613 12.9840 -34.6601 6.5954 0.9922 0.9890 1.0105 0.0048 0.0042 0.0004
+ - cost(#9)=-0.021348 [o=70 t=6]
+ -- Parameters = 57.5182 -1.2167 31.3834 -3.1385 -6.4630 27.0882 0.9509 0.9600 1.0545 -0.0046 -0.0050 0.0018
+ - cost(#10)=-0.020365 [o=47 t=4]
+ -- Parameters = 93.8947 -16.8226 35.2226 -31.5273 -22.8180 -10.7062 1.0354 1.0034 1.0034 0.0014 0.0303 -0.0042
+ - cost(#11)=-0.020501 [o=82 t=8]
+ -- Parameters = 59.3527 -8.7942 35.5393 -0.4460 -10.9955 20.7763 0.9824 0.9968 1.0118 -0.0001 0.0040 0.0003
+ - cost(#12)=-0.019270 [o=8 t=12]
+ -- Parameters = 67.0466 25.4498 -36.2010 44.5865 10.3420 7.3508 0.9920 0.9925 0.9907 -0.0030 0.0097 -0.0013
+ - cost(#13)=-0.018687 [o=30 t=18]
+ -- Parameters = 76.8296 -29.5672 29.8179 7.7623 -31.6844 20.2664 1.0082 1.0127 0.9864 -0.0014 0.0005 0.0007
+ - cost(#14)=-0.018998 [o=66 t=14]
+ -- Parameters = 86.0281 -38.7103 31.4910 -15.5881 -29.0130 6.7000 0.9929 1.0130 1.0213 0.0001 -0.0051 -0.0003
+ - cost(#15)=-0.017926 [o=9 t=23]
+ -- Parameters = 59.2112 -18.8959 27.3255 17.5466 -23.8663 32.5254 1.0045 0.9977 1.0572 0.0021 0.0030 -0.0004
+ - cost(#16)=-0.017203 [o=10 t=13]
+ -- Parameters = 54.6265 21.9881 29.5423 7.9969 -18.9127 26.4641 0.9450 0.9612 1.0510 0.0170 0.0039 0.0054
+ - cost(#17)=-0.017320 [o=32 t=27]
+ -- Parameters = 99.9600 31.6076 32.7940 -18.4650 -12.2970 -8.8602 1.0083 0.9838 0.9964 0.0083 -0.0141 -0.0089
+ - cost(#18)=-0.017315 [o=13 t=17]
+ -- Parameters = 98.5322 -31.1941 32.5155 -8.2373 -30.6176 -19.7030 1.0034 0.9975 0.9975 0.0078 0.0168 -0.0003
+ - cost(#19)=-0.016978 [o=61 t=15]
+ -- Parameters = 106.4878 18.6940 -34.9053 -39.9529 7.0115 -8.8179 1.0029 0.9899 0.9854 0.0023 -0.0012 0.0008
+ - cost(#20)=-0.015227 [o=46 t=16]
+ -- Parameters = 102.1209 -17.4818 36.9526 -14.6687 -35.3503 -23.0428 0.9932 1.0040 0.9933 -0.0030 -0.0016 -0.0039
+ - cost(#21)=-0.014806 [o=7 t=19]
+ -- Parameters = 92.2734 -23.6804 30.5074 -14.8764 -44.9932 -2.9733 1.0087 1.0352 1.0012 -0.0006 -0.0016 0.0027
+ - cost(#22)=-0.013310 [o=68 t=10]
+ -- Parameters = 55.1230 21.9602 28.9433 11.4549 -22.5690 28.3615 0.9643 0.9963 1.0085 0.0125 -0.0032 -0.0007
+ - cost(#23)=-0.014448 [o=21 t=28]
+ -- Parameters = 89.4450 -24.1551 31.0190 -10.1488 -44.4515 3.7800 0.9949 1.0045 0.9993 0.0022 0.0015 -0.0014
+ - cost(#24)=-0.013159 [o=72 t=9]
+ -- Parameters = 56.4200 6.6527 36.2981 13.3174 -29.2528 26.0149 1.0047 0.9951 0.9983 0.0133 0.0007 -0.0013
+ - cost(#25)=-0.014262 [o=2 t=11]
+ -- Parameters = 56.0463 0.5252 28.6334 19.6722 -21.4917 33.0869 0.9954 1.0098 0.9985 -0.0000 0.0001 -0.0004
+ - cost(#26)=-0.011968 [o=76 t=21]
+ -- Parameters = 56.1829 -14.9541 34.3799 25.7761 -39.2599 41.1881 1.0091 0.9923 0.9997 -0.0032 0.0057 -0.0034
+ - cost(#27)=-0.012288 [o=60 t=25]
+ -- Parameters = 113.1542 -6.2886 36.6353 -15.4288 -26.3551 -36.7076 0.9998 0.9898 1.0012 -0.0005 0.0017 0.0021
+ - cost(#28)=-0.011942 [o=16 t=22]
+ -- Parameters = 67.1621 23.8101 36.5434 40.9681 -18.8296 41.9114 1.0107 0.9951 0.9915 0.0002 -0.0025 0.0057
+ - cost(#29)=-0.011327 [o=28 t=20]
+ -- Parameters = 115.3924 0.2828 36.8483 -18.3169 -25.4448 -38.4442 0.9889 0.9951 0.9915 -0.0013 0.0001 -0.0002
+ - cost(#30)=-0.010604 [o=34 t=24]
+ -- Parameters = 65.3401 25.8786 28.1359 -40.8710 -7.6475 -4.7579 1.0014 1.0007 1.0049 -0.0004 0.0183 -0.0004
+ - cost(#31)=-0.107347 [o=-2 t=-2]
+ -- Parameters = 86.1075 -4.6492 -2.5412 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.303988 * [o=-1 t=-1]
+ -- Parameters = 86.0580 -3.6960 0.4015 -0.1350 -0.7547 0.0144 0.9853 1.0272 0.9785 0.0019 -0.0007 -0.0270
+ - cost(#2)=-0.303975 [o=0 t=0]
+ -- Parameters = 86.0416 -3.7953 0.4441 -0.1073 -0.6294 0.0975 0.9827 1.0296 0.9769 0.0013 -0.0040 -0.0253
+ - cost(#3)=-0.027265 [o=50 t=7]
+ -- Parameters = 83.4834 -26.5942 35.9404 19.1229 -27.5320 5.2315 0.9742 1.0017 0.9536 -0.0037 0.0079 0.0008
+ - cost(#4)=-0.023762 [o=5 t=3]
+ -- Parameters = 51.9333 -10.7547 -28.5482 32.6881 31.2869 8.2766 0.9837 0.9890 0.9887 0.0057 0.0009 0.0131
+ - cost(#5)=-0.023361 [o=1 t=5]
+ -- Parameters = 92.5673 -38.2289 28.6071 -8.2633 -35.9615 -7.0918 1.0048 1.0104 1.0526 0.0060 0.0146 -0.0042
+ - cost(#6)=-0.022894 [o=11 t=26]
+ -- Parameters = 81.6773 -30.7569 35.9595 18.4664 -22.0700 -0.2408 1.0542 0.9597 0.9609 0.0325 -0.0026 -0.0046
+ - cost(#7)=-0.022644 [o=23 t=1]
+ -- Parameters = 83.0431 -16.2110 37.4581 -14.2031 -15.6657 6.3461 0.9908 1.0008 1.0182 0.0012 0.0002 0.0108
+ - cost(#8)=-0.022504 [o=41 t=2]
+ -- Parameters = 82.6923 -35.7447 30.5519 13.0370 -34.7160 6.6385 0.9908 0.9890 1.0103 0.0055 0.0041 0.0007
+ - cost(#9)=-0.021936 [o=70 t=6]
+ -- Parameters = 57.7302 -1.5602 31.7773 -3.1322 -6.2540 27.1023 0.9419 0.9628 1.0499 -0.0054 -0.0085 0.0024
+ - cost(#10)=-0.022081 [o=47 t=4]
+ -- Parameters = 94.4424 -18.2697 36.6174 -31.6243 -22.4845 -11.3250 1.0344 0.9904 1.0309 -0.0029 0.0354 -0.0060
+ - cost(#11)=-0.020553 [o=82 t=8]
+ -- Parameters = 59.3335 -8.8144 35.5396 -0.4707 -10.9816 20.7600 0.9813 0.9969 1.0144 -0.0003 0.0039 0.0006
+ - cost(#12)=-0.021331 [o=8 t=12]
+ -- Parameters = 65.0080 25.6589 -36.8964 44.6738 10.1620 8.4735 0.9880 0.9911 0.9890 -0.0034 0.0121 -0.0043
+ - cost(#13)=-0.020698 [o=30 t=18]
+ -- Parameters = 75.7341 -30.8533 30.8070 8.6597 -31.2125 21.2932 1.0334 1.0103 0.9930 0.0026 -0.0014 0.0024
+ - cost(#14)=-0.019049 [o=66 t=14]
+ -- Parameters = 86.0315 -38.7397 31.4657 -15.5627 -28.9085 6.8250 0.9932 1.0127 1.0217 0.0004 -0.0051 -0.0003
+ - cost(#15)=-0.018060 [o=9 t=23]
+ -- Parameters = 59.2304 -18.9401 27.2658 17.5402 -23.9066 32.5488 1.0044 1.0006 1.0579 0.0013 0.0036 -0.0005
+ - cost(#16)=-0.017657 [o=10 t=13]
+ -- Parameters = 54.7736 21.8581 29.4250 7.8393 -18.6273 26.2843 0.9391 0.9582 1.0611 0.0168 0.0023 0.0045
+ - cost(#17)=-0.017485 [o=32 t=27]
+ -- Parameters = 99.9839 31.6770 32.8548 -18.4678 -12.3851 -8.8002 1.0099 0.9832 0.9942 0.0084 -0.0143 -0.0109
+ - cost(#18)=-0.017320 [o=13 t=17]
+ -- Parameters = 98.5167 -31.2034 32.5196 -8.2222 -30.6227 -19.7010 1.0034 0.9976 0.9975 0.0079 0.0168 -0.0003
+ - cost(#19)=-0.017030 [o=61 t=15]
+ -- Parameters = 106.5577 18.7696 -34.9081 -39.9751 7.0864 -8.8359 1.0039 0.9902 0.9855 0.0024 -0.0011 0.0007
+ - cost(#20)=-0.015275 [o=46 t=16]
+ -- Parameters = 102.1139 -17.4371 36.9277 -14.6374 -35.4198 -22.9950 0.9931 1.0031 0.9933 -0.0030 -0.0009 -0.0044
+ - cost(#21)=-0.015328 [o=7 t=19]
+ -- Parameters = 92.1915 -23.3807 30.5415 -14.8208 -44.9722 -3.0731 1.0151 1.0310 1.0039 -0.0018 -0.0020 0.0031
+ - cost(#22)=-0.013917 [o=68 t=10]
+ -- Parameters = 55.2044 22.0465 29.1052 11.0075 -22.5821 27.9420 0.9601 0.9937 1.0148 0.0120 0.0062 -0.0036
+ - cost(#23)=-0.014585 [o=21 t=28]
+ -- Parameters = 89.3675 -24.1417 31.1107 -10.1441 -44.4284 3.7214 0.9959 1.0041 0.9946 0.0014 0.0017 -0.0022
+ - cost(#24)=-0.014339 [o=72 t=9]
+ -- Parameters = 56.4413 6.4815 36.4342 12.7310 -29.6773 25.5880 1.0021 1.0204 0.9938 0.0139 0.0010 -0.0029
+ - cost(#25)=-0.014375 [o=2 t=11]
+ -- Parameters = 56.0668 0.4341 28.7234 19.5668 -21.4290 32.9896 0.9944 1.0130 0.9976 0.0000 -0.0000 -0.0017
+ - cost(#26)=-0.012036 [o=76 t=21]
+ -- Parameters = 56.0972 -14.8794 34.3992 25.7807 -39.2727 41.1926 1.0101 0.9926 0.9998 -0.0036 0.0063 -0.0035
+ - cost(#27)=-0.013229 [o=60 t=25]
+ -- Parameters = 113.0041 -6.1321 36.1318 -15.2735 -26.1562 -35.8207 0.9903 0.9916 1.0230 -0.0016 0.0007 -0.0015
+ - cost(#28)=-0.011980 [o=16 t=22]
+ -- Parameters = 67.1721 23.7993 36.5783 40.9813 -18.8305 41.9159 1.0104 0.9950 0.9907 0.0006 -0.0025 0.0065
+ - cost(#29)=-0.011505 [o=28 t=20]
+ -- Parameters = 115.4237 0.1987 36.8108 -18.3986 -25.3671 -38.5177 0.9888 0.9953 0.9871 -0.0022 0.0002 0.0003
+ - cost(#30)=-0.010767 [o=34 t=24]
+ -- Parameters = 65.3219 25.8255 28.0583 -40.8673 -7.7463 -4.7872 1.0010 1.0008 1.0041 -0.0005 0.0256 -0.0002
+ - cost(#31)=-0.301283 [o=-2 t=-2]
+ -- Parameters = 85.9829 -3.9222 0.2806 -0.0182 -0.6848 -0.0844 0.9869 1.0325 0.9843 0.0047 0.0039 -0.0229
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.303988
+ - Initial fine Parameters = 86.0580 -3.6960 0.4015 -0.1350 -0.7547 0.0144 0.9853 1.0272 0.9785 0.0019 -0.0007 -0.0270
+ - Finalish cost = -0.306916 ; 621 funcs
+ - ini Finalish Parameters = 86.0440 -3.5976 0.6814 -0.1782 -1.3209 0.2872 0.9860 1.0253 0.9765 -0.0016 -0.0106 -0.0443
RMS[0] = 0.320196 0.16682 ITER = 7/50
0.320196
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - Final cost = -0.306923 ; 499 funcs
+ Final fine fit Parameters:
x-shift= 86.0459 y-shift= -3.6118 z-shift= 0.6810 ... enorm= 86.1243 mm
z-angle= -0.1801 x-angle= -1.3192 y-angle= 0.2800 ... total= 1.3610 deg
x-scale= 0.9861 y-scale= 1.0256 z-scale= 0.9765 ... vol3D= 0.9876 = base bigger than source
y/x-shear= -0.0018 z/x-shear= -0.0104 z/y-shear= -0.0442
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.0459 -3.6118 0.6810 -0.1801 -1.3192 0.2800 0.9861 1.0256 0.9765 -0.0018 -0.0104 -0.0442
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Unloading unneeded data
++ Output dataset ./sub-05_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-05_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 19.2
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/__tt_sub-05_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/sub-05_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-05_T1w_ns+orig sub-05_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-05_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-05_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-05_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-05_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
#Script is running (command trimmed):
3dinfo ./__tt_sub-06_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-04_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-04_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-04_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-04_T1w_ns.skl+orig.BRIK
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/__tt_sub-06_T1w_ns+orig is not oblique
#Script is running (command trimmed):
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 355.565613
+ Used gradual clip level = 333.532806 .. 383.478394
++ Output dataset ././__ats_tmp___pad15_pre.sub-04_T1w_ns+orig.BRIK
+ Number voxels above clip level = 39664
+ Clustering voxels ...
+ Largest cluster has 39107 voxels
+ Clustering voxels ...
+ Largest cluster has 38079 voxels
+ Filled 393 voxels in small holes; now have 38472 voxels
+ Clustering voxels ...
+ Largest cluster has 38472 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 125368 voxels
+ Mask now has 38472 voxels
++ 38472 voxels in the mask [out of 163840: 23.48%]
++ first 9 x-planes are zero [from L]
++ last 12 x-planes are zero [from R]
++ first 1 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 4 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 863.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/863.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-06_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-06_T1w_ns+orig -prefix ./sub-06_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-06_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ output dataset: ./__ats_tmp___upad15_pre.sub-04_T1w_ns+orig.BRIK
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-06_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-04_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-04_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-04_T1w_ns+orig __ats_tmp___upad15_pre.sub-04_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-04_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-04_T1w_ns+tlrc.HEAD -> sub-04_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-04_T1w_ns+tlrc.BRIK -> sub-04_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-04_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-04_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-04.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-04.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
Performing center alignment with @Align_Centers
+ Automask has 37190 voxels
+ 5738 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-05_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-05_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-05_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-05_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-05_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-05_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-05_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Center distance of 0.000000 mm
++ 1516645 voxels in -source_automask+2
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=7 ytop=3
++ Zero-pad: zbot=7 ztop=4
++ 38472 voxels [15.9%] in weight mask
++ Output dataset ./__tt_sub-06_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 38472
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 30.070 34.594 27.451 (index)
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
+ source center of mass = 88.048 117.316 123.399 (index)
+ source-target CM = 85.926 -4.425 -2.123 (xyz)
+ estimated center of mass shifts = 85.926 -4.425 -2.123
++ shift param auto-range: 25.3..146.6 -74.7..65.9 -66.3..62.1
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 75.925575 .. 95.925575 center = 85.925575
+ Range param#2 [y-shift] = -14.425499 .. 5.574501 center = -4.425499
+ Range param#3 [z-shift] = -12.123459 .. 7.876541 center = -2.123459
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 45.925575 .. 125.925575 center = 85.925575
+ Range param#2 [y-shift] = -44.425499 .. 35.574501 center = -4.425499
+ Range param#3 [z-shift] = -42.123459 .. 37.876541 center = -2.123459
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000089
++ Final parameter search ranges:
+ x-shift = 45.926 .. 125.926
+ y-shift = -44.425 .. 35.575
+ z-shift = -42.123 .. 37.877
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1516645 [out of 11534336] voxels
+ base mask has 51740 [out of 241536] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
Resampling ...
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=46 usiz=159.5
+ - copying weight image
+ - using 38472 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 32381 total points stored in 65 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1622.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-05_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-05_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/111.00=3.7e+02
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=+0.162 pitch=-0.157 yaw=-0.052 dS=-2.394 dL=-0.016 dP=-0.479
++ Mean: roll=+0.271 pitch=+0.192 yaw=+0.046 dS=-1.558 dL=+0.098 dP=+0.105
++ Max : roll=+0.331 pitch=+0.688 yaw=+0.120 dS=-0.822 dL=+0.153 dP=+0.531
++ Max displacements (mm) for each sub-brick:
2.69(0.00) 2.60(0.16) 2.65(0.12) 2.51(0.18) 2.46(0.10) 2.40(0.08) 2.42(0.05) 2.43(0.04) 2.36(0.08) 2.41(0.07) 2.38(0.06) 2.35(0.11) 2.38(0.05) 2.38(0.10) 2.42(0.09) 2.42(0.05) 2.37(0.10) 2.36(0.07) 2.27(0.11) 2.28(0.10) 2.26(0.10) 2.31(0.08) 2.26(0.08) 2.31(0.08) 2.27(0.06) 2.26(0.06) 2.31(0.07) 2.15(0.16) 2.21(0.09) 2.14(0.10) 2.13(0.06) 2.18(0.11) 2.17(0.09) 2.17(0.10) 2.18(0.05) 2.21(0.12) 2.20(0.05) 2.23(0.10) 2.26(0.05) 2.27(0.13) 2.20(0.08) 2.16(0.08) 2.14(0.12) 2.11(0.12) 2.14(0.14) 2.14(0.06) 2.14(0.05) 2.14(0.07) 2.14(0.04) 2.13(0.02) 2.13(0.06) 2.14(0.06) 2.11(0.13) 2.11(0.03) 2.05(0.12) 2.10(0.13) 2.06(0.06) 1.92(0.16) 2.00(0.10) 1.96(0.05) 1.96(0.05) 1.96(0.05) 1.85(0.13) 1.88(0.07) 1.92(0.10) 1.82(0.13) 1.85(0.10) 1.75(0.15) 1.88(0.13) 1.96(0.12) 1.92(0.06) 1.89(0.08) 1.89(0.02) 1.70(0.20) 1.94(0.24) 1.94(0.02) 1.96(0.06) 1.87(0.10) 1.89(0.09) 1.80(0.12) 2.04(0.25) 1.99(0.09) 2.07(0.09) 1.94(0.13) 1.94(0.04) 1.89(0.10) 2.01(0.14) 1.93(0.10) 2.00(0.09) 1.98(0.06) 2.00(0.08) 2.03(0.06) 2.01(0.08) 2.02(0.07) 1.94(0.13) 2.05(0.12) 2.00(0.05) 2.01(0.10) 1.98(0.08) 1.97(0.06) 1.90(0.10) 2.01(0.12) 1.91(0.11) 2.03(0.12) 2.02(0.05) 2.01(0.05) 1.95(0.12) 2.06(0.12) 1.98(0.09) 2.05(0.08) 2.09(0.06) 2.04(0.07) 2.09(0.10) 2.01(0.10) 1.99(0.05) 2.03(0.08) 2.03(0.08) 1.92(0.13) 2.05(0.18) 1.98(0.08) 1.89(0.09) 1.91(0.03) 1.91(0.06) 1.95(0.06) 1.86(0.09) 1.90(0.07) 1.84(0.11) 1.92(0.10) 1.89(0.10) 1.99(0.14) 1.95(0.06) 1.92(0.05) 1.97(0.06) 1.93(0.06) 1.97(0.05) 1.97(0.09) 1.97(0.05) 1.99(0.06) 1.91(0.10) 2.02(0.11) 1.97(0.06) 2.05(0.08) 2.10(0.09) 2.10(0.08) 2.18(0.09) 1.98(0.21)
++ Max displacement in automask = 2.69 (mm) at sub-brick 0
++ Max delta displ in automask = 0.25 (mm) at sub-brick 80
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-04.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
+ - Test (64+191)*64 params [top5=*o+-.]:#oooooo..+-o-+.+o+o+..+oooo++-o.-$o.-..o-o..+o+-.- + - best 88 costs found:
o= 0 v=-0.132055: 85.93 -4.43 -2.12 0.00 0.00 0.00 [grid]
o= 1 v=-0.037324: 65.67 22.86 -36.61 37.90 5.03 3.08 [rand]
o= 2 v=-0.032195: 65.67 22.86 -36.61 37.90 -5.03 3.08 [rand]
RMS[0] = 0.346218 0.175881 ITER = 7/50
o= 3 v=-0.029058: 70.28 -13.18 -35.96 23.16 17.17 -3.59 [rand]
o= 4 v=-0.027935: 65.67 22.86 -36.61 37.90 5.03 -3.08 [rand]
o= 5 v=-0.024923: 65.67 22.86 -36.61 37.90 -5.03 -3.08 [rand]
o= 6 v=-0.024545: 70.28 -13.18 -35.96 23.16 17.17 3.59 [rand]
0.346218
o= 7 v=-0.021835: 74.44 -20.73 -35.88 19.17 13.96 2.56 [rand]
o= 8 v=-0.021547: 74.44 -20.73 -35.88 19.17 13.96 -2.56 [rand]
o= 9 v=-0.020495: 69.15 16.35 -31.40 12.96 3.05 -9.17 [rand]
o=10 v=-0.019646: 68.87 18.27 -26.37 12.15 -12.44 -39.75 [rand]
o=11 v=-0.019565: 61.55 25.70 -35.57 27.62 21.02 5.11 [rand]
o=12 v=-0.018739: 74.44 11.88 -35.88 19.17 13.96 -2.56 [rand]
o=13 v=-0.017960: 69.15 16.35 -31.40 12.96 -3.05 -9.17 [rand]
o=14 v=-0.017774: 74.44 11.88 -35.88 19.17 -13.96 2.56 [rand]
o=15 v=-0.017539: 74.44 11.88 -35.88 19.17 13.96 2.56 [rand]
o=16 v=-0.017502: 59.26 22.24 -28.79 30.00 7.50 -7.50 [grid]
o=17 v=-0.017183: 122.52 -31.72 29.11 -25.97 -37.53 -40.21 [rand]
o=18 v=-0.017130: 61.55 25.70 -35.57 27.62 21.02 -5.11 [rand]
o=19 v=-0.016639: 66.81 18.99 -35.42 37.88 -13.88 -18.42 [rand]
o=20 v=-0.016480: 52.90 -9.44 34.69 11.98 -4.28 19.20 [rand]
o=21 v=-0.016357: 118.13 -33.47 34.02 -40.41 -19.09 -40.75 [rand]
o=22 v=-0.015563: 83.63 2.75 -33.82 -10.97 18.37 -5.04 [rand]
o=23 v=-0.015382: 69.15 16.35 -31.40 12.96 -3.05 9.17 [rand]
o=24 v=-0.015049: 66.81 18.99 -35.42 37.88 13.88 -18.42 [rand]
o=25 v=-0.015021: 79.26 -11.09 -28.79 -30.00 7.50 -7.50 [grid]
o=26 v=-0.014986: 61.55 -34.55 -35.57 27.62 21.02 5.11 [rand]
o=27 v=-0.014553: 69.15 16.35 -31.40 12.96 3.05 9.17 [rand]
o=28 v=-0.014314: 77.26 -28.58 34.45 -27.41 -42.10 24.35 [rand]
o=29 v=-0.014223: 74.44 11.88 -35.88 19.17 -13.96 -2.56 [rand]
o=30 v=-0.014172: 71.78 23.70 -25.58 27.82 2.77 -20.28 [rand]
o=31 v=-0.013992: 94.59 -28.58 34.45 -27.41 -42.10 -24.35 [rand]
o=32 v=-0.013626: 99.92 11.11 -37.81 -14.61 39.00 28.71 [rand]
o=33 v=-0.013600: 52.90 0.59 34.69 11.98 4.28 19.20 [rand]
o=34 v=-0.013291: 77.41 -33.43 -33.49 -12.98 30.57 -5.92 [rand]
o=35 v=-0.013258: 72.59 -28.67 28.67 6.84 -32.76 20.90 [rand]
o=36 v=-0.013243: 112.59 2.24 -28.79 -30.00 -7.50 30.00 [grid]
o=37 v=-0.013143: 99.26 19.82 -32.91 -6.84 32.76 20.90 [rand]
o=38 v=-0.013023: 99.09 -42.31 35.07 14.03 -8.39 -25.20 [rand]
o=39 v=-0.012881: 92.34 29.52 -38.70 -19.28 27.13 15.13 [rand]
o=40 v=-0.012712: 74.44 11.88 -35.88 -19.17 13.96 2.56 [rand]
o=41 v=-0.012676: 63.46 24.72 -32.15 27.43 11.56 -19.13 [rand]
o=42 v=-0.012555: 121.54 -8.84 -38.99 -37.76 24.22 -3.86 [rand]
o=43 v=-0.012491: 121.54 -0.01 -38.99 -37.76 24.22 3.86 [rand]
o=44 v=-0.012465: 112.59 2.24 -28.79 -30.00 7.50 7.50 [grid]
o=45 v=-0.012437: 99.92 -19.96 33.56 14.61 -39.00 -28.71 [rand]
o=46 v=-0.012424: 79.51 29.52 -38.70 19.28 -27.13 -15.13 [rand]
o=47 v=-0.012219: 60.96 -33.48 33.32 17.69 -14.58 34.92 [rand]
o=48 v=-0.012113: 81.79 -25.00 -32.18 -19.78 42.00 3.76 [rand]
o=49 v=-0.012092: 104.67 -40.82 -35.17 24.90 4.74 -17.23 [rand]
o=50 v=-0.011977: 79.26 -31.09 -28.79 -30.00 30.00 -7.50 [grid]
o=51 v=-0.011943: 79.26 -31.09 -28.79 -30.00 30.00 30.00 [grid]
o=52 v=-0.011928: 59.08 -17.52 30.46 -39.07 -28.37 8.01 [rand]
o=53 v=-0.011851: 81.79 -25.00 -32.18 -19.78 42.00 -3.76 [rand]
o=54 v=-0.011776: 74.44 11.88 -35.88 -19.17 13.96 -2.56 [rand]
o=55 v=-0.011715: 83.63 2.75 -33.82 -10.97 18.37 5.04 [rand]
o=56 v=-0.011669: 79.26 -31.09 -28.79 30.00 7.50 7.50 [grid]
o=57 v=-0.011668: 105.04 18.99 -35.42 -37.88 13.88 18.42 [rand]
o=58 v=-0.011552: 72.22 -7.11 -36.42 41.38 19.47 -31.64 [rand]
o=59 v=-0.011544: 59.00 23.71 35.82 15.05 -4.71 12.78 [rand]
o=60 v=-0.011542: 97.42 -20.73 -35.88 -19.17 13.96 2.56 [rand]
o=61 v=-0.011500: 75.47 22.84 -23.62 34.95 -5.03 -42.69 [rand]
o=62 v=-0.011479: 79.51 -38.37 -38.70 -19.28 27.13 15.13 [rand]
o=63 v=-0.011456: 52.90 -9.44 34.69 11.98 4.28 19.20 [rand]
o=64 v=-0.011445: 63.46 24.72 -32.15 27.43 11.56 19.13 [rand]
o=65 v=-0.011395: 70.28 4.33 -35.96 23.16 17.17 -3.59 [rand]
o=66 v=-0.011377: 118.95 0.59 34.69 -11.98 -4.28 -19.20 [rand]
o=67 v=-0.011377: 69.13 14.04 -34.48 10.10 -16.32 -39.31 [rand]
o=68 v=-0.011317: 99.26 -28.67 28.67 -6.84 -32.76 -20.90 [rand]
o=69 v=-0.011279: 82.16 -27.46 -12.24 23.55 35.33 17.13 [rand]
o=70 v=-0.011211: 89.69 -27.46 -12.24 23.55 35.33 17.13 [rand]
o=71 v=-0.011204: 60.55 -31.68 35.38 41.23 -14.90 41.72 [rand]
o=72 v=-0.011177: 71.93 11.11 -37.81 -14.61 -39.00 -28.71 [rand]
o=73 v=-0.011170: 121.54 -8.84 -38.99 -37.76 24.22 3.86 [rand]
o=74 v=-0.011120: 92.34 29.52 -38.70 -19.28 -27.13 -15.13 [rand]
o=75 v=-0.011114: 72.59 19.82 -32.91 -6.84 -32.76 -20.90 [rand]
o=76 v=-0.011067: 72.51 -27.11 29.21 4.37 -20.12 15.12 [rand]
Warping has converged.
o=77 v=-0.011048: 101.22 16.46 -32.51 -14.56 29.40 15.92 [rand]
o=78 v=-0.010959: 92.98 -24.24 -37.87 25.15 27.05 -27.90 [rand]
o=79 v=-0.010924: 79.26 -31.09 -28.79 30.00 30.00 -7.50 [grid]
o=80 v=-0.010896: 68.40 24.70 -21.14 22.81 23.42 -9.07 [rand]
o=81 v=-0.010824: 99.92 -19.96 33.56 -14.61 -39.00 -28.71 [rand]
o=82 v=-0.010803: 92.34 29.52 34.45 19.28 -27.13 -15.13 [rand]
o=83 v=-0.010773: 79.26 22.24 -28.79 30.00 7.50 -30.00 [grid]
o=84 v=-0.010771: 71.78 23.70 -25.58 27.82 -2.77 -20.28 [rand]
o=85 v=-0.010769: 97.42 -20.73 -35.88 -19.17 13.96 -2.56 [rand]
o=86 v=-0.010725: 99.26 -28.67 28.67 6.84 -32.76 -20.90 [rand]
o=87 v=-0.010715: 92.34 -38.37 -38.70 -19.28 27.13 -15.13 [rand]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-05_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-05_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-05_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-05_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-05_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-05_T1w_ns+orig.BRIK
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-04_T1w_ns+tlrc::WARP_DATA -I sub-04_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-05_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
+ - A little optimization:*[#16324=-0.16143] *[#16333=-0.186333] *[#16335=-0.200609] *[#16337=-0.206972] *[#16338=-0.212445] *[#16339=-0.216884] *[#16344=-0.218577] *[#16347=-0.218752] *[#16349=-0.218922] *[#16350=-0.21905] *[#16351=-0.219058] *[#16352=-0.21922] *[#16357=-0.219243] *[#16359=-0.219257] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.219257: 86.04 -4.17 0.32 -0.11 0.72 -0.49 [grid] [f=39]
o= 1 v=-0.043809: 66.11 23.30 -35.76 37.26 4.06 5.37 [rand] [f=35]
o= 2 v=-0.045247: 65.49 25.30 -34.55 40.43 4.84 4.21 [rand] [f=44]
o= 3 v=-0.032773: 74.24 -12.50 -35.02 22.19 15.80 -3.81 [rand] [f=44]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
o= 4 v=-0.046332: 65.60 23.96 -32.52 37.59 5.17 -0.87 [rand] [f=62]
++ Authored by: RW Cox
o= 5 v=-0.046148: 65.02 23.85 -34.49 36.38 6.76 3.04 [rand] [f=64]
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-05_T1w_ns+orig.HEAD
o= 6 v=-0.031542: 73.04 -13.77 -35.69 24.51 11.44 3.32 [rand] [f=50]
o= 7 v=-0.027774: 74.40 -16.75 -35.99 19.77 13.79 2.76 [rand] [f=30]
+ changing dataset view code
o= 8 v=-0.030705: 73.90 -14.87 -34.86 19.86 16.13 -2.23 [rand] [f=53]
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
o= 9 v=-0.026349: 67.69 16.05 -30.89 18.00 3.08 -8.51 [rand] [f=45]
Setting parent with 3drefit -wset sub-05_T1w_ns+orig __ats_tmp___upad15_pre.sub-05_T1w_ns+tlrc
o=10 v=-0.024515: 70.87 17.29 -26.97 17.45 -17.07 -41.49 [rand] [f=44]
o=11 v=-0.029500: 62.52 22.26 -37.87 26.76 14.90 4.35 [rand] [f=53]
o=12 v=-0.026773: 73.13 11.49 -36.18 17.90 7.73 -1.66 [rand] [f=58]
o=13 v=-0.039095: 65.36 16.32 -29.56 23.61 4.17 -3.86 [rand] [f=83]
o=14 v=-0.038198: 74.78 15.77 -39.21 17.06 -7.76 -3.71 [rand] [f=47]
o=15 v=-0.034195: 71.98 13.77 -34.85 21.59 5.31 4.83 [rand] [f=67]
o=16 v=-0.045297: 65.24 22.11 -31.83 35.33 5.06 -1.39 [grid] [f=70]
o=17 v=-0.027946: 121.45 -41.25 30.19 -32.75 -40.78 -32.75 [rand] [f=49]
o=18 v=-0.040122: 65.94 21.17 -33.95 33.81 9.50 2.59 [rand] [f=58]
o=19 v=-0.025124: 67.59 22.77 -37.92 38.44 -13.69 -17.99 [rand] [f=45]
o=20 v=-0.021463: 53.24 -9.32 35.80 12.90 -4.54 23.63 [rand] [f=43]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
o=21 v=-0.025965: 120.83 -33.54 33.54 -41.36 -23.01 -31.85 [rand] [f=57]
++ Authored by: RW Cox
o=22 v=-0.030680: 80.96 5.13 -38.84 -10.50 15.91 -4.17 [rand] [f=55]
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-05_T1w_ns+tlrc
o=23 v=-0.034671: 79.70 16.57 -38.48 2.61 -1.80 2.63 [rand] [f=81]
+ setting Warp parent
o=24 v=-0.025819: 70.05 19.43 -33.96 38.37 15.71 -17.03 [rand] [f=55]
++ 3drefit processed 1 datasets
o=25 v=-0.023807: 81.13 -11.32 -33.46 -24.61 6.54 -8.71 [grid] [f=65]
o=26 v=-0.018538: 61.90 -34.75 -34.87 27.32 24.97 4.55 [rand] [f=51]
o=27 v=-0.033531: 73.42 16.50 -35.79 21.13 2.71 4.74 [rand] [f=48]
o=28 v=-0.020206: 77.06 -28.14 34.24 -26.59 -37.89 21.59 [rand] [f=77]
o=29 v=-0.038008: 77.30 17.40 -39.51 16.13 -10.83 -1.73 [rand] [f=48]
o=30 v=-0.027814: 70.00 17.97 -23.99 26.74 2.77 -19.25 [rand] [f=49]
o=31 v=-0.023039: 94.00 -27.49 35.22 -30.71 -36.47 -24.78 [rand] [f=66]
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
o=32 v=-0.018123: 97.21 11.16 -37.72 -16.53 41.06 27.52 [rand] [f=58]
++ Warning: ignoring +tlrc on new_prefix.
o=33 v=-0.018110: 52.18 -0.14 35.48 16.99 3.23 18.88 [rand] [f=51]
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-05_T1w_ns+tlrc.HEAD -> sub-05_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-05_T1w_ns+tlrc.BRIK -> sub-05_T1w_ns+tlrc.BRIK
o=34 v=-0.021553: 83.75 -30.38 -35.61 -14.11 30.63 -5.84 [rand] [f=67]
o=35 v=-0.016863: 72.97 -33.84 29.59 6.77 -32.59 19.33 [rand] [f=51]
Cleanup ...
o=36 v=-0.018326: 111.03 2.48 -28.08 -34.28 -8.26 27.24 [grid] [f=56]
o=37 v=-0.021167: 97.63 18.82 -35.09 -4.84 38.50 15.49 [rand] [f=74]
o=38 v=-0.018027: 99.28 -43.30 35.09 9.76 -2.27 -24.45 [rand] [f=54]
o=39 v=-0.024352: 90.58 29.69 -40.17 -18.55 21.33 11.03 [rand] [f=70]
o=40 v=-0.032580: 77.26 8.02 -40.17 -13.62 10.11 -1.47 [rand] [f=54]
o=41 v=-0.039627: 65.23 22.05 -28.96 33.67 5.66 -8.61 [rand] [f=65]
o=42 v=-0.016791: 124.58 -7.78 -41.38 -37.00 22.83 -4.33 [rand] [f=51]
o=43 v=-0.019456: 125.88 0.50 -38.24 -37.75 24.17 3.51 [rand] [f=42]
o=44 v=-0.018211: 111.33 4.87 -29.70 -32.39 7.13 12.11 [grid] [f=51]
o=45 v=-0.020715: 103.71 -21.12 35.31 15.37 -39.20 -26.54 [rand] [f=62]
o=46 v=-0.018145: 78.84 34.69 -38.84 16.36 -29.41 -15.87 [rand] [f=65]
o=47 v=-0.016611: 59.08 -33.01 34.92 19.29 -16.22 34.82 [rand] [f=48]
o=48 v=-0.017471: 81.76 -31.78 -31.64 -24.68 39.27 -0.00 [rand] [f=59]
o=49 v=-0.014885: 104.46 -44.25 -35.12 25.02 5.04 -17.15 [rand] [f=46]
o=50 v=-0.014296: 79.53 -35.04 -29.20 -29.77 30.60 -7.56 [grid] [f=43]
o=51 v=-0.017193: 78.29 -35.10 -29.65 -31.46 28.89 29.38 [grid] [f=48]
o=52 v=-0.015060: 58.89 -17.33 30.11 -38.07 -26.60 13.18 [rand] [f=47]
o=53 v=-0.016952: 82.26 -30.54 -31.52 -19.94 41.82 -3.91 [rand] [f=33]
o=54 v=-0.037592: 80.34 5.11 -38.59 -18.51 14.38 -2.67 [rand] [f=72]
o=55 v=-0.033046: 81.02 4.64 -37.96 -17.38 16.74 -1.70 [rand] [f=59]
o=56 v=-0.019055: 82.67 -31.88 -27.91 31.21 9.48 7.85 [grid] [f=53]
o=57 v=-0.018705: 108.15 16.00 -33.84 -38.90 7.38 15.40 [rand] [f=75]
o=58 v=-0.017537: 68.95 -10.28 -36.34 41.00 25.02 -30.15 [rand] [f=41]
o=59 v=-0.022138: 55.30 26.73 36.13 14.33 -6.11 17.86 [rand] [f=53]
3dAllineate -base sub-04_T1w_ns+tlrc -input pb00.sub-04.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
o=60 v=-0.016212: 93.04 -21.01 -35.41 -17.84 13.07 2.98 [rand] [f=54]
o=61 v=-0.025541: 70.81 19.11 -22.60 33.64 -8.09 -42.87 [rand] [f=67]
o=62 v=-0.018477: 79.12 -29.54 -40.42 -23.03 28.05 18.43 [rand] [f=56]
o=63 v=-0.020055: 49.49 -9.01 34.44 10.95 2.95 19.22 [rand] [f=39]
o=64 v=-0.046979: 66.96 28.42 -38.28 42.96 8.07 8.88 [rand] [f=67]
o=65 v=-0.028225: 68.99 -9.72 -32.61 27.07 19.72 -1.96 [rand] [f=45]
o=66 v=-0.020326: 118.18 -2.30 37.09 -14.46 -3.21 -22.57 [rand] [f=61]
o=67 v=-0.022600: 72.73 14.19 -33.03 9.25 -17.34 -38.90 [rand] [f=54]
o=68 v=-0.016500: 98.75 -24.76 28.04 -6.40 -32.87 -21.24 [rand] [f=40]
o=69 v=-0.021018: 85.66 -28.15 -11.48 23.63 32.08 10.66 [rand] [f=59]
o=70 v=-0.019243: 86.04 -29.17 -13.80 22.56 32.61 13.24 [rand] [f=48]
o=71 v=-0.015927: 61.20 -28.36 35.50 40.81 -15.56 41.31 [rand] [f=43]
o=72 v=-0.030042: 70.96 18.74 -38.41 -10.63 -44.30 -36.27 [rand] [f=58]
o=73 v=-0.016948: 125.89 -8.38 -38.49 -38.13 24.11 3.59 [rand] [f=41]
o=74 v=-0.035622: 86.66 31.11 -39.65 -5.99 -14.17 -5.22 [rand] [f=63]
o=75 v=-0.028918: 71.25 17.67 -36.64 -9.33 -40.98 -33.95 [rand] [f=73]
o=76 v=-0.019927: 71.55 -27.21 37.23 -0.42 -20.68 11.86 [rand] [f=46]
o=77 v=-0.019985: 98.77 14.86 -38.42 -13.24 26.77 18.90 [rand] [f=64]
o=78 v=-0.015855: 97.45 -25.13 -40.21 24.59 24.00 -27.98 [rand] [f=57]
o=79 v=-0.017383: 80.28 -29.57 -35.43 29.08 30.38 -2.41 [grid] [f=63]
o=80 v=-0.022970: 66.84 21.93 -25.33 21.93 21.80 -12.21 [rand] [f=64]
o=81 v=-0.020420: 98.28 -20.84 37.87 -16.21 -39.18 -28.81 [rand] [f=37]
o=82 v=-0.015135: 92.94 33.31 35.11 18.88 -27.87 -14.96 [rand] [f=62]
o=83 v=-0.023307: 75.37 23.75 -30.42 28.67 4.45 -32.90 [grid] [f=43]
o=84 v=-0.024877: 70.51 17.69 -26.61 30.29 -3.31 -21.55 [rand] [f=58]
o=85 v=-0.012847: 98.11 -24.86 -35.52 -19.63 14.17 -2.74 [rand] [f=35]
o=86 v=-0.018006: 96.30 -28.64 28.43 6.11 -32.27 -17.86 [rand] [f=62]
o=87 v=-0.021161: 90.84 -38.56 -34.65 -20.11 26.84 -14.91 [rand] [f=62]
+ - saving # 0 for use with twobest
+ - saving #64 for use with twobest
+ - saving # 4 for use with twobest
+ - skip # 5 for twobest: too close to set # 4
+ - skip #16 for twobest: too close to set # 4
+ - skip # 2 for twobest: too close to set # 5
cat_matvec sub-05_T1w_ns+tlrc::WARP_DATA -I
+ - skip # 1 for twobest: too close to set # 5
+ - skip #18 for twobest: too close to set # 4
+ - saving #41 for use with twobest
+ - saving #13 for use with twobest
+ - saving #14 for use with twobest
+ - skip #29 for twobest: too close to set #14
+ - saving #54 for use with twobest
+ - saving #74 for use with twobest
+ - saving #23 for use with twobest
+ - saving #15 for use with twobest
+ - skip #27 for twobest: too close to set #15
+ - skip #55 for twobest: too close to set #54
+ - saving # 3 for use with twobest
+ - saving #40 for use with twobest
+ - saving # 6 for use with twobest
+ - skip # 8 for twobest: too close to set # 3
+ - saving #22 for use with twobest
+ - saving #72 for use with twobest
+ - saving #11 for use with twobest
+ - skip #75 for twobest: too close to set #72
+ - saving #65 for use with twobest
+ - saving #17 for use with twobest
+ - saving #30 for use with twobest
+ - saving # 7 for use with twobest
+ - saving #12 for use with twobest
+ - saving # 9 for use with twobest
+ - saving #21 for use with twobest
+ - saving #24 for use with twobest
+ - saving #61 for use with twobest
+ - saving #19 for use with twobest
+ - saving #84 for use with twobest
+ - saving #10 for use with twobest
+ - saving #39 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-04.r01.tcat+orig.HEAD
++ Base dataset: ./sub-04_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
if ( ! -f sub-05_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-05.r01.tcat+orig
+ !source mask fill: ubot=46 usiz=159.5
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
+ - retaining old weight image
+ - using 38472 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32264 total points stored in 66 'TOHD(17.3748)' bloks (0 duplicates)
++ Reading input dataset ./pb00.sub-05.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
*[#21213=-0.270708] *[#21221=-0.280742] *[#21235=-0.28232] *[#21238=-0.285243] *[#21243=-0.286176] *[#21245=-0.286191] *[#21247=-0.28745] *[#21248=-0.287776] *[#21252=-0.288994] *[#21255=-0.289846] *[#21256=-0.289968] *[#21258=-0.290512] *[#21261=-0.291422] *[#21266=-0.291439] *[#21270=-0.291817] *[#21273=-0.29228] *[#21276=-0.292445] *[#21277=-0.292571] *[#21280=-0.29264] *[#21281=-0.292927] *[#21282=-0.293255] *[#21283=-0.293402] *[#21286=-0.293517] *[#21287=-0.293771] *[#21288=-0.29401] *[#21289=-0.294173] *[#21292=-0.294307] *[#21293=-0.29465] *[#21294=-0.295081] *[#21296=-0.295234] *[#21299=-0.295505] *[#21300=-0.295961] *[#21301=-0.296177] *[#21303=-0.296515] *[#21304=-0.296569] *[#21307=-0.296746] *[#21308=-0.297017] *[#21309=-0.297286] *[#21310=-0.297328]
+ - param set #1 has cost=-0.297328 [o=0 t=0]
+ -- Parameters = 86.1318 -4.8659 0.0080 -0.0612 0.2899 -0.8074 0.9860 1.0324 0.9764 0.0002 0.0058 -0.0014
+ - param set #2 has cost=-0.039781 [o=64 t=1]
+ -- Parameters = 67.4132 28.5074 -37.4688 41.8316 7.4775 9.0457 0.9939 0.9985 0.9874 0.0023 0.0015 0.0098
+ - param set #3 has cost=-0.035330 [o=4 t=2]
+ -- Parameters = 65.2811 23.7634 -32.3264 37.8010 5.0870 -0.7163 0.9999 1.0047 1.0035 0.0125 -0.0008 -0.0007
+ - param set #4 has cost=-0.032177 [o=41 t=3]
+ -- Parameters = 65.4769 22.2517 -28.6832 33.5540 5.7511 -9.1109 1.0052 0.9930 0.9951 0.0006 -0.0003 -0.0011
+ - param set #5 has cost=-0.028350 [o=13 t=4]
+ -- Parameters = 65.3981 16.3708 -29.6188 23.6824 4.2071 -3.8489 1.0000 1.0003 1.0013 -0.0003 0.0009 -0.0008
+ - param set #6 has cost=-0.030200 [o=14 t=5]
+ -- Parameters = 74.6710 15.6951 -39.1803 16.9548 -7.7806 -3.6350 1.0010 1.0015 1.0000 -0.0017 0.0003 0.0006
+ - param set #7 has cost=-0.027572 [o=54 t=6]
+ -- Parameters = 80.2217 5.1126 -38.4746 -18.6080 14.4735 -2.5131 1.0033 1.0007 1.0019 0.0003 0.0001 0.0000
+ - param set #8 has cost=-0.029317 [o=74 t=7]
+ -- Parameters = 86.5984 31.2392 -39.5214 -5.7584 -14.2095 -4.9205 1.0044 1.0004 1.0023 0.0001 0.0009 -0.0004
+ - param set #9 has cost=-0.030766 [o=23 t=8]
+ -cmass x y z shifts = 5.425 -8.412 -9.735
+ -- Parameters = 77.9310 16.1949 -39.1458 1.7602 -2.4052 -0.6297 0.9630 0.9957 1.0043 -0.0003 -0.0015 -0.0030
+ shift search range is +/- = 57.780 69.336 57.780
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
+ Automask has 41011 voxels
+ 6244 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
+ - param set #10 has cost=-0.027949 [o=15 t=9]
+ -- Parameters = 72.2412 13.8793 -34.8165 21.5040 5.0016 4.7532 0.9961 0.9953 0.9985 0.0011 -0.0015 0.0002
+ - param set #11 has cost=-0.026207 [o=3 t=10]
+ -- Parameters = 74.2158 -12.4422 -35.0712 22.3160 15.6999 -3.8293 1.0079 0.9972 0.9996 -0.0005 0.0014 -0.0009
+ - param set #12 has cost=-0.025269 [o=40 t=11]
+ -- Parameters = 76.9884 8.0452 -40.3365 -13.4105 9.9293 -1.4377 1.0004 0.9935 1.0016 -0.0016 0.0030 -0.0002
+ - param set #13 has cost=-0.023215 [o=6 t=12]
+ -- Parameters = 73.0090 -13.7469 -35.7102 24.5533 11.4627 3.3518 1.0001 0.9996 1.0005 -0.0001 -0.0001 0.0001
+ - param set #14 has cost=-0.025379 [o=22 t=13]
+ -- Parameters = 80.4491 6.4645 -38.9286 -13.6104 15.0535 -3.4856 0.9933 0.9966 0.9999 0.0003 0.0021 -0.0006
+ - param set #15 has cost=-0.024776 [o=72 t=14]
+ -- Parameters = 70.8819 18.6372 -38.3427 -10.6223 -44.4524 -36.3289 0.9966 0.9999 1.0058 0.0137 -0.0004 0.0007
+ - param set #16 has cost=-0.024302 [o=11 t=15]
+ -- Parameters = 61.9268 21.8066 -37.9964 27.0722 12.8956 3.0357 0.9762 0.9843 0.9907 -0.0019 -0.0020 -0.0005
+ - param set #17 has cost=-0.027640 [o=65 t=16]
+ -- Parameters = 69.6953 -9.7770 -33.9472 27.4481 18.8804 -5.8763 0.9962 0.9842 0.9760 -0.0021 0.0023 0.0034
+ - param set #18 has cost=-0.021211 [o=17 t=17]
+ -- Parameters = 121.4419 -41.2429 30.2765 -32.8427 -40.8786 -32.8466 0.9961 0.9997 0.9976 -0.0000 0.0013 -0.0008
+ - param set #19 has cost=-0.023068 [o=30 t=18]
+ -- Parameters = 69.0969 18.5837 -23.9468 30.5282 2.0516 -19.5787 0.9915 0.9942 0.9958 0.0018 0.0011 0.0027
+ - param set #20 has cost=-0.021818 [o=7 t=19]
+ -- Parameters = 74.0888 -16.5011 -36.0601 20.0485 13.6846 2.6046 0.9999 0.9939 0.9984 0.0017 0.0002 -0.0016
+ - param set #21 has cost=-0.027874 [o=12 t=20]
+ -- Parameters = 72.9574 11.7635 -35.5821 18.1087 -1.7818 4.9854 0.9849 0.9721 1.0030 0.0048 -0.0030 -0.0070
+ - param set #22 has cost=-0.026141 [o=9 t=21]
+ -- Parameters = 66.0028 15.9751 -31.4171 21.3344 4.6534 -0.8176 0.9909 1.0060 0.9904 -0.0008 0.0040 0.0005
+ - param set #23 has cost=-0.022264 [o=21 t=22]
+ -- Parameters = 120.3743 -34.0291 33.2328 -41.8254 -23.5419 -31.8346 0.9980 0.9947 0.9933 -0.0001 0.0006 0.0006
+ - param set #24 has cost=-0.022496 [o=24 t=23]
+ -- Parameters = 69.4379 19.5743 -34.0388 38.0189 15.7291 -16.7522 0.9896 0.9819 1.0021 -0.0012 -0.0012 0.0117
+ - param set #25 has cost=-0.020163 [o=61 t=24]
+ -- Parameters = 70.9464 19.0621 -22.3284 33.6890 -8.0131 -42.9399 1.0007 0.9979 1.0020 0.0002 -0.0013 0.0007
+ - param set #26 has cost=-0.024276 [o=19 t=25]
+ -- Parameters = 72.4836 23.0564 -38.0548 37.9934 -15.8564 -19.7546 0.9844 0.9819 0.9832 -0.0014 0.0024 -0.0021
+ - param set #27 has cost=-0.019828 [o=84 t=26]
+ -- Parameters = 70.4013 17.4948 -26.6845 30.1384 -3.6087 -21.4898 1.0026 0.9954 1.0045 -0.0007 0.0006 -0.0004
+ - param set #28 has cost=-0.020907 [o=10 t=27]
+ -- Parameters = 70.9497 17.3606 -26.9706 17.4853 -17.2073 -41.5377 1.0070 0.9993 1.0045 -0.0011 -0.0000 0.0002
+ - param set #29 has cost=-0.022600 [o=39 t=28]
+ -- Parameters = 90.3470 29.6895 -39.7688 -18.8453 19.1765 5.5574 0.9940 0.9862 0.9697 -0.0011 -0.0019 -0.0014
+ - param set #30 has cost=-0.290725 [o=-1 t=-1]
+ -- Parameters = 86.1125 -4.7784 -0.1287 0.2445 0.6225 -1.0106 0.9913 1.0411 0.9832 0.0122 0.0147 0.0150
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0741
+ --- dist(#3,#1) = 0.468
+ --- dist(#4,#1) = 0.421
+ --- dist(#5,#1) = 0.374
+ --- dist(#6,#1) = 0.489
+ --- dist(#7,#1) = 0.49
+ --- dist(#8,#1) = 0.494
+ --- dist(#9,#1) = 0.37
+ --- dist(#10,#1) = 0.435
+ --- dist(#11,#1) = 0.445
+ --- dist(#12,#1) = 0.424
+ --- dist(#13,#1) = 0.481
+ --- dist(#14,#1) = 0.438
+ --- dist(#15,#1) = 0.393
+ --- dist(#16,#1) = 0.487
+ --- dist(#17,#1) = 0.504
+ --- dist(#18,#1) = 0.497
+ --- dist(#19,#1) = 0.475
+ --- dist(#20,#1) = 0.476
+ --- dist(#21,#1) = 0.446
+ --- dist(#22,#1) = 0.34
+ --- dist(#23,#1) = 0.497
+ --- dist(#24,#1) = 0.426
+ --- dist(#25,#1) = 0.464
+ --- dist(#26,#1) = 0.451
+ --- dist(#27,#1) = 0.457
+ --- dist(#28,#1) = 0.453
+ --- dist(#29,#1) = 0.468
+ --- dist(#30,#1) = 0.336
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=46 usiz=159.5
+ - retaining old weight image
+ - using 38472 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31819 total points stored in 65 'TOHD(17.2644)' bloks (0 duplicates)
*[#24087=-0.332439] *[#24109=-0.334014] *[#24110=-0.33476] *[#24113=-0.335099] *[#24114=-0.335564] *[#24115=-0.336036] *[#24116=-0.336107] *[#24117=-0.336164] *[#24120=-0.336379] *[#24127=-0.336531] *[#24129=-0.337033] *[#24131=-0.337333] *[#24138=-0.337573] *[#24142=-0.337576] *[#24146=-0.337714] *[#24152=-0.337716] *[#24153=-0.337732] *[#24158=-0.337793] *[#24159=-0.33784] *[#24164=-0.337867] *[#24165=-0.337903] *[#24166=-0.337938] *[#24169=-0.337948] *[#24173=-0.337959] *[#24175=-0.337964] *[#24176=-0.337968] *[#24179=-0.337977] *[#24180=-0.337984] *[#24181=-0.337986] *[#24183=-0.337993] *[#24184=-0.338003] *[#24185=-0.338023] *[#24186=-0.338032] *[#24187=-0.338034] *[#24188=-0.338039] *[#24193=-0.338058] *[#24195=-0.338065] *[#24196=-0.338081]
+ - param set #1 has cost=-0.338081 [o=0 t=0]
+ -- Parameters = 86.0719 -4.7322 -0.2043 0.0164 -0.1554 -0.6991 0.9879 1.0355 0.9705 -0.0066 0.0065 -0.0041
+ - param set #2 has cost=-0.330350 [o=-1 t=-1]
+ -- Parameters = 86.0818 -4.5923 -0.0937 0.4062 0.5391 -0.9507 0.9854 1.0298 0.9801 0.0106 0.0131 0.0077
+ - param set #3 has cost=-0.032499 [o=64 t=1]
+ -- Parameters = 67.3387 28.4855 -37.5574 41.7162 7.6330 9.0692 0.9961 0.9976 0.9878 0.0013 0.0016 0.0185
+ - param set #4 has cost=-0.028778 [o=4 t=2]
+ -- Parameters = 65.4349 23.7602 -32.1482 37.8756 5.0091 -0.5435 1.0062 1.0040 1.0053 0.0138 -0.0007 0.0005
+ - param set #5 has cost=-0.026053 [o=41 t=3]
+ -- Parameters = 65.4729 22.2425 -28.6147 33.6264 5.6790 -9.1427 1.0047 0.9951 0.9954 0.0002 -0.0006 -0.0016
+ - param set #6 has cost=-0.026453 [o=23 t=8]
+ -- Parameters = 77.0476 16.3790 -39.2158 4.5102 -3.1577 -0.4748 0.9601 0.9971 1.0062 -0.0002 -0.0011 -0.0030
+ - param set #7 has cost=-0.025124 [o=14 t=5]
+ -- Parameters = 74.5449 15.7172 -39.1958 16.9559 -7.8047 -3.6295 1.0020 1.0015 1.0002 -0.0017 0.0004 0.0007
+ - param set #8 has cost=-0.029414 [o=74 t=7]
+ -- Parameters = 87.4539 30.2811 -39.2960 -6.9629 -14.1270 -4.1662 1.0038 0.9766 0.9982 0.0061 0.0143 -0.0042
+ - param set #9 has cost=-0.022286 [o=13 t=4]
+ -- Parameters = 65.3616 16.4713 -29.6830 23.7458 4.2733 -3.8320 1.0002 1.0007 1.0022 -0.0009 0.0008 -0.0012
+ - param set #10 has cost=-0.023560 [o=15 t=9]
+ -- Parameters = 72.2858 14.1459 -34.9066 21.4612 5.1780 4.6900 0.9903 0.9931 0.9952 0.0014 -0.0017 -0.0001
+ - param set #11 has cost=-0.024137 [o=12 t=20]
+ -- Parameters = 72.9241 11.7032 -35.6185 18.0582 -1.8232 5.0118 0.9852 0.9720 1.0029 0.0048 -0.0029 -0.0071
+ - param set #12 has cost=-0.022554 [o=65 t=16]
+ -- Parameters = 69.8614 -9.9156 -33.9215 27.5943 18.7498 -5.9206 0.9963 0.9805 0.9783 0.0006 0.0051 0.0023
+ - param set #13 has cost=-0.022224 [o=54 t=6]
+ -- Parameters = 80.2152 5.0879 -38.4556 -18.6237 14.5005 -2.4773 1.0025 1.0013 1.0020 0.0006 -0.0003 0.0002
+ - param set #14 has cost=-0.020520 [o=3 t=10]
+ -- Parameters = 73.4718 -12.6863 -35.0250 22.3907 15.7361 -3.6360 1.0013 0.9998 0.9916 -0.0033 0.0020 -0.0001
+ - param set #15 has cost=-0.021704 [o=9 t=21]
+ -- Parameters = 65.9948 16.4017 -30.9694 21.1724 4.9177 -0.5746 0.9940 1.0063 0.9956 -0.0008 0.0121 0.0007
+ - param set #16 has cost=-0.020750 [o=22 t=13]
+ -- Parameters = 80.3024 6.6280 -38.9585 -13.6246 15.2571 -3.2554 0.9930 0.9971 1.0004 0.0020 0.0022 -0.0006
+ - param set #17 has cost=-0.020196 [o=40 t=11]
+ -- Parameters = 76.9633 8.0450 -40.1580 -13.5125 9.9561 -1.3767 1.0024 0.9944 1.0094 -0.0027 0.0034 -0.0001
+ - param set #18 has cost=-0.020988 [o=72 t=14]
+ -- Parameters = 70.7760 18.4914 -38.4616 -10.5781 -44.5415 -36.2702 0.9969 1.0029 1.0065 0.0108 -0.0024 0.0019
+ - param set #19 has cost=-0.020033 [o=11 t=15]
+ -- Parameters = 61.5614 22.1545 -38.0919 27.2557 13.1193 3.2031 0.9815 0.9841 0.9896 -0.0026 -0.0030 -0.0015
+ - param set #20 has cost=-0.019581 [o=19 t=25]
+ -- Parameters = 72.4419 23.0872 -38.0267 37.8996 -15.9176 -19.7231 0.9841 0.9832 0.9830 -0.0000 0.0027 -0.0023
+ - param set #21 has cost=-0.018519 [o=6 t=12]
+ -- Parameters = 72.9216 -13.7863 -35.6588 24.5373 11.3968 3.4179 0.9986 0.9993 1.0012 -0.0006 -0.0004 0.0010
+ - param set #22 has cost=-0.019430 [o=30 t=18]
+ -- Parameters = 69.0917 18.6219 -23.9418 30.5466 2.0865 -19.5694 0.9920 0.9949 0.9956 0.0027 0.0009 0.0023
+ - param set #23 has cost=-0.019817 [o=39 t=28]
+ -- Parameters = 90.3367 29.6708 -39.7934 -18.8728 19.0854 5.5997 0.9934 0.9870 0.9698 -0.0006 -0.0016 -0.0011
+ - param set #24 has cost=-0.019374 [o=24 t=23]
+ -- Parameters = 69.3021 19.6492 -33.9776 38.2211 15.6856 -16.7290 0.9916 0.9760 1.0026 -0.0002 0.0006 0.0116
+ - param set #25 has cost=-0.018824 [o=21 t=22]
+ -- Parameters = 120.4674 -34.0472 33.1131 -41.8390 -23.7920 -32.1300 0.9918 0.9964 0.9863 -0.0000 0.0018 0.0008
+ - param set #26 has cost=-0.017345 [o=7 t=19]
+ -- Parameters = 74.1180 -16.4735 -36.0837 20.0032 13.5851 2.6289 0.9997 0.9909 1.0000 0.0031 0.0022 -0.0019
+ - param set #27 has cost=-0.024436 [o=17 t=17]
+ -- Parameters = 120.7040 -41.2570 29.3889 -34.4992 -40.1308 -30.3254 0.9234 0.9322 0.9079 -0.0014 0.0077 -0.0103
+ - param set #28 has cost=-0.016244 [o=10 t=27]
+ -- Parameters = 70.6672 17.5519 -26.8621 17.4809 -17.1231 -41.4149 1.0135 0.9965 1.0051 -0.0028 -0.0018 -0.0004
+ - param set #29 has cost=-0.016709 [o=61 t=24]
+ -- Parameters = 71.1109 18.8284 -22.3325 33.5124 -8.1701 -43.1118 1.0058 0.9989 1.0066 -0.0011 -0.0010 -0.0001
+ - param set #30 has cost=-0.017532 [o=84 t=26]
+ -- Parameters = 70.3593 17.5648 -26.6703 30.2102 -3.6059 -21.4719 1.0013 0.9947 1.0050 -0.0005 0.0005 -0.0008
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0774
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.489
+ --- dist(#5,#1) = 0.421
+ --- dist(#6,#1) = 0.488
+ --- dist(#7,#1) = 0.373
+ --- dist(#8,#1) = 0.487
+ --- dist(#9,#1) = 0.457
+ --- dist(#10,#1) = 0.443
+ --- dist(#11,#1) = 0.434
+ --- dist(#12,#1) = 0.421
+ --- dist(#13,#1) = 0.368
+ --- dist(#14,#1) = 0.478
+ --- dist(#15,#1) = 0.385
+ --- dist(#16,#1) = 0.493
+ --- dist(#17,#1) = 0.484
+ --- dist(#18,#1) = 0.435
+ --- dist(#19,#1) = 0.499
+ --- dist(#20,#1) = 0.474
+ --- dist(#21,#1) = 0.495
+ --- dist(#22,#1) = 0.473
+ --- dist(#23,#1) = 0.339
+ --- dist(#24,#1) = 0.424
+ --- dist(#25,#1) = 0.465
+ --- dist(#26,#1) = 0.443
+ --- dist(#27,#1) = 0.335
+ --- dist(#28,#1) = 0.448
+ --- dist(#29,#1) = 0.471
+ --- dist(#30,#1) = 0.452
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=46 usiz=159.5
+ - retaining old weight image
+ - using 38472 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31989 total points stored in 66 'TOHD(17.1973)' bloks (0 duplicates)
*[#27186=-0.344111] *[#27208=-0.344468] *[#27211=-0.344898] *[#27214=-0.345074] *[#27215=-0.345084] *[#27217=-0.345144] *[#27218=-0.345242] *[#27221=-0.345287] *[#27222=-0.345351] *[#27223=-0.345438] *[#27228=-0.345542] *[#27243=-0.34559] *[#27247=-0.345601] *[#27248=-0.345615] *[#27250=-0.345654] *[#27252=-0.34566] *[#27253=-0.34567] *[#27254=-0.345682] *[#27255=-0.345693] *[#27257=-0.345704] *[#27258=-0.345714] *[#27259=-0.345728] *[#27260=-0.345747] *[#27261=-0.345758] *[#27262=-0.345759] *[#27263=-0.34577] *[#27265=-0.345784] *[#27266=-0.345785] *[#27268=-0.345786] *[#27270=-0.34579] *[#27271=-0.345799] *[#27272=-0.345807] *[#27273=-0.345824] *[#27274=-0.345827] *[#27275=-0.345843] *[#27276=-0.345878] *[#27277=-0.345912] *[#27278=-0.34595] *[#27281=-0.346033] *[#27282=-0.346043] *[#27283=-0.346082] *[#27284=-0.346117] *[#27289=-0.34616] *[#27293=-0.346161] *[#27294=-0.346177] *[#27295=-0.346182] *[#27296=-0.346182] *[#27298=-0.346194] *[#27301=-0.346202] *[#27302=-0.346205] *[#27303=-0.346217] *[#27304=-0.346222] *[#27306=-0.346225]
+ - param set #1 has cost=-0.346225 [o=0 t=0]
+ -- Parameters = 86.0092 -4.7193 -0.1840 0.0802 -0.6440 -0.6101 0.9922 1.0341 0.9762 -0.0063 0.0065 -0.0138
+ - param set #2 has cost=-0.345873 [o=-1 t=-1]
+ -- Parameters = 85.9876 -4.7599 -0.1238 0.1885 -0.7792 -0.4840 0.9923 1.0333 0.9781 -0.0035 0.0023 -0.0170
+ - param set #3 has cost=-0.026991 [o=64 t=1]
+ -- Parameters = 67.3392 28.5605 -37.5391 41.7147 7.6860 8.9952 0.9964 0.9963 0.9899 0.0009 0.0013 0.0186
+ - param set #4 has cost=-0.027376 [o=74 t=7]
+ -- Parameters = 87.4141 30.2891 -39.3244 -6.9542 -14.1410 -4.1183 1.0041 0.9762 0.9955 0.0067 0.0136 -0.0044
+ - param set #5 has cost=-0.023018 [o=4 t=2]
+ -- Parameters = 65.5606 23.7458 -32.0908 37.9299 4.9735 -0.6101 1.0076 1.0041 1.0067 0.0145 -0.0007 0.0005
+ - param set #6 has cost=-0.023337 [o=23 t=8]
+ -- Parameters = 77.3465 16.9018 -38.8605 4.4142 -2.8887 -0.7429 0.9599 0.9931 1.0038 -0.0002 -0.0017 -0.0049
+ - param set #7 has cost=-0.021409 [o=41 t=3]
+ -- Parameters = 65.7081 22.2649 -28.5861 33.6063 5.6494 -9.1576 1.0047 0.9946 0.9965 0.0001 -0.0005 -0.0019
+ - param set #8 has cost=-0.023588 [o=14 t=5]
+ -- Parameters = 73.6803 16.1054 -38.9380 16.5473 -7.6753 -2.6601 0.9727 1.0031 1.0063 -0.0023 -0.0003 -0.0028
+ - param set #9 has cost=-0.020316 [o=17 t=17]
+ -- Parameters = 120.6949 -41.4107 29.4949 -34.3125 -40.0569 -30.3693 0.9213 0.9338 0.9095 -0.0012 0.0137 -0.0099
+ - param set #10 has cost=-0.021290 [o=12 t=20]
+ -- Parameters = 72.8048 11.2429 -35.6995 17.8722 -2.3568 5.1614 0.9893 0.9725 1.0076 0.0053 -0.0037 -0.0074
+ - param set #11 has cost=-0.019713 [o=15 t=9]
+ -- Parameters = 72.4543 14.2053 -34.8442 21.4951 5.1406 4.6484 0.9873 0.9909 0.9961 0.0009 -0.0014 0.0004
+ - param set #12 has cost=-0.018944 [o=65 t=16]
+ -- Parameters = 69.9531 -9.9037 -34.2405 27.7053 19.4308 -5.8660 0.9995 0.9764 0.9815 0.0074 0.0066 0.0027
+ - param set #13 has cost=-0.019156 [o=13 t=4]
+ -- Parameters = 64.9646 16.2966 -29.5952 23.6199 4.3918 -3.6354 0.9760 1.0100 1.0087 -0.0023 -0.0011 0.0014
+ - param set #14 has cost=-0.017647 [o=54 t=6]
+ -- Parameters = 80.3008 5.0756 -38.2655 -18.7597 14.6951 -2.4583 1.0032 1.0035 1.0008 0.0000 -0.0013 -0.0006
+ - param set #15 has cost=-0.018595 [o=9 t=21]
+ -- Parameters = 66.0200 16.4685 -30.9586 21.2262 5.1934 -0.5059 0.9961 1.0060 0.9944 -0.0013 0.0111 0.0071
+ - param set #16 has cost=-0.017821 [o=72 t=14]
+ -- Parameters = 70.6824 18.3391 -38.3817 -10.5271 -44.5951 -36.1687 0.9958 1.0040 1.0066 0.0102 -0.0021 0.0019
+ - param set #17 has cost=-0.017057 [o=22 t=13]
+ -- Parameters = 80.3181 6.5936 -39.0380 -13.6746 15.2203 -3.0308 0.9921 0.9970 1.0005 0.0024 0.0022 -0.0005
+ - param set #18 has cost=-0.018131 [o=3 t=10]
+ -- Parameters = 73.0379 -12.4889 -34.9286 22.2350 15.8433 -3.8630 0.9788 1.0008 0.9952 -0.0035 0.0023 -0.0009
+ - param set #19 has cost=-0.020256 [o=40 t=11]
+ -- Parameters = 77.4621 9.1483 -41.0941 -10.4806 10.5919 -0.8414 1.0092 0.9943 1.0069 -0.0013 0.0035 -0.0005
+ - param set #20 has cost=-0.015972 [o=11 t=15]
+ -- Parameters = 61.2939 22.1059 -37.8988 27.1648 13.0760 3.2489 0.9851 0.9835 0.9914 -0.0030 -0.0039 -0.0023
+ - param set #21 has cost=-0.017862 [o=39 t=28]
+ -- Parameters = 90.2094 29.6743 -39.9323 -18.9471 18.8658 5.8258 0.9955 0.9870 0.9676 -0.0003 -0.0016 -0.0012
+ - param set #22 has cost=-0.016598 [o=19 t=25]
+ -- Parameters = 72.4189 23.0813 -38.0120 37.8974 -15.9184 -19.7048 0.9840 0.9831 0.9830 -0.0000 0.0027 -0.0023
+ - param set #23 has cost=-0.015722 [o=30 t=18]
+ -- Parameters = 69.0432 18.6183 -24.0638 30.5725 2.0435 -19.5258 0.9939 0.9944 0.9945 0.0030 0.0011 0.0024
+ - param set #24 has cost=-0.016454 [o=24 t=23]
+ -- Parameters = 69.5318 19.7446 -34.2407 38.4266 15.1332 -17.2546 0.9954 0.9650 0.9992 -0.0001 0.0104 0.0135
+ - param set #25 has cost=-0.016888 [o=21 t=22]
+ -- Parameters = 120.4425 -34.0477 33.0641 -41.8288 -23.5972 -32.2381 0.9913 0.9950 0.9861 0.0002 0.0027 0.0061
+ - param set #26 has cost=-0.016666 [o=6 t=12]
+ -- Parameters = 72.5753 -14.5513 -35.3208 24.5763 8.4733 3.7769 1.0006 0.9950 1.0110 0.0004 -0.0005 0.0018
+ - param set #27 has cost=-0.013961 [o=84 t=26]
+ -- Parameters = 70.3541 17.5682 -26.6270 30.2870 -3.7479 -21.5689 0.9996 0.9964 1.0049 0.0046 0.0004 -0.0010
+ - param set #28 has cost=-0.014898 [o=7 t=19]
+ -- Parameters = 74.1295 -16.3872 -36.2110 19.9510 13.4401 2.6377 0.9972 0.9888 0.9997 0.0025 0.0074 -0.0016
+ - param set #29 has cost=-0.016026 [o=61 t=24]
+ -- Parameters = 71.0594 16.7544 -22.5131 33.5734 -8.5559 -43.2616 1.0310 1.0030 0.9799 -0.0005 -0.0077 0.0015
+ - param set #30 has cost=-0.013667 [o=10 t=27]
+ -- Parameters = 70.6642 17.7215 -26.9460 17.6386 -17.0338 -41.4205 1.0131 0.9950 1.0045 -0.0027 -0.0012 0.0058
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0187 XXX
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.484
+ --- dist(#5,#1) = 0.483
+ --- dist(#6,#1) = 0.421
+ --- dist(#7,#1) = 0.373
+ --- dist(#8,#1) = 0.444
+ --- dist(#9,#1) = 0.459
+ --- dist(#10,#1) = 0.511
+ --- dist(#11,#1) = 0.433
+ --- dist(#12,#1) = 0.368
+ --- dist(#13,#1) = 0.426
+ --- dist(#14,#1) = 0.385
+ --- dist(#15,#1) = 0.434
+ --- dist(#16,#1) = 0.497
+ --- dist(#17,#1) = 0.488
+ --- dist(#18,#1) = 0.476
+ --- dist(#19,#1) = 0.486
+ --- dist(#20,#1) = 0.466
+ --- dist(#21,#1) = 0.439
+ --- dist(#22,#1) = 0.473
+ --- dist(#23,#1) = 0.426
+ --- dist(#24,#1) = 0.474
+ --- dist(#25,#1) = 0.471
+ --- dist(#26,#1) = 0.339
+ --- dist(#27,#1) = 0.45
+ --- dist(#28,#1) = 0.336
+ --- dist(#29,#1) = 0.453
+ - cast out 1 parameter set for being too close to best set
+ - Total coarse refinement net CPU time = 0.0 s; 9466 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=46 usiz=159.5
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 30 cases
+ 31780 total points stored in 66 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.343483 * [o=0 t=0]
+ -- Parameters = 86.0092 -4.7193 -0.1840 0.0802 -0.6440 -0.6101 0.9922 1.0341 0.9762 -0.0063 0.0065 -0.0138
+ - cost(#2)=-0.024773 [o=74 t=7]
+ -- Parameters = 87.4141 30.2891 -39.3244 -6.9542 -14.1410 -4.1183 1.0041 0.9762 0.9955 0.0067 0.0136 -0.0044
+ - cost(#3)=-0.024933 [o=64 t=1]
+ -- Parameters = 67.3392 28.5605 -37.5391 41.7147 7.6860 8.9952 0.9964 0.9963 0.9899 0.0009 0.0013 0.0186
+ - cost(#4)=-0.021618 [o=14 t=5]
+ -- Parameters = 73.6803 16.1054 -38.9380 16.5473 -7.6753 -2.6601 0.9727 1.0031 1.0063 -0.0023 -0.0003 -0.0028
+ - cost(#5)=-0.021245 [o=23 t=8]
+ -- Parameters = 77.3465 16.9018 -38.8605 4.4142 -2.8887 -0.7429 0.9599 0.9931 1.0038 -0.0002 -0.0017 -0.0049
+ - cost(#6)=-0.021236 [o=4 t=2]
+ -- Parameters = 65.5606 23.7458 -32.0908 37.9299 4.9735 -0.6101 1.0076 1.0041 1.0067 0.0145 -0.0007 0.0005
+ - cost(#7)=-0.020130 [o=41 t=3]
+ -- Parameters = 65.7081 22.2649 -28.5861 33.6063 5.6494 -9.1576 1.0047 0.9946 0.9965 0.0001 -0.0005 -0.0019
+ - cost(#8)=-0.019897 [o=12 t=20]
+ -- Parameters = 72.8048 11.2429 -35.6995 17.8722 -2.3568 5.1614 0.9893 0.9725 1.0076 0.0053 -0.0037 -0.0074
+ - cost(#9)=-0.018775 [o=17 t=17]
+ -- Parameters = 120.6949 -41.4107 29.4949 -34.3125 -40.0569 -30.3693 0.9213 0.9338 0.9095 -0.0012 0.0137 -0.0099
+ - cost(#10)=-0.020278 [o=40 t=11]
+ -- Parameters = 77.4621 9.1483 -41.0941 -10.4806 10.5919 -0.8414 1.0092 0.9943 1.0069 -0.0013 0.0035 -0.0005
+ - cost(#11)=-0.017781 [o=15 t=9]
+ -- Parameters = 72.4543 14.2053 -34.8442 21.4951 5.1406 4.6484 0.9873 0.9909 0.9961 0.0009 -0.0014 0.0004
+ - cost(#12)=-0.017983 [o=13 t=4]
+ -- Parameters = 64.9646 16.2966 -29.5952 23.6199 4.3918 -3.6354 0.9760 1.0100 1.0087 -0.0023 -0.0011 0.0014
+ - cost(#13)=-0.017649 [o=65 t=16]
+ -- Parameters = 69.9531 -9.9037 -34.2405 27.7053 19.4308 -5.8660 0.9995 0.9764 0.9815 0.0074 0.0066 0.0027
+ - cost(#14)=-0.017169 [o=9 t=21]
+ -- Parameters = 66.0200 16.4685 -30.9586 21.2262 5.1934 -0.5059 0.9961 1.0060 0.9944 -0.0013 0.0111 0.0071
+ - cost(#15)=-0.017790 [o=3 t=10]
+ -- Parameters = 73.0379 -12.4889 -34.9286 22.2350 15.8433 -3.8630 0.9788 1.0008 0.9952 -0.0035 0.0023 -0.0009
+ - cost(#16)=-0.016590 [o=39 t=28]
+ -- Parameters = 90.2094 29.6743 -39.9323 -18.9471 18.8658 5.8258 0.9955 0.9870 0.9676 -0.0003 -0.0016 -0.0012
+ - cost(#17)=-0.016600 [o=72 t=14]
+ -- Parameters = 70.6824 18.3391 -38.3817 -10.5271 -44.5951 -36.1687 0.9958 1.0040 1.0066 0.0102 -0.0021 0.0019
+ - cost(#18)=-0.017654 [o=54 t=6]
+ -- Parameters = 80.3008 5.0756 -38.2655 -18.7597 14.6951 -2.4583 1.0032 1.0035 1.0008 0.0000 -0.0013 -0.0006
+ - cost(#19)=-0.017565 [o=22 t=13]
+ -- Parameters = 80.3181 6.5936 -39.0380 -13.6746 15.2203 -3.0308 0.9921 0.9970 1.0005 0.0024 0.0022 -0.0005
+ - cost(#20)=-0.016830 [o=21 t=22]
+ -- Parameters = 120.4425 -34.0477 33.0641 -41.8288 -23.5972 -32.2381 0.9913 0.9950 0.9861 0.0002 0.0027 0.0061
+ - cost(#21)=-0.015950 [o=6 t=12]
+ -- Parameters = 72.5753 -14.5513 -35.3208 24.5763 8.4733 3.7769 1.0006 0.9950 1.0110 0.0004 -0.0005 0.0018
+ - cost(#22)=-0.015967 [o=19 t=25]
+ -- Parameters = 72.4189 23.0813 -38.0120 37.8974 -15.9184 -19.7048 0.9840 0.9831 0.9830 -0.0000 0.0027 -0.0023
+ - cost(#23)=-0.014986 [o=24 t=23]
+ -- Parameters = 69.5318 19.7446 -34.2407 38.4266 15.1332 -17.2546 0.9954 0.9650 0.9992 -0.0001 0.0104 0.0135
+ - cost(#24)=-0.014755 [o=61 t=24]
+ -- Parameters = 71.0594 16.7544 -22.5131 33.5734 -8.5559 -43.2616 1.0310 1.0030 0.9799 -0.0005 -0.0077 0.0015
+ - cost(#25)=-0.015095 [o=11 t=15]
+ -- Parameters = 61.2939 22.1059 -37.8988 27.1648 13.0760 3.2489 0.9851 0.9835 0.9914 -0.0030 -0.0039 -0.0023
+ - cost(#26)=-0.014600 [o=30 t=18]
+ -- Parameters = 69.0432 18.6183 -24.0638 30.5725 2.0435 -19.5258 0.9939 0.9944 0.9945 0.0030 0.0011 0.0024
+ - cost(#27)=-0.014531 [o=7 t=19]
+ -- Parameters = 74.1295 -16.3872 -36.2110 19.9510 13.4401 2.6377 0.9972 0.9888 0.9997 0.0025 0.0074 -0.0016
+ - cost(#28)=-0.012870 [o=84 t=26]
+ -- Parameters = 70.3541 17.5682 -26.6270 30.2870 -3.7479 -21.5689 0.9996 0.9964 1.0049 0.0046 0.0004 -0.0010
+ - cost(#29)=-0.012413 [o=10 t=27]
+ -- Parameters = 70.6642 17.7215 -26.9460 17.6386 -17.0338 -41.4205 1.0131 0.9950 1.0045 -0.0027 -0.0012 0.0058
+ - cost(#30)=-0.184701 [o=-2 t=-2]
+ -- Parameters = 85.9256 -4.4255 -2.1235 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 30 cases
+ - cost(#1)=-0.343691 * [o=0 t=0]
+ -- Parameters = 86.0174 -4.7121 -0.2082 0.0771 -0.6886 -0.5639 0.9911 1.0335 0.9769 -0.0064 0.0062 -0.0139
+ - cost(#2)=-0.024883 [o=74 t=7]
+ -- Parameters = 87.3978 30.3733 -39.3387 -6.9651 -14.0612 -4.0797 1.0043 0.9760 0.9926 0.0069 0.0137 -0.0044
+ - cost(#3)=-0.025416 [o=64 t=1]
+ -- Parameters = 67.3432 28.6034 -37.5384 41.5289 7.6880 9.0428 0.9950 0.9946 0.9916 -0.0065 0.0017 0.0186
+ - cost(#4)=-0.023484 [o=14 t=5]
+ -- Parameters = 74.0346 16.3431 -39.1605 16.2805 -7.2460 -1.0110 0.9675 1.0084 1.0015 -0.0043 -0.0038 -0.0033
+ - cost(#5)=-0.021359 [o=23 t=8]
+ -- Parameters = 77.4224 16.8979 -38.9079 4.4465 -2.8862 -0.8547 0.9613 0.9954 1.0040 0.0006 -0.0021 -0.0052
+ - cost(#6)=-0.021440 [o=4 t=2]
+ -- Parameters = 65.5651 23.7503 -32.0199 37.9880 5.0648 -0.5729 1.0081 1.0042 1.0074 0.0227 -0.0009 0.0008
+ - cost(#7)=-0.020292 [o=41 t=3]
+ -- Parameters = 65.7415 22.2869 -28.5425 33.5763 5.6863 -9.1654 1.0059 0.9944 1.0011 0.0008 0.0007 -0.0015
+ - cost(#8)=-0.019942 [o=12 t=20]
+ -- Parameters = 72.8161 11.2337 -35.7095 17.8893 -2.3374 5.1537 0.9892 0.9737 1.0076 0.0057 -0.0037 -0.0074
+ - cost(#9)=-0.019000 [o=17 t=17]
+ -- Parameters = 120.5448 -41.4025 29.5340 -33.7751 -40.0432 -29.9795 0.9156 0.9332 0.9028 -0.0008 0.0167 -0.0106
+ - cost(#10)=-0.020447 [o=40 t=11]
+ -- Parameters = 77.5162 9.0762 -41.0567 -10.5338 10.6445 -0.8267 1.0092 0.9962 1.0073 -0.0021 0.0034 -0.0004
+ - cost(#11)=-0.021410 [o=15 t=9]
+ -- Parameters = 75.8390 17.3423 -34.9030 21.3770 5.5737 4.9256 0.9792 0.9958 0.9975 0.0016 -0.0007 0.0036
+ - cost(#12)=-0.018163 [o=13 t=4]
+ -- Parameters = 64.8229 16.2660 -29.5810 23.6992 4.2631 -3.6516 0.9758 1.0099 1.0091 -0.0028 -0.0014 0.0009
+ - cost(#13)=-0.017931 [o=65 t=16]
+ -- Parameters = 69.9138 -9.8590 -33.9870 27.7780 19.5540 -5.9913 0.9925 0.9790 0.9851 0.0084 0.0087 0.0041
+ - cost(#14)=-0.017394 [o=9 t=21]
+ -- Parameters = 65.9975 16.4795 -30.9009 21.2225 5.2972 -0.5801 0.9946 1.0084 0.9937 -0.0023 0.0111 0.0089
+ - cost(#15)=-0.017894 [o=3 t=10]
+ -- Parameters = 73.0708 -12.4074 -34.8032 22.2573 15.8719 -3.9049 0.9785 1.0002 0.9952 -0.0025 0.0022 -0.0017
+ - cost(#16)=-0.016829 [o=39 t=28]
+ -- Parameters = 90.2267 29.5518 -39.8530 -19.0186 18.7733 5.8209 0.9934 0.9888 0.9693 0.0008 -0.0011 0.0061
+ - cost(#17)=-0.018764 [o=72 t=14]
+ -- Parameters = 70.0136 16.8300 -39.4725 -9.2499 -44.9985 -36.8251 0.9874 0.9983 1.0040 0.0043 -0.0036 0.0037
+ - cost(#18)=-0.017683 [o=54 t=6]
+ -- Parameters = 80.3170 5.0694 -38.2816 -18.7457 14.6955 -2.4658 1.0037 1.0033 1.0007 0.0007 -0.0013 -0.0004
+ - cost(#19)=-0.017610 [o=22 t=13]
+ -- Parameters = 80.2411 6.5986 -39.0005 -13.6599 15.2136 -3.0032 0.9912 0.9976 1.0009 0.0031 0.0018 -0.0003
+ - cost(#20)=-0.016963 [o=21 t=22]
+ -- Parameters = 120.4445 -34.0975 33.1048 -41.7215 -23.5205 -32.3114 0.9925 0.9922 0.9844 0.0002 0.0042 0.0067
+ - cost(#21)=-0.016066 [o=6 t=12]
+ -- Parameters = 72.5017 -14.5278 -35.3541 24.5767 8.4251 3.8248 1.0016 0.9935 1.0111 -0.0001 -0.0005 0.0023
+ - cost(#22)=-0.016014 [o=19 t=25]
+ -- Parameters = 72.4258 23.0939 -37.9572 37.8854 -15.8953 -19.7325 0.9838 0.9827 0.9833 -0.0002 0.0028 -0.0033
+ - cost(#23)=-0.015274 [o=24 t=23]
+ -- Parameters = 69.5474 19.6486 -34.3038 38.3950 15.0327 -17.3136 0.9954 0.9652 0.9986 0.0003 0.0180 0.0139
+ - cost(#24)=-0.014986 [o=61 t=24]
+ -- Parameters = 71.0588 16.7338 -22.4895 33.5807 -8.5529 -43.2697 1.0269 1.0029 0.9800 -0.0077 -0.0075 0.0013
+ - cost(#25)=-0.015140 [o=11 t=15]
+ -- Parameters = 61.2649 22.0816 -37.8493 27.1782 13.0301 3.3111 0.9849 0.9833 0.9913 -0.0029 -0.0045 -0.0024
+ - cost(#26)=-0.018278 [o=30 t=18]
+ -- Parameters = 68.8675 16.9891 -22.0428 30.2832 2.8211 -18.5739 0.9946 1.0028 0.9807 0.0029 0.0022 0.0103
+ - cost(#27)=-0.016613 [o=7 t=19]
+ -- Parameters = 72.1181 -15.2475 -36.0244 19.4467 12.8226 3.1586 0.9865 0.9885 0.9984 -0.0045 0.0062 -0.0012
+ - cost(#28)=-0.013007 [o=84 t=26]
+ -- Parameters = 70.2366 17.4814 -26.5970 30.3142 -3.6900 -21.4837 1.0003 1.0003 1.0062 0.0026 0.0004 0.0000
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
+ - cost(#29)=-0.012516 [o=10 t=27]
+ -- Parameters = 70.6268 17.8060 -26.9689 17.6197 -16.9789 -41.4464 1.0132 0.9927 1.0038 -0.0024 -0.0003 0.0056
+ - cost(#30)=-0.341904 [o=-2 t=-2]
+ -- Parameters = 85.9187 -4.6619 -0.2747 0.2497 -0.3036 -0.4489 0.9903 1.0342 0.9755 0.0008 0.0021 -0.0046
+ - case #1 [o=0 t=0] is now the best
+ - Initial cost = -0.343691
+ - Initial fine Parameters = 86.0174 -4.7121 -0.2082 0.0771 -0.6886 -0.5639 0.9911 1.0335 0.9769 -0.0064 0.0062 -0.0139
+ - Finalish cost = -0.343753 ; 211 funcs
+ - ini Finalish Parameters = 86.0139 -4.7314 -0.2228 0.1008 -0.7197 -0.5897 0.9904 1.0336 0.9761 -0.0058 0.0061 -0.0142
+ - Final cost = -0.343754 ; 276 funcs
+ Final fine fit Parameters:
x-shift= 86.0143 y-shift= -4.7300 z-shift= -0.2226 ... enorm= 86.1446 mm
z-angle= 0.1017 x-angle= -0.7210 y-angle= -0.5902 ... total= 0.9375 deg
x-scale= 0.9904 y-scale= 1.0336 z-scale= 0.9761 ... vol3D= 0.9992 = base bigger than source
y/x-shear= -0.0058 z/x-shear= 0.0060 z/y-shear= -0.0142
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.0143 -4.7300 -0.2226 0.1017 -0.7210 -0.5902 0.9904 1.0336 0.9761 -0.0058 0.0060 -0.0142
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 6.2
++ ###########################################################
3dAllineate -base sub-04_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ Unloading unneeded data
++ Output dataset ./sub-06_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-06_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 18.8
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/__tt_sub-06_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/sub-06_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-06_T1w_ns+orig sub-06_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 5.487 -17.680 -9.546
+ shift search range is +/- = 57.780 69.336 57.780
+ 9.5% 25.5% 16.5%
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-06_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-06_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-06_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-06_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
Performing center alignment with @Align_Centers
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.036 pitch=-0.727 yaw=-0.182 dS=-0.799 dL=-0.160 dP=+0.206
++ Mean: roll=+0.034 pitch=-0.450 yaw=+0.016 dS=-0.506 dL=-0.079 dP=+0.311
++ Max : roll=+0.139 pitch=-0.065 yaw=+0.139 dS=-0.307 dL=+0.059 dP=+0.379
++ Max displacements (mm) for each sub-brick:
1.89(0.00) 1.82(0.08) 1.81(0.08) 1.81(0.03) 1.82(0.03) 1.79(0.04) 1.73(0.07) 1.75(0.04) 1.79(0.12) 1.81(0.02) 1.79(0.04) 1.73(0.11) 1.73(0.05) 1.74(0.03) 1.71(0.08) 1.73(0.06) 1.72(0.05) 1.69(0.04) 1.45(0.33) 1.58(0.30) 1.62(0.14) 1.57(0.09) 1.58(0.06) 1.64(0.13) 1.64(0.07) 1.62(0.07) 1.63(0.03) 1.55(0.09) 1.58(0.05) 1.54(0.11) 1.55(0.11) 1.58(0.09) 1.60(0.02) 1.56(0.09) 1.58(0.03) 1.63(0.14) 1.59(0.08) 1.56(0.05) 1.56(0.06) 1.55(0.03) 1.60(0.07) 1.55(0.10) 1.55(0.06) 1.59(0.08) 1.57(0.07) 1.55(0.04) 1.56(0.06) 1.50(0.06) 1.53(0.03) 1.50(0.04) 1.50(0.07) 1.49(0.13) 1.49(0.11) 1.54(0.06) 1.53(0.09) 1.50(0.07) 1.53(0.08) 1.46(0.08) 1.48(0.05) 1.42(0.07) 1.46(0.07) 1.46(0.03) 1.43(0.07) 1.40(0.07) 1.43(0.12) 1.39(0.11) 1.37(0.10) 1.35(0.11) 1.36(0.10) 1.33(0.08) 1.32(0.06) 1.31(0.03) 1.29(0.09) 1.24(0.05) 1.25(0.06) 1.21(0.06) 1.26(0.07) 1.25(0.06) 1.23(0.05) 1.27(0.04) 1.25(0.04) 1.23(0.06) 1.20(0.08) 1.19(0.03) 1.24(0.05) 1.15(0.15) 1.03(0.33) 0.86(0.47) 0.90(0.41) 0.91(0.10) 0.87(0.06) 0.87(0.07) 0.87(0.03) 0.87(0.10) 0.89(0.10) 0.83(0.08) 0.85(0.08) 0.84(0.07) 0.85(0.04) 0.85(0.04) 0.91(0.07) 0.87(0.06) 0.90(0.07) 0.87(0.05) 0.93(0.06) 0.90(0.06) 0.90(0.22) 0.97(0.09) 0.91(0.07) 0.94(0.07) 0.94(0.09) 0.94(0.12) 0.93(0.07) 0.94(0.04) 0.95(0.07) 0.93(0.10) 0.88(0.11) 0.96(0.09) 0.92(0.05) 0.93(0.05) 0.88(0.09) 0.89(0.03) 0.86(0.05) 0.91(0.06) 0.87(0.05) 0.87(0.05) 0.90(0.06) 0.85(0.07) 0.85(0.05) 0.82(0.07) 0.86(0.10) 0.87(0.05) 0.84(0.04) 0.85(0.08) 0.80(0.06) 0.81(0.03) 0.79(0.05) 0.83(0.09) 0.87(0.07) 0.85(0.07) 0.80(0.06) 0.77(0.09) 0.79(0.06) 0.73(0.07) 0.76(0.04) 0.71(0.05)
++ Max displacement in automask = 1.89 (mm) at sub-brick 0
++ Max delta displ in automask = 0.47 (mm) at sub-brick 87
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-05.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-06_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-06_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-06_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-06_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-06_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-06_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-06_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.1
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-04.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-04.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
Center distance of 0.000008 mm
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./rm.epi.all1+orig.BRIK
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
cat_matvec -ONELINE sub-05_T1w_ns+tlrc::WARP_DATA -I sub-05_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
+ Automask has 36983 voxels
+ 5737 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
3dAllineate -base sub-05_T1w_ns+tlrc -input pb00.sub-05.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
Resampling ...
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-05.r01.tcat+orig.HEAD
++ Base dataset: ./sub-05_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 5.052 5.769 -22.159
+ shift search range is +/- = 57.780 69.336 57.780
+ 8.7% 8.3% 38.4%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1476.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-06_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-06_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/118.00=4e+02
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.9
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-05_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 2.944 -6.532 -22.972
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.1% 9.4% 39.8%
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.052 pitch=-0.028 yaw=-0.192 dS=-0.331 dL=-0.160 dP=-0.098
++ Mean: roll=-0.005 pitch=+0.098 yaw=+0.053 dS=-0.038 dL=-0.065 dP=+0.084
++ Max : roll=+0.077 pitch=+0.287 yaw=+0.302 dS=+0.141 dL=+0.027 dP=+0.305
++ Max displacements (mm) for each sub-brick:
0.70(0.00) 0.60(0.11) 0.62(0.05) 0.49(0.17) 0.38(0.26) 0.42(0.11) 0.40(0.04) 0.44(0.06) 0.42(0.04) 0.48(0.08) 0.45(0.07) 0.43(0.07) 0.48(0.07) 0.45(0.05) 0.49(0.10) 0.46(0.06) 0.44(0.06) 0.47(0.09) 0.47(0.06) 0.45(0.10) 0.41(0.07) 0.43(0.05) 0.33(0.11) 0.41(0.10) 0.37(0.08) 0.33(0.03) 0.33(0.06) 0.25(0.08) 0.29(0.07) 0.26(0.09) 0.29(0.06) 0.31(0.05) 0.27(0.05) 0.26(0.06) 0.21(0.11) 0.26(0.08) 0.29(0.07) 0.25(0.06) 0.28(0.04) 0.22(0.14) 0.24(0.10) 0.25(0.07) 0.23(0.05) 0.23(0.05) 0.22(0.05) 0.20(0.05) 0.22(0.03) 0.16(0.09) 0.17(0.05) 0.05(0.13) 0.09(0.09) 0.05(0.11) 0.05(0.07) 0.00(0.05) 0.09(0.09) 0.06(0.04) 0.06(0.03) 0.15(0.13) 0.17(0.08) 0.16(0.12) 0.10(0.08) 0.10(0.11) 0.10(0.07) 0.13(0.09) 0.11(0.10) 0.12(0.13) 0.14(0.10) 0.10(0.06) 0.21(0.13) 0.16(0.07) 0.16(0.08) 0.18(0.09) 0.17(0.09) 0.28(0.17) 0.22(0.11) 0.23(0.05) 0.23(0.09) 0.25(0.05) 0.26(0.06) 0.32(0.17) 0.41(0.09) 0.35(0.07) 0.32(0.07) 0.29(0.07) 0.26(0.06) 0.22(0.09) 0.17(0.07) 0.20(0.06) 0.22(0.08) 0.32(0.19) 0.38(0.08) 0.42(0.15) 0.36(0.11) 0.35(0.12) 0.27(0.10) 0.44(0.19) 0.32(0.15) 0.44(0.16) 0.35(0.11) 0.31(0.08) 0.33(0.07) 0.29(0.08) 0.39(0.12) 0.37(0.03) 0.39(0.07) 0.39(0.11) 0.36(0.09) 0.48(0.13) 0.42(0.08) 0.49(0.10) 0.43(0.09) 0.47(0.06) 0.51(0.09) 0.45(0.11) 0.53(0.10) 0.44(0.13) 0.53(0.10) 0.49(0.07) 0.49(0.08) 0.61(0.13) 0.53(0.10) 0.61(0.08) 0.59(0.08) 0.63(0.05) 0.66(0.08) 0.62(0.08) 0.72(0.11) 0.65(0.09) 0.74(0.10) 0.68(0.08) 0.77(0.09) 0.70(0.12) 0.77(0.13) 0.75(0.11) 0.87(0.23) 0.82(0.12) 0.83(0.09) 0.85(0.13) 0.79(0.19) 0.84(0.06) 0.78(0.06) 0.82(0.10) 0.75(0.10) 0.77(0.11) 0.78(0.10) 0.80(0.15)
++ Max displacement in automask = 0.87 (mm) at sub-brick 134
++ Max delta displ in automask = 0.26 (mm) at sub-brick 4
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-04.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
RMS[0] = 0.282796 0.164272 ITER = 7/50
0.282796
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-06_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-06_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-06_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-06_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-06_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-06_T1w_ns+orig.BRIK
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-04_T1w_ns+tlrc::WARP_DATA -I sub-04_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-06_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-06_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-06_T1w_ns+orig __ats_tmp___upad15_pre.sub-06_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-06_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-06_T1w_ns+tlrc.HEAD -> sub-06_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-06_T1w_ns+tlrc.BRIK -> sub-06_T1w_ns+tlrc.BRIK
Cleanup ...
3dAllineate -base sub-04_T1w_ns+tlrc -input pb00.sub-04.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.0
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
cat_matvec sub-06_T1w_ns+tlrc::WARP_DATA -I
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-04.r02.tcat+orig.HEAD
++ Base dataset: ./sub-04_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
if ( ! -f sub-06_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-06.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-06.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ -cmass x y z shifts = 5.557 -8.297 -11.120
+ shift search range is +/- = 57.780 69.336 57.780
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
+ Automask has 40877 voxels
+ 6169 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-05.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-05.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40836 voxels
+ 6159 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.8
++ ###########################################################
3dAllineate -base sub-04_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 5.487 -17.680 -9.546
+ shift search range is +/- = 57.780 69.336 57.780
+ 9.5% 25.5% 16.5%
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.032 pitch=-0.184 yaw=-0.203 dS=-0.162 dL=-0.046 dP=-0.096
++ Mean: roll=+0.067 pitch=+0.039 yaw=-0.031 dS=+0.115 dL=+0.048 dP=-0.015
++ Max : roll=+0.143 pitch=+0.131 yaw=+0.148 dS=+0.312 dL=+0.143 dP=+0.067
++ Max displacements (mm) for each sub-brick:
0.41(0.00) 0.35(0.12) 0.29(0.13) 0.34(0.10) 0.31(0.05) 0.28(0.08) 0.21(0.08) 0.26(0.09) 0.28(0.08) 0.23(0.09) 0.21(0.12) 0.16(0.09) 0.15(0.11) 0.16(0.07) 0.12(0.06) 0.19(0.10) 0.12(0.12) 0.08(0.07) 0.07(0.05) 0.10(0.05) 0.10(0.02) 0.16(0.09) 0.13(0.05) 0.08(0.10) 0.10(0.14) 0.00(0.10) 0.09(0.09) 0.12(0.08) 0.12(0.10) 0.12(0.07) 0.07(0.10) 0.14(0.09) 0.08(0.09) 0.10(0.08) 0.09(0.07) 0.12(0.05) 0.07(0.05) 0.13(0.07) 0.08(0.08) 0.10(0.03) 0.11(0.06) 0.11(0.09) 0.16(0.10) 0.15(0.06) 0.19(0.09) 0.15(0.09) 0.19(0.10) 0.17(0.05) 0.20(0.06) 0.24(0.08) 0.26(0.06) 0.18(0.13) 0.18(0.03) 0.17(0.04) 0.18(0.06) 0.17(0.07) 0.19(0.09) 0.18(0.06) 0.14(0.11) 0.18(0.11) 0.21(0.05) 0.25(0.07) 0.20(0.08) 0.19(0.06) 0.25(0.07) 0.17(0.10) 0.23(0.09) 0.22(0.05) 0.20(0.08) 0.24(0.07) 0.21(0.08) 0.24(0.05) 0.22(0.06) 0.26(0.05) 0.24(0.08) 0.23(0.06) 0.23(0.05) 0.25(0.08) 0.26(0.06) 0.23(0.07) 0.26(0.07) 0.26(0.02) 0.29(0.05) 0.28(0.07) 0.23(0.08) 0.24(0.05) 0.23(0.03) 0.26(0.03) 0.28(0.06) 0.35(0.22) 0.34(0.18) 0.33(0.33) 0.39(0.10) 0.32(0.11) 0.48(0.21) 0.54(0.09) 0.45(0.20) 0.42(0.11) 0.41(0.09) 0.40(0.06) 0.39(0.07) 0.39(0.09) 0.42(0.07) 0.42(0.09) 0.40(0.04) 0.40(0.07) 0.38(0.10) 0.37(0.05) 0.37(0.05) 0.34(0.08) 0.36(0.09) 0.31(0.06) 0.38(0.11) 0.32(0.11) 0.37(0.08) 0.33(0.06) 0.34(0.05) 0.29(0.08) 0.36(0.09) 0.37(0.09) 0.40(0.11) 0.41(0.09) 0.42(0.10) 0.40(0.05) 0.38(0.08) 0.39(0.04) 0.41(0.05) 0.40(0.05) 0.46(0.10) 0.44(0.06) 0.45(0.09) 0.45(0.07) 0.44(0.09) 0.48(0.12) 0.45(0.09) 0.51(0.10) 0.53(0.09) 0.53(0.03) 0.52(0.09) 0.50(0.07) 0.50(0.10) 0.49(0.07) 0.51(0.06) 0.54(0.05) 0.53(0.08) 0.53(0.05)
++ Max displacement in automask = 0.54 (mm) at sub-brick 143
++ Max delta displ in automask = 0.33 (mm) at sub-brick 91
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-06.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.061 pitch=-0.553 yaw=-0.248 dS=-0.521 dL=-0.077 dP=-0.085
++ Mean: roll=+0.017 pitch=-0.207 yaw=-0.073 dS=-0.103 dL=+0.005 dP=-0.001
++ Max : roll=+0.134 pitch=+0.059 yaw=+0.035 dS=+0.078 dL=+0.258 dP=+0.121
++ Max displacements (mm) for each sub-brick:
1.22(0.00) 1.18(0.09) 1.17(0.08) 1.02(0.16) 1.03(0.11) 0.93(0.11) 0.94(0.05) 0.94(0.06) 0.87(0.07) 0.87(0.01) 0.82(0.06) 0.83(0.04) 0.86(0.06) 0.79(0.09) 0.78(0.05) 0.75(0.05) 0.74(0.05) 0.79(0.06) 0.74(0.08) 0.71(0.04) 0.72(0.02) 0.66(0.06) 0.72(0.06) 0.70(0.04) 0.65(0.06) 0.58(0.10) 0.63(0.07) 0.57(0.07) 0.58(0.02) 0.53(0.06) 0.54(0.03) 0.54(0.05) 0.51(0.12) 0.58(0.20) 0.65(0.14) 0.59(0.45) 0.95(0.82) 0.81(0.52) 0.85(0.15) 0.68(0.20) 0.70(0.16) 0.67(0.07) 0.62(0.06) 0.65(0.11) 0.62(0.07) 0.65(0.06) 0.63(0.09) 0.77(0.17) 0.68(0.12) 0.61(0.10) 0.61(0.02) 0.58(0.05) 0.59(0.02) 0.64(0.06) 0.59(0.07) 0.57(0.03) 0.55(0.05) 0.57(0.09) 0.60(0.04) 0.52(0.09) 0.54(0.04) 0.50(0.07) 0.47(0.09) 0.45(0.09) 0.43(0.08) 0.46(0.05) 0.39(0.10) 0.46(0.08) 0.41(0.06) 0.42(0.07) 0.39(0.04) 0.34(0.06) 0.38(0.04) 0.34(0.05) 0.37(0.03) 0.36(0.06) 0.36(0.05) 0.34(0.05) 0.36(0.10) 0.37(0.06) 0.36(0.02) 0.36(0.03) 0.34(0.03) 0.34(0.05) 0.35(0.06) 0.33(0.03) 0.34(0.03) 0.31(0.08) 0.31(0.10) 0.31(0.12) 0.32(0.10) 0.28(0.11) 0.32(0.09) 0.29(0.03) 0.31(0.05) 0.34(0.07) 0.25(0.10) 0.23(0.08) 0.26(0.05) 0.20(0.06) 0.28(0.10) 0.24(0.06) 0.22(0.03) 0.23(0.03) 0.19(0.05) 0.21(0.05) 0.21(0.05) 0.18(0.08) 0.13(0.08) 0.16(0.07) 0.10(0.05) 0.14(0.05) 0.15(0.05) 0.11(0.07) 0.15(0.04) 0.12(0.08) 0.11(0.09) 0.07(0.05) 0.11(0.05) 0.10(0.09) 0.07(0.05) 0.00(0.07) 0.06(0.06) 0.06(0.05) 0.07(0.06) 0.08(0.05) 0.05(0.07) 0.05(0.06) 0.09(0.07) 0.14(0.06) 0.20(0.19) 0.12(0.16) 0.19(0.11) 0.14(0.27) 0.08(0.15) 0.10(0.06) 0.09(0.04) 0.10(0.06) 0.11(0.05) 0.09(0.08) 0.10(0.04) 0.07(0.03) 0.13(0.07) 0.15(0.04) 0.11(0.08) 0.14(0.07)
++ Max displacement in automask = 1.22 (mm) at sub-brick 0
++ Max delta displ in automask = 0.82 (mm) at sub-brick 36
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-05.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.3
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-06_T1w_ns+tlrc::WARP_DATA -I sub-06_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-06_T1w_ns+tlrc -input pb00.sub-06.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-06.r01.tcat+orig.HEAD
++ Base dataset: ./sub-06_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 7.048 -9.018 -32.550
+ shift search range is +/- = 57.780 69.336 57.780
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
+ 12.2% 13.0% 56.3%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-05_T1w_ns+tlrc::WARP_DATA -I sub-05_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-04.r01.volreg
3dAllineate -base sub-05_T1w_ns+tlrc -input pb00.sub-05.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-05.r02.tcat+orig.HEAD
++ Base dataset: ./sub-05_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 4.966 6.114 -22.483
+ shift search range is +/- = 57.780 69.336 57.780
+ 8.6% 8.8% 38.9%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-06_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ Output dataset ./pb01.sub-04.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-04.r02.volreg
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 3.185 -17.918 -33.371
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.5% 25.8% 57.8%
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.2
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-05_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 2.944 -6.532 -22.972
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.1% 9.4% 39.8%
++ Output dataset ./pb01.sub-04.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-04_T1w_ns+tlrc::WARP_DATA -I sub-04_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-04_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-04_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
+ -cmass x y z shifts = 5.507 -8.270 -11.113
+ shift search range is +/- = 57.780 69.336 57.780
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.7
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
3dcopy sub-04_T1w_ns+tlrc anat_final.sub-04
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-04+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-04+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-06.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -0.207 2.286 -3.943
+ shift search range is +/- = 73.188 86.670 75.114
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
++ Reading input dataset ./pb00.sub-06.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.5
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.170307
sp = 0.333985
mi = 2.57557
crM = 0.0480106
nmi = 0.845798
je = 2.57557
hel = -0.0915264
crA = 0.219726
crU = 0.23613
lss = 0.829693
lpc = 0.201004
lpa = 0.798995
lpc+ = 0.360288
lpa+ = 0.958279
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.9
++ ###########################################################
3dAllineate -source sub-04_T1w+orig -master anat_final.sub-04+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
+ Automask has 41021 voxels
+ 6223 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-04_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.1
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-04.r01.blur pb01.sub-04.r01.volreg+tlrc
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-05.r01.volreg
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-04.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-04.r02.blur pb01.sub-04.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-04.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-04.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-04.r01.blur+tlrc
++ Forming automask
+ Fixed clip level = 328.779022
+ Used gradual clip level = 321.974487 .. 340.750275
+ Number voxels above clip level = 74084
+ Clustering voxels ...
+ Largest cluster has 72949 voxels
+ Clustering voxels ...
+ Largest cluster has 72886 voxels
+ Filled 105 voxels in small holes; now have 72991 voxels
+ Clustering voxels ...
+ Largest cluster has 72990 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 198643 voxels
+ Mask now has 72990 voxels
++ 72990 voxels in the mask [out of 271633: 26.87%]
++ first 6 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 2 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-04.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-04.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 331.756439
+ Used gradual clip level = 324.587799 .. 343.809326
+ Number voxels above clip level = 74185
+ Clustering voxels ...
+ Largest cluster has 73110 voxels
+ Clustering voxels ...
+ Largest cluster has 72863 voxels
+ Filled 101 voxels in small holes; now have 72964 voxels
+ Clustering voxels ...
+ Largest cluster has 72964 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 198669 voxels
+ Mask now has 72964 voxels
++ 72964 voxels in the mask [out of 271633: 26.86%]
++ first 6 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 2 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-04
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 73360 survived, 198273 were zero
++ writing result full_mask.sub-04...
++ Output dataset ./full_mask.sub-04+tlrc.BRIK
3dresample -master full_mask.sub-04+tlrc -input sub-04_T1w_ns+tlrc -prefix rm.resam.anat
++ Output dataset ./pb01.sub-05.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-05.r02.volreg
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-04
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 78037 survived, 193596 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-04...
++ Output dataset ./mask_anat.sub-04+tlrc.BRIK
3dmask_tool -input full_mask.sub-04+tlrc mask_anat.sub-04+tlrc -inter -prefix mask_epi_anat.sub-04
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 12689 clipped, 69354 survived, 189590 were zero
++ writing result mask_epi_anat.sub-04...
++ Output dataset ./mask_epi_anat.sub-04+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-04+tlrc mask_anat.sub-04+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-04+tlrc.BRIK B=./mask_anat.sub-04+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
73360 78037 82043 69354 4006 8683 5.4607 11.1268 1.0610 0.9154 1.0705
3ddot -dodice full_mask.sub-04+tlrc mask_anat.sub-04+tlrc
tee out.mask_ae_dice.txt
0.916187
3dresample -master full_mask.sub-04+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-04+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.109 pitch=-0.263 yaw=-0.798 dS=-0.627 dL=-0.144 dP=-0.373
++ Mean: roll=+0.028 pitch=-0.126 yaw=-0.555 dS=+0.388 dL=-0.031 dP=-0.265
++ Max : roll=+0.083 pitch=+0.384 yaw=-0.431 dS=+0.671 dL=+0.068 dP=-0.021
++ Max displacements (mm) for each sub-brick:
1.58(0.00) 1.18(0.55) 1.03(0.26) 0.91(0.18) 0.90(0.08) 0.83(0.12) 0.78(0.21) 0.80(0.22) 0.77(0.10) 1.88(1.43) 1.14(0.96) 1.00(0.32) 0.94(0.13) 0.88(0.08) 0.87(0.07) 0.92(0.16) 0.92(0.11) 0.99(0.09) 0.97(0.11) 0.96(0.08) 0.98(0.11) 1.08(0.13) 1.05(0.06) 1.08(0.06) 1.07(0.06) 1.11(0.04) 1.13(0.07) 1.12(0.05) 1.12(0.06) 1.12(0.02) 1.19(0.09) 1.20(0.05) 1.20(0.04) 1.21(0.01) 1.22(0.05) 1.24(0.07) 1.20(0.06) 1.25(0.08) 1.26(0.07) 1.25(0.09) 1.22(0.06) 1.27(0.09) 1.26(0.05) 1.23(0.05) 1.26(0.05) 1.20(0.08) 1.21(0.05) 1.22(0.06) 1.22(0.04) 1.20(0.03) 1.25(0.06) 1.28(0.08) 1.19(0.13) 1.19(0.13) 1.14(0.08) 1.22(0.12) 1.23(0.06) 1.22(0.08) 1.30(0.09) 1.25(0.12) 1.30(0.08) 1.29(0.05) 1.33(0.05) 1.35(0.05) 1.37(0.06) 1.41(0.05) 1.32(0.09) 1.33(0.14) 1.36(0.11) 1.37(0.04) 1.41(0.07) 1.39(0.09) 1.35(0.07) 1.38(0.08) 1.39(0.06) 1.42(0.06) 1.38(0.10) 1.36(0.06) 1.39(0.07) 1.42(0.08) 1.46(0.10) 1.48(0.08) 1.50(0.08) 1.40(0.12) 1.45(0.07) 1.45(0.04) 1.44(0.05) 1.48(0.06) 1.35(0.17) 1.40(0.15) 1.36(0.10) 1.42(0.10) 1.43(0.11) 1.38(0.08) 1.34(0.06) 1.34(0.07) 1.32(0.11) 1.31(0.07) 1.33(0.06) 1.25(0.11) 1.26(0.06) 1.28(0.07) 1.21(0.08) 1.18(0.11) 1.19(0.09) 1.17(0.08) 1.14(0.08) 1.18(0.09) 1.16(0.06) 1.20(0.04) 1.22(0.04) 1.15(0.16) 1.14(0.11) 1.19(0.08) 1.23(0.08) 1.23(0.03) 1.20(0.06) 1.17(0.15) 1.17(0.06) 1.20(0.05) 1.15(0.07) 1.17(0.06) 1.10(0.08) 1.10(0.08) 1.10(0.12) 1.07(0.06) 1.02(0.07) 1.01(0.02) 1.00(0.05) 1.01(0.04) 1.01(0.12) 0.99(0.19) 0.92(0.21) 0.91(0.14) 0.95(0.18) 0.97(0.14) 0.93(0.11) 0.92(0.06) 0.92(0.07) 0.93(0.09) 0.95(0.10) 0.96(0.05) 0.95(0.06) 0.93(0.06) 0.94(0.04) 0.94(0.08)
++ Max displacement in automask = 1.88 (mm) at sub-brick 9
++ Max delta displ in automask = 1.43 (mm) at sub-brick 9
0.954796
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-04.r01.blur+tlrc
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-06.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-04.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-04.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb01.sub-05.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-05_T1w_ns+tlrc::WARP_DATA -I sub-05_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-05_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-05_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ Output dataset ./rm.epi.all1+orig.BRIK
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 4.955 6.082 -22.628
+ shift search range is +/- = 57.780 69.336 57.780
+ 8.6% 8.8% 39.2%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
cat_matvec -ONELINE sub-06_T1w_ns+tlrc::WARP_DATA -I sub-06_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-05_T1w_ns+tlrc anat_final.sub-05
3dAllineate -base sub-06_T1w_ns+tlrc -input pb00.sub-06.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-06.r02.tcat+orig.HEAD
++ Base dataset: ./sub-06_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-05+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-05+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 7.107 -8.817 -32.825
+ shift search range is +/- = 57.780 69.336 57.780
+ 12.3% 12.7% 56.8%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = 0.279 1.170 -1.578
+ shift search range is +/- = 70.299 84.744 73.188
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.161421
sp = 0.314375
mi = 2.59313
crM = 0.0455354
nmi = 0.844678
je = 2.59313
hel = -0.0921928
crA = 0.213903
crU = 0.228712
lss = 0.838579
lpc = 0.326093
lpa = 0.673907
lpc+ = 0.486573
lpa+ = 0.834387
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.8
++ ###########################################################
3dAllineate -source sub-05_T1w+orig -master anat_final.sub-05+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-05_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.2
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-05.r01.blur pb01.sub-05.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-05.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-05.r02.blur pb01.sub-05.r02.volreg+tlrc
++ Output dataset ./pb03.sub-04.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-04.r02.blur+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-04.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-04.r02.scale
..................................................................................................................................................
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
-- Wrote edited dataset: ./pb02.sub-05.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-05.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-05.r01.blur+tlrc
++ Forming automask
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-06_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
+ Fixed clip level = 357.094666
+ Used gradual clip level = 338.113312 .. 381.064087
+ Number voxels above clip level = 72363
+ Clustering voxels ...
+ Largest cluster has 71780 voxels
+ Clustering voxels ...
+ Largest cluster has 71529 voxels
+ Filled 110 voxels in small holes; now have 71639 voxels
+ Clustering voxels ...
+ Largest cluster has 71638 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 199995 voxels
+ Mask now has 71638 voxels
++ 71638 voxels in the mask [out of 271633: 26.37%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-05.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-05.r02.blur+tlrc
++ Forming automask
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 3.185 -17.918 -33.371
+ shift search range is +/- = 57.780 69.336 57.780
+ 5.5% 25.8% 57.8%
+ Fixed clip level = 358.343018
+ Used gradual clip level = 339.054138 .. 382.411041
+ Number voxels above clip level = 72246
+ Clustering voxels ...
+ Largest cluster has 71564 voxels
+ Clustering voxels ...
+ Largest cluster has 71511 voxels
+ Filled 124 voxels in small holes; now have 71635 voxels
+ Clustering voxels ...
+ Largest cluster has 71635 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 199998 voxels
+ Mask now has 71635 voxels
++ 71635 voxels in the mask [out of 271633: 26.37%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-05
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 71803 survived, 199830 were zero
++ writing result full_mask.sub-05...
++ Output dataset ./full_mask.sub-05+tlrc.BRIK
3dresample -master full_mask.sub-05+tlrc -input sub-05_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-05
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76811 survived, 194822 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-05...
++ Output dataset ./mask_anat.sub-05+tlrc.BRIK
3dmask_tool -input full_mask.sub-05+tlrc mask_anat.sub-05+tlrc -inter -prefix mask_epi_anat.sub-05
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 12570 clipped, 68022 survived, 191041 were zero
++ writing result mask_epi_anat.sub-05...
++ Output dataset ./mask_epi_anat.sub-05+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-05+tlrc mask_anat.sub-05+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-05+tlrc.BRIK B=./mask_anat.sub-05+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
71803 76811 80592 68022 3781 8789 5.2658 11.4424 1.1123 0.9630 0.9737
3ddot -dodice full_mask.sub-05+tlrc mask_anat.sub-05+tlrc
tee out.mask_ae_dice.txt
0.915418
3dresample -master full_mask.sub-05+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-05+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.954347
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-05.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-05.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-05.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.4
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./pb03.sub-04.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-04
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 0
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-04_censor.1D -show_trs_uncensored encoded
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-06.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-04.r01.scale+tlrc.HEAD pb03.sub-04.r02.scale+tlrc.HEAD -censor motion_sub-04_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-04 -errts errts.sub-04 -bucket stats.sub-04
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-04.r01.scale+tlrc.HEAD pb03.sub-04.r02.scale+tlrc.HEAD
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
++ STAT automask has 255329 voxels (out of 271633 = 94.0%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (no censoring)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-04.r01.scale+tlrc.HEAD pb03.sub-04.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-04_REML -Rvar stats.sub-04_REMLvar \
-Rfitts fitts.sub-04_REML -Rerrts errts.sub-04_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (292x20): 5.57979 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (292x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (292x18): 5.57289 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (292x12): 4.63313 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (292x6): 1 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 3.62289e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.70 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=3.773
++ Job #2: processing voxels 67908 to 101861; elapsed time=3.786
++ Job #3: processing voxels 101862 to 135815; elapsed time=3.799
++ Job #4: processing voxels 135816 to 169769; elapsed time=3.812
++ Job #5: processing voxels 169770 to 203723; elapsed time=3.825
++ Job #6: processing voxels 203724 to 237677; elapsed time=3.840
++ Job #7: processing voxels 237678 to 271632; elapsed time=3.854
++ Job #0: processing voxels 0 to 33953; elapsed time=3.864
++ Output dataset ./pb01.sub-06.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-06.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb03.sub-05.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-05.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-05.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-05.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ voxel loop:0123456789.0123456789.0123456789.0123456789.012345678++ Job #7 finished; elapsed time=7.972
++ Job #3 finished; elapsed time=7.991
9.
++ Job #0 waiting for children to finish; elapsed time=8.024
++ Job #1 finished; elapsed time=8.032
++ Job #4 finished; elapsed time=8.042
++ Job #5 finished; elapsed time=8.055
++ Job #6 finished; elapsed time=8.100
++ Job #2 finished; elapsed time=8.112
++ Job #0 now finishing up; elapsed time=8.119
++ Smallest FDR q [0 Full_Fstat] = 3.28937e-12
++ Output dataset ./pb01.sub-06.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-06_T1w_ns+tlrc::WARP_DATA -I sub-06_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-06_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-06_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 7.046 -9.036 -32.411
+ shift search range is +/- = 57.780 69.336 57.780
+ 12.2% 13.0% 56.1%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-06_T1w_ns+tlrc anat_final.sub-06
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-06+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-06+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = 0.108 1.926 -0.604
+ shift search range is +/- = 70.299 83.781 69.336
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
++ Smallest FDR q [2 congruent#0_Tstat] = 7.3004e-11
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.116006
sp = 0.318701
mi = 2.5712
crM = 0.0219631
nmi = 0.820325
je = 2.5712
hel = -0.117789
crA = 0.148904
crU = 0.16337
lss = 0.883994
lpc = 0.478835
lpa = 0.521165
lpc+ = 0.580931
lpa+ = 0.623262
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.8
++ ###########################################################
3dAllineate -source sub-06_T1w+orig -master anat_final.sub-06+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-06_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
++ Smallest FDR q [3 congruent_Fstat] = 7.30061e-11
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./pb03.sub-05.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-05
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.3
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-06.r01.blur pb01.sub-06.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 3
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-05_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-05.r01.scale+tlrc.HEAD pb03.sub-05.r02.scale+tlrc.HEAD -censor motion_sub-05_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-05 -errts errts.sub-05 -bucket stats.sub-05
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-05.r01.scale+tlrc.HEAD pb03.sub-05.r02.scale+tlrc.HEAD
++ Smallest FDR q [5 incongruent#0_Tstat] = 9.90019e-12
..................................................................................................................................................
++ Smallest FDR q [6 incongruent_Fstat] = 9.90035e-12
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
-- Wrote edited dataset: ./pb02.sub-06.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-06.r02.blur pb01.sub-06.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
++ STAT automask has 249116 voxels (out of 271633 = 91.7%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (before censor) ; 289 (after)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
-- Wrote edited dataset: ./pb02.sub-06.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-06.r01.blur+tlrc
++ Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.0453437
++ Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.0453468
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-06.r01.blur+tlrc
++ Forming automask
+ Fixed clip level = 356.218719
+ Used gradual clip level = 328.784546 .. 380.542053
+ Number voxels above clip level = 72168
+ Clustering voxels ...
+ Largest cluster has 71711 voxels
+ Clustering voxels ...
+ Largest cluster has 70955 voxels
+ Filled 155 voxels in small holes; now have 71110 voxels
+ Filled 2 voxels in large holes; now have 71112 voxels
+ Clustering voxels ...
+ Largest cluster has 71112 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200521 voxels
+ Mask now has 71112 voxels
++ 71112 voxels in the mask [out of 271633: 26.18%]
++ first 7 x-planes are zero [from L]
++ last 7 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-06.r02.blur+tlrc
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-06.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 355.373657
+ Used gradual clip level = 328.181641 .. 379.672516
+ Number voxels above clip level = 72175
+ Clustering voxels ...
+ Largest cluster has 71625 voxels
+ Clustering voxels ...
+ Largest cluster has 70914 voxels
+ Filled 154 voxels in small holes; now have 71068 voxels
+ Filled 2 voxels in large holes; now have 71070 voxels
+ Clustering voxels ...
+ Largest cluster has 71068 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200565 voxels
++ Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.0453437
+ Mask now has 71068 voxels
++ 71068 voxels in the mask [out of 271633: 26.16%]
++ first 7 x-planes are zero [from L]
++ last 7 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-05.r01.scale+tlrc.HEAD pb03.sub-05.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-05_REML -Rvar stats.sub-05_REMLvar \
-Rfitts fitts.sub-05_REML -Rerrts errts.sub-05_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (289x20): 5.09936 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (289x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (289x18): 5.08759 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (289x12): 2.99081 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (289x6): 1.00988 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 3.5638e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.63 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=4.376
++ Job #2: processing voxels 67908 to 101861; elapsed time=4.389
++ Job #3: processing voxels 101862 to 135815; elapsed time=4.401
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ Job #4: processing voxels 135816 to 169769; elapsed time=4.414
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-06
++ Job #5: processing voxels 169770 to 203723; elapsed time=4.427
++ Job #6: processing voxels 203724 to 237677; elapsed time=4.439
++ Job #7: processing voxels 237678 to 271632; elapsed time=4.452
++ Job #0: processing voxels 0 to 33953; elapsed time=4.462
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.0453468
++ Wrote bucket dataset into ./stats.sub-04+tlrc.BRIK
++ frac 0 over 2 volumes gives min count 0
+ created 9 FDR curves in bucket header
++ voxel limits: 0 clipped, 71257 survived, 200376 were zero
++ writing result full_mask.sub-06...
++ Output dataset ./full_mask.sub-06+tlrc.BRIK
3dresample -master full_mask.sub-06+tlrc -input sub-06_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-06
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76377 survived, 195256 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-06...
++ Output dataset ./mask_anat.sub-06+tlrc.BRIK
3dmask_tool -input full_mask.sub-06+tlrc mask_anat.sub-06+tlrc -inter -prefix mask_epi_anat.sub-06
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 10158 clipped, 68738 survived, 192737 were zero
++ writing result mask_epi_anat.sub-06...
++ Output dataset ./mask_epi_anat.sub-06+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-06+tlrc mask_anat.sub-06+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-06+tlrc.BRIK B=./mask_anat.sub-06+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
71257 76377 78896 68738 2519 7639 3.5351 10.0017 1.0743 0.9683 1.0262
3ddot -dodice full_mask.sub-06+tlrc mask_anat.sub-06+tlrc
tee out.mask_ae_dice.txt
++ Wrote 3D+time dataset into ./fitts.sub-04+tlrc.BRIK
0.931195
3dresample -master full_mask.sub-06+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-06+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.956218
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-06.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ voxel loop:0123456789.0123456789.0123456789.0123456789.0123456789.
++ Job #0 waiting for children to finish; elapsed time=8.905
++ Job #2 finished; elapsed time=8.978
++ Job #4 finished; elapsed time=9.000
++ Job #6 finished; elapsed time=9.017
++ Wrote 3D+time dataset into ./errts.sub-04+tlrc.BRIK
++ Job #7 finished; elapsed time=9.071
++ Program finished; elapsed time=21.308
++ Job #3 finished; elapsed time=9.103
++ Job #5 finished; elapsed time=9.105
++ Job #1 finished; elapsed time=9.177
++ Job #0 now finishing up; elapsed time=9.183
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
++ Smallest FDR q [0 Full_Fstat] = 1.86374e-12
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-06.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-06.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 0 : 0.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 20 : 6.8%
final DF : 272 : 93.2%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-04.r01.scale+tlrc.HEAD pb03.sub-04.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-04_REML -Rvar stats.sub-04_REMLvar -Rfitts fitts.sub-04_REML -Rerrts errts.sub-04_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ Smallest FDR q [2 congruent#0_Tstat] = 9.5233e-11
++ FDR automask has 255329 voxels (out of 271633 = 94.0%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (292x20): 5.57979 ++ VERY GOOD ++
++ Smallest FDR q [3 congruent_Fstat] = 9.52309e-11
+ masked off 29496 voxels for being all zero; 242137 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.36
+ X matrix: 79.110% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=292 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.38
+ average case bandwidth = 16.10
++ Smallest FDR q [5 incongruent#0_Tstat] = 3.87704e-12
++ Smallest FDR q [6 incongruent_Fstat] = 3.87679e-12
++ Output dataset ./pb03.sub-06.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-06.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.205914 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.205448 ==> few true single voxel detections
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-06.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-06.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.205914 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.205448 ==> few true single voxel detections
++ Wrote bucket dataset into ./stats.sub-05+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Wrote 3D+time dataset into ./fitts.sub-05+tlrc.BRIK
++ Wrote 3D+time dataset into ./errts.sub-05+tlrc.BRIK
++ Program finished; elapsed time=19.346
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 3 : 1.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 23 : 7.9%
final DF : 269 : 92.1%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-05.r01.scale+tlrc.HEAD pb03.sub-05.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-05_REML -Rvar stats.sub-05_REMLvar -Rfitts fitts.sub-05_REML -Rerrts errts.sub-05_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ Output dataset ./pb03.sub-06.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-06
++ FDR automask has 249116 voxels (out of 271633 = 91.7%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (289x20): 5.09936 ++ VERY GOOD ++
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 3
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-06_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-06.r01.scale+tlrc.HEAD pb03.sub-06.r02.scale+tlrc.HEAD -censor motion_sub-06_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-06 -errts errts.sub-06 -bucket stats.sub-06
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-06.r01.scale+tlrc.HEAD pb03.sub-06.r02.scale+tlrc.HEAD
+ masked off 51071 voxels for being all zero; 220562 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.58
+ X matrix: 79.100% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=289 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.61
+ average case bandwidth = 15.93
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
++ STAT automask has 251031 voxels (out of 271633 = 92.4%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (before censor) ; 289 (after)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-06.r01.scale+tlrc.HEAD pb03.sub-06.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-06_REML -Rvar stats.sub-06_REMLvar \
-Rfitts fitts.sub-06_REML -Rerrts errts.sub-06_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (289x20): 4.26535 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (289x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (289x18): 4.25387 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (289x12): 3.35701 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (289x6): 1.03058 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 2.22876e-15 ++ VERY GOOD ++
++ Matrix setup time = 1.32 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=4.385
++ Job #2: processing voxels 67908 to 101861; elapsed time=4.403
++ Job #3: processing voxels 101862 to 135815; elapsed time=4.418
++ Job #4: processing voxels 135816 to 169769; elapsed time=4.433
++ Job #5: processing voxels 169770 to 203723; elapsed time=4.459
++ Job #6: processing voxels 203724 to 237677; elapsed time=4.486
++ Job #7: processing voxels 237678 to 271632; elapsed time=4.498
++ Job #0: processing voxels 0 to 33953; elapsed time=4.499
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.01234
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=20.37
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ voxel loop:0123456789.0123456789.0123456789.0123456789.0123456789.
++ Job #0 waiting for children to finish; elapsed time=10.644
++ Job #1 finished; elapsed time=12.527
++ Job #3 finished; elapsed time=12.659
++ Job #5 finished; elapsed time=12.703
++ Job #7 finished; elapsed time=13.026
++ Job #4 finished; elapsed time=13.123
++ Job #2 finished; elapsed time=13.264
++ Job #6 finished; elapsed time=13.404
++ Job #0 now finishing up; elapsed time=13.411
++ Smallest FDR q [0 Full_Fstat] = 8.10361e-12
++ Smallest FDR q [2 congruent#0_Tstat] = 1.03514e-07
++ Smallest FDR q [3 congruent_Fstat] = 1.03513e-07
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.0
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=19.62
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.6808e-11
++ Smallest FDR q [6 incongruent_Fstat] = 2.68075e-11
*+ WARNING: Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.235974 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.235438 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.235974 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.235438 ==> few true single voxel detections
++ Wrote bucket dataset into ./stats.sub-06+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Wrote 3D+time dataset into ./fitts.sub-06+tlrc.BRIK
++ Wrote 3D+time dataset into ./errts.sub-06+tlrc.BRIK
++ Program finished; elapsed time=26.075
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 3 : 1.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 23 : 7.9%
final DF : 269 : 92.1%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-06.r01.scale+tlrc.HEAD pb03.sub-06.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-06_REML -Rvar stats.sub-06_REMLvar -Rfitts fitts.sub-06_REML -Rerrts errts.sub-06_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ FDR automask has 251031 voxels (out of 271633 = 92.4%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (289x20): 4.26535 ++ VERY GOOD ++
+ masked off 34505 voxels for being all zero; 237128 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.73
+ X matrix: 79.100% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=289 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.76
+ average case bandwidth = 16.01
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=50.75
++ Output dataset ./stats.sub-04_REMLvar+tlrc.BRIK
++ Output dataset ./fitts.sub-04_REML+tlrc.BRIK
++ Output dataset ./errts.sub-04_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-04_REML+tlrc
++ Smallest FDR q [0 Full_Fstat] = 7.44306e-13
++ Smallest FDR q [2 congruent#0_Tstat] = 3.89421e-12
++ Smallest FDR q [3 congruent_Fstat] = 3.89421e-12
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=46.84
++ Output dataset ./stats.sub-05_REMLvar+tlrc.BRIK
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.0123
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=18.03
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ Smallest FDR q [5 incongruent#0_Tstat] = 3.92754e-12
++ Smallest FDR q [6 incongruent_Fstat] = 3.92754e-12
++ Output dataset ./fitts.sub-05_REML+tlrc.BRIK
++ Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.0456481
++ Output dataset ./errts.sub-05_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-05_REML+tlrc
++ Smallest FDR q [9 incongruent-congruent_Fstat] = 0.0456481
++ Smallest FDR q [0 Full_Fstat] = 4.53624e-12
++ Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.0456481
++ Smallest FDR q [2 congruent#0_Tstat] = 1.23399e-09
++ Smallest FDR q [12 congruent-incongruent_Fstat] = 0.0456481
+ Added 9 FDR curves to dataset stats.sub-04_REML+tlrc
++ Output dataset ./stats.sub-04_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=62.69
++ Smallest FDR q [3 congruent_Fstat] = 1.23398e-09
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-04 pb03.sub-04.r01.scale+tlrc.HEAD pb03.sub-04.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.8 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-04+tlrc[0..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Smallest FDR q [5 incongruent#0_Tstat] = 1.37298e-11
++ Smallest FDR q [6 incongruent_Fstat] = 1.37298e-11
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-04_REML+tlrc[0..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-04
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-04+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-04_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.927655 ==> few true single voxel detections
++ Authored by: RW Cox
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.927654 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.927655 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.927654 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-05_REML+tlrc
++ Output dataset ./stats.sub-05_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=57.94
++ Output dataset ./rm.errts.unit+tlrc.BRIK
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-05 pb03.sub-05.r01.scale+tlrc.HEAD pb03.sub-05.r02.scale+tlrc.HEAD
3dmaskave -quiet -mask full_mask.sub-04+tlrc rm.errts.unit+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 73360 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.0709615
3dmaskave -quiet -mask full_mask.sub-04+tlrc errts.sub-04_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 73360 voxels survive the mask
++ elapsed time = 0.6 s
3dTcorr1D -prefix corr_brain errts.sub-04_REML+tlrc mean.errts.1D
3dTstat -mean -prefix rm.signal.all all_runs.sub-05+tlrc[0..179,183..291]
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-04_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-04_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-05_REML+tlrc[0..179,183..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: THD_Tcorr1D: 29496 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-04.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-05
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-04+tlrc[0..145]
++ Output dataset ./TSNR.sub-05+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-05_REML+tlrc
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Number of voxels in mask = 73360
++ detrending start: 11 baseline funcs, 146 time points
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-05+tlrc rm.errts.unit+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 71803 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.0654473
3dmaskave -quiet -mask full_mask.sub-05+tlrc errts.sub-05_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ detrending done (0.00 CPU s thus far)
+++ 71803 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-05_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-05_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-05_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ start ACF calculations out to radius = 17.77 mm
*+ WARNING: THD_Tcorr1D: 51071 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-05.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-05+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-04+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 73360
++ detrending start: 11 baseline funcs, 146 time points
++ start ACF calculations out to radius = 17.45 mm
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=43.45
++ Output dataset ./stats.sub-06_REMLvar+tlrc.BRIK
++ Output dataset ./fitts.sub-06_REML+tlrc.BRIK
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.82 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ Output dataset ./errts.sub-06_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-06_REML+tlrc
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
++ Smallest FDR q [0 Full_Fstat] = 9.43382e-12
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..179,183..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-05+tlrc[146..179,183..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 143 time points
++ Smallest FDR q [2 congruent#0_Tstat] = 3.67971e-08
++ Smallest FDR q [3 congruent_Fstat] = 3.67971e-08
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.56 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ Smallest FDR q [5 incongruent#0_Tstat] = 5.60502e-11
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ Smallest FDR q [6 incongruent_Fstat] = 5.60502e-11
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.853374 3.41358 13.2351 8.49432
average epits ACF blurs: 0.853374 3.41358 13.2351 8.49432
echo 0.853374 3.41358 13.2351 8.49432 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-04+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 73360
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.536653 ==> few true single voxel detections
++ detrending start: 11 baseline funcs, 146 time points
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.536653 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.536653 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.536653 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-06_REML+tlrc
++ Output dataset ./stats.sub-06_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=55.49
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-06 pb03.sub-06.r01.scale+tlrc.HEAD pb03.sub-06.r02.scale+tlrc.HEAD
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ detrending done (0.00 CPU s thus far)
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.838488 3.33348 13.2488 8.38009
average epits ACF blurs: 0.838488 3.33348 13.2488 8.38009
echo 0.838488 3.33348 13.2488 8.38009 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ elapsed time = 0.7 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-06+tlrc[0..153,157..291]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Authored by: KR Hammett & RW Cox
++ start ACF calculations out to radius = 17.68 mm
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-05+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 146 time points
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-06_REML+tlrc[0..153,157..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-06
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-06+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-06_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.37 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-06+tlrc rm.errts.unit+tlrc
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 71257 voxels survive the mask
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-04+tlrc[146..291]
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.0449711
3dmaskave -quiet -mask full_mask.sub-06+tlrc errts.sub-06_REML+tlrc
++ Number of voxels in mask = 73360
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 71257 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-06_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-06_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-06_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ detrending start: 11 baseline funcs, 146 time points
*+ WARNING: THD_Tcorr1D: 34505 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
+ detrending done (0.00 CPU s thus far)
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-06.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..179,183..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-05+tlrc[146..179,183..291]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-06+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
++ Number of voxels in mask = 71257
++ detrending start: 11 baseline funcs, 146 time points
++ start ACF calculations out to radius = 17.82 mm
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 143 time points
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.38 mm
++ start ACF calculations out to radius = 17.51 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.862478 3.41262 13.4671 8.47297
average errts ACF blurs: 0.862478 3.41262 13.4671 8.47297
echo 0.862478 3.41262 13.4671 8.47297 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-04_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 73360
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..153,157..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-06+tlrc[146..153,157..291]
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ Number of voxels in mask = 71257
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.851748 3.33325 13.6677 8.35231
average errts ACF blurs: 0.851748 3.33325 13.6677 8.35231
echo 0.851748 3.33325 13.6677 8.35231 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ detrending start: 11 baseline funcs, 143 time points
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-05_REML+tlrc[0..145]
+ detrending done (0.00 CPU s thus far)
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 146 time points
++ start ACF calculations out to radius = 17.65 mm
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.30 mm
++ start ACF calculations out to radius = 17.34 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-04+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-04_REML+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 73360
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.865616 3.34069 12.6094 8.29885
average epits ACF blurs: 0.865616 3.34069 12.6094 8.29885
echo 0.865616 3.34069 12.6094 8.29885 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-06+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71257
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ detrending start: 11 baseline funcs, 146 time points
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..179,183..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-05+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-05_REML+tlrc[146..179,183..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71803
+ detrending done (0.00 CPU s thus far)
*+ WARNING: removed 5 voxels from mask because they are constant in time
++ detrending start: 11 baseline funcs, 143 time points
++ start ACF calculations out to radius = 17.77 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.34 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.48 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.863972 3.4079 13.4515 8.45709
average err_reml ACF blurs: 0.863972 3.4079 13.4515 8.45709
echo 0.863972 3.4079 13.4515 8.45709 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-04 -dsets pb00.sub-04.r01.tcat+orig.HEAD pb00.sub-04.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-04+tlrc.HEAD -ss_review_dset out.ss_review.sub-04.txt -write_uvars_json out.ss_review_uvars.json
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-04.txt
subject ID : sub-04
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-04+tlrc.HEAD
final stats dset : stats.sub-04_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
motion limit : 0.3
if ( 146..153,157..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-06+tlrc[146..153,157..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
num TRs above mot limit : 0
++ Number of voxels in mask = 71257
average motion (per TR) : 0.0651394
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ detrending start: 11 baseline funcs, 143 time points
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
average censored motion : 0.0651394
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.853893 3.3299 13.7531 8.34173
average err_reml ACF blurs: 0.853893 3.3299 13.7531 8.34173
echo 0.853893 3.3299 13.7531 8.34173 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-05 -dsets pb00.sub-05.r01.tcat+orig.HEAD pb00.sub-05.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-05+tlrc.HEAD -ss_review_dset out.ss_review.sub-05.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 2.6206
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
max censored displacement : 2.6206
average outlier frac (TR) : 0.000910993
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-05.txt
subject ID : sub-05
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-05+tlrc.HEAD
final stats dset : stats.sub-05_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs above mot limit : 2
average motion (per TR) : 0.0532986
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
average censored motion : 0.0504625
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.22172
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.22172
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
average outlier frac (TR) : 0.00152065
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
++ start ACF calculations out to radius = 17.28 mm
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
censor fraction : 0.000000
num regs of interest : 2
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 143
num TRs per run (censored): 0 3
fraction censored per run : 0 0.0205479
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 289
degrees of freedom used : 20
degrees of freedom left : 269
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 3
censor fraction : 0.010274
num regs of interest : 2
ave mot per sresp (orig) : 0.063507 0.067320
ave mot per sresp (cens) : 0.063507 0.067320
num TRs per stim (orig) : 120 120
num TRs censored per stim : 3 0
fraction TRs censored : 0.025 0.000
TSNR average : 125.258
global correlation (GCOR) : 0.0709615
anat/EPI mask Dice coef : 0.916187
anat/templ mask Dice coef : 0.954796
maximum F-stat (masked) : 98.597
blur estimates (ACF) : 0.863972 3.4079 13.4515
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 37 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
tcsh @ss_review_html
tee out.review_html
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.874602 3.34156 12.8084 8.28139
average errts ACF blurs: 0.874602 3.34156 12.8084 8.28139
echo 0.874602 3.34156 12.8084 8.28139 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
ave mot per sresp (orig) : 0.064327 0.045496
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
ave mot per sresp (cens) : 0.057653 0.045496
TSNR average : 133.994
global correlation (GCOR) : 0.0654473
anat/EPI mask Dice coef : 0.915418
anat/templ mask Dice coef : 0.954347
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-06_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
maximum F-stat (masked) : 73.2756
blur estimates (ACF) : 0.853893 3.3299 13.7531
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
++ Number of voxels in mask = 71257
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ detrending start: 11 baseline funcs, 146 time points
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 38 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 820.000000 -func_range 820.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-04/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-04/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ Using blowup factor: 4
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-04/media/__tmp_chauf_9WfgA46eR8A
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 820.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_9WfgA46eR8A/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_9WfgA46eR8A/tmp_olay.nii
++ User-entered function range value value (820.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=7..55 y=3..60 z=0..34
++ 3dAutobox: output dataset = QC_sub-04/media/__tmp_chauf_9WfgA46eR8A/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-04/media/__tmp_chauf_9WfgA46eR8A/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_0aBtO75zWU9HNKNpgYLMWQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_0aBtO75zWU9HNKNpgYLMWQ+orig.BRIK
+ detrending done (0.00 CPU s thus far)
++ 99470 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 8.837349 4.391563 -10
++ Will have the ref box central gapord: 7 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :982
[1] 2525529
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 867.000000 -func_range 867.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-05/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-05/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-05/media/__tmp_chauf_EC51bBVAmgB
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 867.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__tmp_chauf_EC51bBVAmgB/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__tmp_chauf_EC51bBVAmgB/tmp_olay.nii
++ User-entered function range value value (867.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ start ACF calculations out to radius = 17.32 mm
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=8..55 y=1..63 z=0..36
++ 3dAutobox: output dataset = QC_sub-05/media/__tmp_chauf_EC51bBVAmgB/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-05/media/__tmp_chauf_EC51bBVAmgB/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_H7Twtn5dYk9gunAcAgOMhA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_H7Twtn5dYk9gunAcAgOMhA+orig.BRIK
++ 111888 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3.668678 12.2349 -19.01205
++ Will have the ref box central gapord: 6 9 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :406
[1] 2525904
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_9WfgA46eR8A++ Writing palette image to QC_sub-04/media/qc_00_vorig_EPI.pbar.jpg
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-04/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 820.000000 -com SET_FUNC_RANGE 820.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 8.837349 4.391563 -10 -com SAVE_JPEG sagittalimage QC_sub-04/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-04/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-04/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-04/media/__tmp_chauf_9WfgA46eR8A
+* Removing temporary image directory 'QC_sub-04/media/__tmp_chauf_9WfgA46eR8A'.
++ DONE (good exit)
see: QC_sub-04/media/qc_00_vorig_EPI*
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..153,157..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-06+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-06_REML+tlrc[146..153,157..291]
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_EC51bBVAmgB++ Writing palette image to QC_sub-05/media/qc_00_vorig_EPI.pbar.jpg
++ Number of voxels in mask = 71257
++ Output dataset ./QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_00_vorig_EPI.axi.jpg'
++ Output dataset ./QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
AFNI QUITTs!
+ shift search range is +/- = 57.780 69.336 57.780
+++ Command Echo:
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:6:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:9:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-05/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 867.000000 -com SET_FUNC_RANGE 867.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3.668678 12.2349 -19.01205 -com SAVE_JPEG sagittalimage QC_sub-05/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-05/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-05/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-05/media/__tmp_chauf_EC51bBVAmgB
+* Removing temporary image directory 'QC_sub-05/media/__tmp_chauf_EC51bBVAmgB'.
[1] Done Xvfb :406 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-05/media/qc_00_vorig_EPI*
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH/eac_1_grid2olay.nii
++ detrending start: 11 baseline funcs, 143 time points
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.3
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 112.021500
+ Used gradual clip level = 106.034698 .. 121.515198
+ Number voxels above clip level = 218979
+ Clustering voxels ...
+ Largest cluster has 218628 voxels
+ Clustering voxels ...
+ Largest cluster has 212358 voxels
+ Filled 12083 voxels in small holes; now have 224441 voxels
+ Filled 10943 voxels in large holes; now have 235384 voxels
+ Clustering voxels ...
+ Largest cluster has 235375 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 666444 voxels
+ Mask now has 236185 voxels
++ 236185 voxels in the mask [out of 902629: 26.17%]
++ first 6 x-planes are zero [from L]
++ last 8 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 8 y-planes are zero [from A]
++ first 2 z-planes are zero [from I]
++ last 12 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 40.694290 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_nPIAiSh9Zoi
++ Output dataset ./QC_sub-05/media/__workdir_EAC_94p9x9HQFSA/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Output dataset ./QC_sub-05/media/__workdir_EAC_94p9x9HQFSA/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__workdir_EAC_94p9x9HQFSA/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.579094, 852.813599]
++ Output dataset QC_sub-05/media/__workdir_EAC_94p9x9HQFSA/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_nPIAiSh9Zoi/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__workdir_EAC_94p9x9HQFSA/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Output dataset ./../__tmp_chauf_nPIAiSh9Zoi/tmp_olay.nii
++ Forming automask
+ Fixed clip level = 110.604599
+ Used gradual clip level = 101.499199 .. 120.995499
+ Number voxels above clip level = 217863
+ Clustering voxels ...
+ Largest cluster has 217639 voxels
++ For olay, the 33%ile value leads to
--> upper range value: 99.312141
+ Clustering voxels ...
+ Largest cluster has 210993 voxels
+ Filled 10186 voxels in small holes; now have 221179 voxels
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_nPIAiSh9Zoi/ulay_box_0.nii
+ Filled 9543 voxels in large holes; now have 230722 voxels
+ Clustering voxels ...
+ Largest cluster has 230710 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 670732 voxels
+ Mask now has 231897 voxels
++ 231897 voxels in the mask [out of 902629: 25.69%]
++ first 5 x-planes are zero [from L]
++ last 5 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_nPIAiSh9Zoi/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_XTC80R-1ghuvQS5JxK1TFg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_XTC80R-1ghuvQS5JxK1TFg+tlrc.BRIK
++ olay_alpha has known value: No
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ My command:
++ Using only the boxes+balls mask
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 38.412792 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ How many coors? 3
+ detrending done (0.00 CPU s thus far)
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
++ Using AFNI ver : AFNI_21.2.00
------------------- end of optionizing -------------------
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
-- trying to start Xvfb :795
[1] 2527366
++ Found input file: eac_6_edgy.nii
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_d0GWh2HPWdO
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.333874, 899.774475]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_d0GWh2HPWdO/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_d0GWh2HPWdO/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 95.911522
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_d0GWh2HPWdO/ulay_box_0.nii
++ start ACF calculations out to radius = 17.25 mm
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_d0GWh2HPWdO/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_l9nR5zXrcomRyaUMLWOOLA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_l9nR5zXrcomRyaUMLWOOLA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :870
[1] 2527676
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_nPIAiSh9Zoi++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.579094 852.813599 -com SET_FUNC_RANGE 99.312141 -com SET_THRESHNEW 40.694290 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_nPIAiSh9Zoi
+* Removing temporary image directory '../__tmp_chauf_nPIAiSh9Zoi'.
[1] Done Xvfb :795 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-04/media/__workdir_EAC_1KJHkTaQRzH*'
++ DONE! Image output:
QC_sub-04/media/qc_01_ve2a_epi2anat
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.875446 3.33871 12.7428 8.27184
average err_reml ACF blurs: 0.875446 3.33871 12.7428 8.27184
echo 0.875446 3.33871 12.7428 8.27184 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-06 -dsets pb00.sub-06.r01.tcat+orig.HEAD pb00.sub-06.r02.tcat+orig.HEAD
++ Output dataset ./QC_sub-04/media/__workdir_EAC_gxBru9QSvNl/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-06+tlrc.HEAD -ss_review_dset out.ss_review.sub-06.txt -write_uvars_json out.ss_review_uvars.json
++ Output dataset ./QC_sub-04/media/__workdir_EAC_gxBru9QSvNl/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-04/media/__workdir_EAC_gxBru9QSvNl/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_d0GWh2HPWdO++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.333874 899.774475 -com SET_FUNC_RANGE 95.911522 -com SET_THRESHNEW 38.412792 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_d0GWh2HPWdO
+* Removing temporary image directory '../__tmp_chauf_d0GWh2HPWdO'.
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-05/media/__workdir_EAC_94p9x9HQFSA*'
++ DONE! Image output:
QC_sub-05/media/qc_01_ve2a_epi2anat
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-06.txt
subject ID : sub-06
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-06+tlrc.HEAD
final stats dset : stats.sub-06_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 2
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
average motion (per TR) : 0.0614535
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_BCZarmP3ijj
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
average censored motion : 0.0570993
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 442.971783]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_BCZarmP3ijj/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__workdir_EAC_QxmXfUnGBpT/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_BCZarmP3ijj/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Output dataset ./QC_sub-05/media/__workdir_EAC_QxmXfUnGBpT/eac_0_cp.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Output dataset ./eac_1_ulay_shrp.nii
max motion displacement : 1.47981
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_BCZarmP3ijj/ulay_box_0.nii
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-05/media/__workdir_EAC_QxmXfUnGBpT/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
+ Largest cluster has 233075 voxels
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+ Clustering non-brain voxels ...
+ Clustering voxels ...
max censored displacement : 1.28192
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
average outlier frac (TR) : 0.0018449
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_BCZarmP3ijj/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_lq5Jg6mlGzdeaRA0wp00KA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_lq5Jg6mlGzdeaRA0wp00KA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :129
[1] 2529614
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_IzkT2TbCQja
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 445.098480]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_IzkT2TbCQja/tmp_ulay.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_IzkT2TbCQja/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_IzkT2TbCQja/ulay_box_0.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 143
num TRs per run (censored): 0 3
fraction censored per run : 0 0.0205479
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_IzkT2TbCQja/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_HJKHeIzIf6Qd_--8m9HSug argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_HJKHeIzIf6Qd_--8m9HSug+tlrc.BRIK
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :300
[1] 2530241
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 289
degrees of freedom used : 20
degrees of freedom left : 269
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 3
censor fraction : 0.010274
num regs of interest : 2
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 3
fraction TRs censored : 0.000 0.025
ave mot per sresp (orig) : 0.063176 0.064382
ave mot per sresp (cens) : 0.063176 0.053776
TSNR average : 141.58
global correlation (GCOR) : 0.0449711
anat/EPI mask Dice coef : 0.931195
anat/templ mask Dice coef : 0.956218
maximum F-stat (masked) : 63.1471
blur estimates (ACF) : 0.875446 3.33871 12.7428
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_BCZarmP3ijj++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 442.971783 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_BCZarmP3ijj
+* Removing temporary image directory '../__tmp_chauf_BCZarmP3ijj'.
[1] Done Xvfb :129 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-04/media/__workdir_EAC_gxBru9QSvNl*'
++ DONE! Image output:
QC_sub-04/media/qc_02_va2t_anat2temp
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 38 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_IzkT2TbCQja++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 445.098480 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_IzkT2TbCQja
+* Removing temporary image directory '../__tmp_chauf_IzkT2TbCQja'.
[1] Done Xvfb :300 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-05/media/__workdir_EAC_QxmXfUnGBpT*'
++ DONE! Image output:
QC_sub-05/media/qc_02_va2t_anat2temp
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-04_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 25.373457 -thr_olay 6.773558 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-04/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-04/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-04_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-04/media/__tmp_chauf_UQi3Od52wWL
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_UQi3Od52wWL/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 873.000000 -func_range 873.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-06/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-06/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-06/media/__tmp_chauf_XX1plqJNbpF
++ Output dataset ./QC_sub-04/media/__tmp_chauf_UQi3Od52wWL/tmp_olay.nii
++ User-entered function range value value (25.373457)
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 873.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./QC_sub-06/media/__tmp_chauf_XX1plqJNbpF/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-04/media/__tmp_chauf_UQi3Od52wWL/ulay_box_0.nii
++ Output dataset ./QC_sub-06/media/__tmp_chauf_XX1plqJNbpF/tmp_olay.nii
++ User-entered function range value value (873.000000)
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-05_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 19.055254 -thr_olay 6.415933 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-05/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-05/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ Found input file: stats.sub-05_REML+tlrc.HEAD
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=6..55 y=0..60 z=0..36
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ 3dAutobox: output dataset = QC_sub-06/media/__tmp_chauf_XX1plqJNbpF/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-04/media/__tmp_chauf_UQi3Od52wWL/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_oW90FKR71aPg0KM6kO0MSA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset /tmp/3dcalc_XYZ_oW90FKR71aPg0KM6kO0MSA+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :357
[1] 2534174
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-06/media/__tmp_chauf_XX1plqJNbpF/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_K4NmTaqOEnudBCKYf3PNHw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_K4NmTaqOEnudBCKYf3PNHw+orig.BRIK
++ Output dataset ./QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 112850 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 6.668678 7.391563 -29.1325
++ Will have the ref box central gapord: 7 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :289
[1] 2534250
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Output dataset ./QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ/tmp_olay.nii
++ User-entered function range value value (19.055254)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_jrotSqf2C12c7trOpK8Nhw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_jrotSqf2C12c7trOpK8Nhw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :960
[1] 2534476
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_XX1plqJNbpF++ Writing palette image to QC_sub-06/media/qc_00_vorig_EPI.pbar.jpg
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_UQi3Od52wWL++ Writing palette image to QC_sub-04/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-06/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 873.000000 -com SET_FUNC_RANGE 873.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 6.668678 7.391563 -29.1325 -com SAVE_JPEG sagittalimage QC_sub-06/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-06/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-06/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-06/media/__tmp_chauf_XX1plqJNbpF
+* Removing temporary image directory 'QC_sub-06/media/__tmp_chauf_XX1plqJNbpF'.
[1] Done Xvfb :289 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-06/media/qc_00_vorig_EPI*
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-04/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 25.373457 -com SET_THRESHNEW 6.773558 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-04/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-04/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-04/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-04/media/__tmp_chauf_UQi3Od52wWL
+* Removing temporary image directory 'QC_sub-04/media/__tmp_chauf_UQi3Od52wWL'.
[1] Done Xvfb :357 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-04/media/qc_03_vstat_Full_Fstat*
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ++ Writing palette image to QC_sub-05/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Output dataset ./QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Output dataset ./QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-05/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 19.055254 -com SET_THRESHNEW 6.415933 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-05/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-05/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-05/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ
+* Removing temporary image directory 'QC_sub-05/media/__tmp_chauf_Wj0B3kPL4MJ'.
[1] + Done Xvfb :960 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-05/media/qc_03_vstat_Full_Fstat*
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 117.508400
+ Used gradual clip level = 105.030899 .. 129.502396
+ Number voxels above clip level = 219723
+ Clustering voxels ...
+ Largest cluster has 219478 voxels
+ Clustering voxels ...
+ Largest cluster has 213505 voxels
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+ Filled 10586 voxels in small holes; now have 224091 voxels
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
+ Filled 8198 voxels in large holes; now have 232289 voxels
+ Clustering voxels ...
+ Largest cluster has 232275 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669725 voxels
+ Mask now has 232904 voxels
++ 232904 voxels in the mask [out of 902629: 25.80%]
++ first 9 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 8 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-04_REML+tlrc.HEAD
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
+ Number of voxels in mask = 73360
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 37.447773 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_zTiucky2kaR
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.437025, 901.367737]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_zTiucky2kaR/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_zTiucky2kaR/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 99.665665
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_zTiucky2kaR/ulay_box_0.nii
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-05_REML+tlrc.HEAD
+ Number of voxels in mask = 71803
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_zTiucky2kaR/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_SayOyrfVcROleVVIR-_WGg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_SayOyrfVcROleVVIR-_WGg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :974
[1] 2536366
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_zTiucky2kaR++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.437025 901.367737 -com SET_FUNC_RANGE 99.665665 -com SET_THRESHNEW 37.447773 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_zTiucky2kaR
+* Removing temporary image directory '../__tmp_chauf_zTiucky2kaR'.
[1] Done Xvfb :974 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-06/media/__workdir_EAC_Y0Ka3q0HjFg*'
++ DONE! Image output:
QC_sub-06/media/qc_01_ve2a_epi2anat
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__workdir_EAC_eB12pEJdrIt/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__workdir_EAC_eB12pEJdrIt/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-06/media/__workdir_EAC_eB12pEJdrIt/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_h9Yk75BK8q2
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 461.532326]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_h9Yk75BK8q2/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_h9Yk75BK8q2/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_h9Yk75BK8q2/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_h9Yk75BK8q2/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_uuFB5i8fS2o9TB6vF4gcFw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
+ 3dGrayplot: Elapsed = 9.6 s
++ Output dataset /tmp/3dcalc_XYZ_uuFB5i8fS2o9TB6vF4gcFw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :257
[1] 2537181
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-04+tlrc.HEAD -olay full_mask.sub-04+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-04/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-04+tlrc.HEAD
++ Found input file: full_mask.sub-04+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_gI1ZptTLJh7
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_gI1ZptTLJh7/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_gI1ZptTLJh7/tmp_olay.nii
++ User-entered function range value value (3.29)
+ 3dGrayplot: Elapsed = 9.3 s
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ pbar name has known extension: jpg
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ My command:
@chauffeur_afni -ulay full_mask.sub-05+tlrc.HEAD -olay full_mask.sub-05+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-05/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ 3dAutobox: output dataset = ./__tmp_chauf_gI1ZptTLJh7/ulay_box_0.nii
++ Found input file: full_mask.sub-05+tlrc.HEAD
++ Found input file: full_mask.sub-05+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_ZdFeiohaBMq
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_gI1ZptTLJh7/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_5EWQpKfeCWDt7cjbc7eSow argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Output dataset ././__tmp_chauf_ZdFeiohaBMq/tmp_ulay.nii
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset /tmp/3dcalc_XYZ_5EWQpKfeCWDt7cjbc7eSow+tlrc.BRIK
++ Output dataset ././__tmp_chauf_ZdFeiohaBMq/tmp_olay.nii
++ User-entered function range value value (3.29)
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :974
[1] 2537671
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_ZdFeiohaBMq/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_ZdFeiohaBMq/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_9xx0YC_5FfmjxgiEahAoyA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_9xx0YC_5FfmjxgiEahAoyA+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :683
[1] 2537884
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_h9Yk75BK8q2++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 461.532326 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_h9Yk75BK8q2
+* Removing temporary image directory '../__tmp_chauf_h9Yk75BK8q2'.
[1] Done Xvfb :257 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-06/media/__workdir_EAC_eB12pEJdrIt*'
++ DONE! Image output:
QC_sub-06/media/qc_02_va2t_anat2temp
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/__tmp_chauf_gI1ZptTLJh7++ Writing palette image to QC_sub-04/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-04/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_gI1ZptTLJh7
+* Removing temporary image directory './__tmp_chauf_gI1ZptTLJh7'.
[1] Done Xvfb :974 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-06_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 16.662317 -thr_olay 4.965799 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-06/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-06/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-06_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ Using opacity: 9
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
++ Making temporary work directory to copy vis files: QC_sub-06/media/__tmp_chauf_1MztDywgtnM
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-04/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/__tmp_chauf_ZdFeiohaBMq++ Writing palette image to QC_sub-05/media/qc_06_mot_grayplot.pbar.jpg
++ Output dataset ./QC_sub-06/media/__tmp_chauf_1MztDywgtnM/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-05/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_ZdFeiohaBMq
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
+* Removing temporary image directory './__tmp_chauf_ZdFeiohaBMq'.
[1] Done Xvfb :683 -screen 0 1024x768x24
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
++ DONE (good exit)
ModuleNotFoundError: No module named 'matplotlib'
see: ./__tmp_ZXCV_img*
++ Output dataset ./QC_sub-06/media/__tmp_chauf_1MztDywgtnM/tmp_olay.nii
++ User-entered function range value value (16.662317)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
++ Dimensions (xyzt): 91 109 91 1
ModuleNotFoundError: No module named 'matplotlib'
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-06/media/__tmp_chauf_1MztDywgtnM/ulay_box_0.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-05/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-06/media/__tmp_chauf_1MztDywgtnM/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ__etLxVXZ4fQNe_Mr4OhU9Q argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ__etLxVXZ4fQNe_Mr4OhU9Q+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :151
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
[1] 2539708
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-04/media/qc_09_regr_df.dat
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-04/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-04/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-05/media/qc_09_regr_df.dat
++ Output dataset ./QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE/ulay_box_0.nii
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-05/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-05/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_DmakVi97YUbafb1r3jSVXQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_DmakVi97YUbafb1r3jSVXQ+tlrc.BRIK
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :690
[1] 2540782
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Output dataset ./QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_b76O9R7-QSSSrmZS-RQ0Lg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_b76O9R7-QSSSrmZS-RQ0Lg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :686
[1] 2541013
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_1MztDywgtnM++ Writing palette image to QC_sub-06/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-06/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 16.662317 -com SET_THRESHNEW 4.965799 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-06/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-06/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-06/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-06/media/__tmp_chauf_1MztDywgtnM
+* Removing temporary image directory 'QC_sub-06/media/__tmp_chauf_1MztDywgtnM'.
[1] Done Xvfb :151 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-06/media/qc_03_vstat_Full_Fstat*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE++ Writing palette image to QC_sub-04/media/qc_10_regr_corr_errts.pbar.jpg
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-06_REML+tlrc.HEAD
+ Number of voxels in mask = 71257
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-04/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-04/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-04/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-04/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE
+* Removing temporary image directory 'QC_sub-04/media/__tmp_chauf_pgFSNQjw4kE'.
[1] Done Xvfb :690 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-04/media/qc_10_regr_corr_errts*
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw++ Writing palette image to QC_sub-05/media/qc_10_regr_corr_errts.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-05/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-05/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-05/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-05/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw
+* Removing temporary image directory 'QC_sub-05/media/__tmp_chauf_8yUWBGxSwkw'.
[1] Done Xvfb :686 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-05/media/qc_10_regr_corr_errts*
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-04+tlrc.HEAD -ulay_range 0% 120% -func_range 166 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 166=red 148=oran-red 131=orange 114=oran-yell 96=yell-oran 79=yellow 62=lt-blue2 36=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-04/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 62 - 166 -prefix QC_sub-04/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-04+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc/tmp_olay.nii
++ User-entered function range value value (166)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc/ulay_box_0.nii
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-05+tlrc.HEAD -ulay_range 0% 120% -func_range 178 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 178=red 158=oran-red 139=orange 119=oran-yell 100=yell-oran 80=yellow 61=lt-blue2 33=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-05/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 61 - 178 -prefix QC_sub-05/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-05+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-05/media/__tmp_chauf_djtauunDaRv
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_vWJiOK1tjvObvCXIYjwh-A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_vWJiOK1tjvObvCXIYjwh-A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
-- trying to start Xvfb :688
[1] 2542992
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Output dataset ./QC_sub-05/media/__tmp_chauf_djtauunDaRv/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-05/media/__tmp_chauf_djtauunDaRv/tmp_olay.nii
++ User-entered function range value value (178)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-05/media/__tmp_chauf_djtauunDaRv/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-05/media/__tmp_chauf_djtauunDaRv/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_LNCcF25RS4iRyQ63iGrPxg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_LNCcF25RS4iRyQ63iGrPxg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :721
[1] 2543226
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc++ Writing palette image to QC_sub-04/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-04/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 166=red 148=oran-red 131=orange 114=oran-yell 96=yell-oran 79=yellow 62=lt-blue2 36=blue 0=none -com PBAR_SAVEIM QC_sub-04/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 166 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-04/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-04/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-04/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc
+* Removing temporary image directory 'QC_sub-04/media/__tmp_chauf_IhiZi0WxBDc'.
[1] Done Xvfb :688 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-04/media/qc_11_regr_tsnr_fin*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-04/media/qc_12_warns_xmat.dat
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/QC_sub-05/media/__tmp_chauf_djtauunDaRv++ Writing palette image to QC_sub-05/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Check for censor fraction warnings (general): QC_sub-04/media/qc_13_warns_cen_total.dat
+ 3dGrayplot: Elapsed = 9.0 s
++ Check for censor fraction warnings (per stim): QC_sub-04/media/qc_14_warns_cen_stim.dat
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-05/media/qc_11_regr_tsnr_fin.axi.jpg'
++ pbar name has known extension: jpg
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 178=red 158=oran-red 139=orange 119=oran-yell 100=yell-oran 80=yellow 61=lt-blue2 33=blue 0=none -com PBAR_SAVEIM QC_sub-05/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 178 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-05/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-05/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-05/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-05/media/__tmp_chauf_djtauunDaRv
+* Removing temporary image directory 'QC_sub-05/media/__tmp_chauf_djtauunDaRv'.
++ My command:
[1] Done Xvfb :721 -screen 0 1024x768x24
@chauffeur_afni -ulay full_mask.sub-06+tlrc.HEAD -olay full_mask.sub-06+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-06/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ DONE (good exit)
see: QC_sub-05/media/qc_11_regr_tsnr_fin*
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-06+tlrc.HEAD
++ Found input file: full_mask.sub-06+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_ZmGRqORMgSJ
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Check basic summary quants from proc in: QC_sub-04/media/qc_16_qsumm_ssrev.dat
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-04
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-04+tlrc.HEAD
final stats dset : stats.sub-04_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 0
average motion (per TR) : 0.0651394
average censored motion : 0.0651394
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 2.6206
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 2.6206
average outlier frac (TR) : 0.000910993
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
++ Output dataset ././__tmp_chauf_ZmGRqORMgSJ/tmp_ulay.nii
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
censor fraction : 0.000000
num regs of interest : 2
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
num TRs per stim (orig) : 120 120
++ Authored by: A cast of thousands
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
ave mot per sresp (orig) : 0.063507 0.067320
ave mot per sresp (cens) : 0.063507 0.067320
TSNR average : 125.258
global correlation (GCOR) : 0.0709615
anat/EPI mask Dice coef : 0.916187
anat/templ mask Dice coef : 0.954796
maximum F-stat (masked) : 98.597
blur estimates (ACF) : 0.863972 3.4079 13.4515
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-04
++ Output dataset ././__tmp_chauf_ZmGRqORMgSJ/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-04/index.html
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-04.results/QC_sub-04/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-04.results/QC_sub-04/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:11:41 UTC 2025
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-05/media/qc_12_warns_xmat.dat
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_ZmGRqORMgSJ/ulay_box_0.nii
++ Check for censor fraction warnings (general): QC_sub-05/media/qc_13_warns_cen_total.dat
++ Check for censor fraction warnings (per stim): QC_sub-05/media/qc_14_warns_cen_stim.dat
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_ZmGRqORMgSJ/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Fo-YziX7FUmd4JQEYEVirw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Fo-YziX7FUmd4JQEYEVirw+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :307
[1] 2544860
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Check basic summary quants from proc in: QC_sub-05/media/qc_16_qsumm_ssrev.dat
echo auto-generated by afni_proc.py, Tue Nov 4 05:11:42 2025
auto-generated by afni_proc.py, Tue Nov 4 05:11:42 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 05:11:42 UTC 2025
afni -ver
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-05
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-05+tlrc.HEAD
final stats dset : stats.sub-05_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 2
average motion (per TR) : 0.0532986
average censored motion : 0.0504625
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.22172
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.22172
average outlier frac (TR) : 0.00152065
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 143
num TRs per run (censored): 0 3
fraction censored per run : 0 0.0205479
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 289
degrees of freedom used : 20
degrees of freedom left : 269
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 3
censor fraction : 0.010274
num regs of interest : 2
num TRs per stim (orig) : 120 120
num TRs censored per stim : 3 0
fraction TRs censored : 0.025 0.000
ave mot per sresp (orig) : 0.064327 0.045496
ave mot per sresp (cens) : 0.057653 0.045496
TSNR average : 133.994
global correlation (GCOR) : 0.0654473
anat/EPI mask Dice coef : 0.915418
anat/templ mask Dice coef : 0.954347
maximum F-stat (masked) : 73.2756
blur estimates (ACF) : 0.853893 3.3299 13.7531
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-05
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-07_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-07.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-07.tcsh |& tee ./afni_pro_glm/output.proc.sub-07.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-07.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-07.tcsh
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
set subj = sub-07
endif
set output_dir = ./afni_pro_glm/sub-07.results
if ( -d ./afni_pro_glm/sub-07.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-05/index.html
mkdir -p ./afni_pro_glm/sub-07.results
mkdir ./afni_pro_glm/sub-07.results/stimuli
cp ./ds000102/sub-07/func/congruent.1D ./ds000102/sub-07/func/incongruent.1D ./afni_pro_glm/sub-07.results/stimuli
3dcopy ds000102/sub-07/anat/sub-07_T1w.nii.gz ./afni_pro_glm/sub-07.results/sub-07_T1w
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-05.results/QC_sub-05/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-05.results/QC_sub-05/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:11:42 UTC 2025
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_pro_glm/sub-07.results/pb00.sub-07.r01.tcat ds000102/sub-07/func/sub-07_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
echo auto-generated by afni_proc.py, Tue Nov 4 05:11:43 2025
auto-generated by afni_proc.py, Tue Nov 4 05:11:43 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 05:11:43 UTC 2025
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-08_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-08.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-08.tcsh |& tee ./afni_pro_glm/output.proc.sub-08.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-08.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-08.tcsh
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
set subj = sub-08
endif
set output_dir = ./afni_pro_glm/sub-08.results
if ( -d ./afni_pro_glm/sub-08.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_pro_glm/sub-08.results
mkdir ./afni_pro_glm/sub-08.results/stimuli
cp ./ds000102/sub-08/func/congruent.1D ./ds000102/sub-08/func/incongruent.1D ./afni_pro_glm/sub-08.results/stimuli
3dcopy ds000102/sub-08/anat/sub-08_T1w.nii.gz ./afni_pro_glm/sub-08.results/sub-08_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.8 s
3dTcat -prefix ./afni_pro_glm/sub-07.results/pb00.sub-07.r02.tcat ds000102/sub-07/func/sub-07_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_pro_glm/sub-08.results/pb00.sub-08.r01.tcat ds000102/sub-08/func/sub-08_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.9 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-07.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-07.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.9 s
3dTcat -prefix ./afni_pro_glm/sub-08.results/pb00.sub-08.r02.tcat ds000102/sub-08/func/sub-08_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-08.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-08.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/__tmp_chauf_ZmGRqORMgSJ++ Writing palette image to QC_sub-06/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-06/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_ZmGRqORMgSJ
+* Removing temporary image directory './__tmp_chauf_ZmGRqORMgSJ'.
[1] Done Xvfb :307 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
++ 39984 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-07.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-06/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 38915 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-08.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-06/media/qc_09_regr_df.dat
++ 39885 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 269
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 02
set minouttr = 123
echo min outlier: run 02, TR 123
tee out.min_outlier.txt
min outlier: run 02, TR 123
3dbucket -prefix vr_base_min_outlier pb00.sub-07.r02.tcat+orig[123]
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-06/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-06/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-06/media/__tmp_chauf_uenAWOWmGM6
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
align_epi_anat.py -anat2epi -anat sub-07_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
++ 38896 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-07_T1w+orig
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-07_T1w*
#Script is running (command trimmed):
3dcopy ./sub-07_T1w+orig ./__tt_sub-07_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 28
++ Output dataset ./QC_sub-06/media/__tmp_chauf_uenAWOWmGM6/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__tmp_chauf_uenAWOWmGM6/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-07_T1w+orig -prefix ./__tt_sub-07_T1w_ns
set minoutrun = 01
set minouttr = 28
echo min outlier: run 01, TR 28
tee out.min_outlier.txt
min outlier: run 01, TR 28
3dbucket -prefix vr_base_min_outlier pb00.sub-08.r01.tcat+orig[28]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-06/media/__tmp_chauf_uenAWOWmGM6/ulay_box_0.nii
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
align_epi_anat.py -anat2epi -anat sub-08_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-06/media/__tmp_chauf_uenAWOWmGM6/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_ciHdcHObuxeMYPpGNppEuQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_ciHdcHObuxeMYPpGNppEuQ+tlrc.BRIK
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-08_T1w+orig
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
#++ Multi-cost is lpc
++ Using only the boxes+balls mask
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
#Script is running:
\rm -f ./__tt_sub-08_T1w*
------------------- end of optionizing -------------------
-- trying to start Xvfb :978
[1] 2548221
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
#Script is running (command trimmed):
3dcopy ./sub-08_T1w+orig ./__tt_sub-08_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-08_T1w+orig -prefix ./__tt_sub-08_T1w_ns
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_uenAWOWmGM6++ Writing palette image to QC_sub-06/media/qc_10_regr_corr_errts.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-06/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-06/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-06/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-06/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-06/media/__tmp_chauf_uenAWOWmGM6
+* Removing temporary image directory 'QC_sub-06/media/__tmp_chauf_uenAWOWmGM6'.
++ DONE (good exit)
see: QC_sub-06/media/qc_10_regr_corr_errts*
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-06+tlrc.HEAD -ulay_range 0% 120% -func_range 186 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 186=red 167=oran-red 148=orange 129=oran-yell 110=yell-oran 91=yellow 72=lt-blue2 46=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-06/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 72 - 186 -prefix QC_sub-06/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-06+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-06/media/__tmp_chauf_32VAFyruEt7
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__tmp_chauf_32VAFyruEt7/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-06/media/__tmp_chauf_32VAFyruEt7/tmp_olay.nii
++ User-entered function range value value (186)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-06/media/__tmp_chauf_32VAFyruEt7/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-06/media/__tmp_chauf_32VAFyruEt7/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Z_4f4tGSsccJeU2dMuJnzg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Z_4f4tGSsccJeU2dMuJnzg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :692
[1] 2549070
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/QC_sub-06/media/__tmp_chauf_32VAFyruEt7++ Writing palette image to QC_sub-06/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-06/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 186=red 167=oran-red 148=orange 129=oran-yell 110=yell-oran 91=yellow 72=lt-blue2 46=blue 0=none -com PBAR_SAVEIM QC_sub-06/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 186 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-06/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-06/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-06/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-06/media/__tmp_chauf_32VAFyruEt7
+* Removing temporary image directory 'QC_sub-06/media/__tmp_chauf_32VAFyruEt7'.
[1] Done Xvfb :692 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-06/media/qc_11_regr_tsnr_fin*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-06/media/qc_12_warns_xmat.dat
++ Check for censor fraction warnings (general): QC_sub-06/media/qc_13_warns_cen_total.dat
++ Check for censor fraction warnings (per stim): QC_sub-06/media/qc_14_warns_cen_stim.dat
++ Check basic summary quants from proc in: QC_sub-06/media/qc_16_qsumm_ssrev.dat
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-06
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-06+tlrc.HEAD
final stats dset : stats.sub-06_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 2
average motion (per TR) : 0.0614535
average censored motion : 0.0570993
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.47981
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.28192
average outlier frac (TR) : 0.0018449
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 143
num TRs per run (censored): 0 3
fraction censored per run : 0 0.0205479
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 289
degrees of freedom used : 20
degrees of freedom left : 269
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 3
censor fraction : 0.010274
num regs of interest : 2
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 3
fraction TRs censored : 0.000 0.025
ave mot per sresp (orig) : 0.063176 0.064382
ave mot per sresp (cens) : 0.063176 0.053776
TSNR average : 141.58
global correlation (GCOR) : 0.0449711
anat/EPI mask Dice coef : 0.931195
anat/templ mask Dice coef : 0.956218
maximum F-stat (masked) : 63.1471
blur estimates (ACF) : 0.875446 3.33871 12.7428
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-06
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-06/index.html
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-06.results/QC_sub-06/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-06.results/QC_sub-06/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:12:04 UTC 2025
echo auto-generated by afni_proc.py, Tue Nov 4 05:12:05 2025
auto-generated by afni_proc.py, Tue Nov 4 05:12:05 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 05:12:05 UTC 2025
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-09_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
** failed to load module matplotlib.pyplot
** warning: -html_review_style pythonic: missing matplotlib library
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: ./afni_pro_glm/proc.sub-09.tcsh
to execute via tcsh:
tcsh -xef ./afni_pro_glm/proc.sub-09.tcsh |& tee ./afni_pro_glm/output.proc.sub-09.tcsh
to execute via bash:
tcsh -xef ./afni_pro_glm/proc.sub-09.tcsh 2>&1 | tee ./afni_pro_glm/output.proc.sub-09.tcsh
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
set subj = sub-09
endif
set output_dir = ./afni_pro_glm/sub-09.results
if ( -d ./afni_pro_glm/sub-09.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_pro_glm/sub-09.results
mkdir ./afni_pro_glm/sub-09.results/stimuli
cp ./ds000102/sub-09/func/congruent.1D ./ds000102/sub-09/func/incongruent.1D ./afni_pro_glm/sub-09.results/stimuli
3dcopy ds000102/sub-09/anat/sub-09_T1w.nii.gz ./afni_pro_glm/sub-09.results/sub-09_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_pro_glm/sub-09.results/pb00.sub-09.r01.tcat ds000102/sub-09/func/sub-09_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.8 s
3dTcat -prefix ./afni_pro_glm/sub-09.results/pb00.sub-09.r02.tcat ds000102/sub-09/func/sub-09_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.8 s
set tr_counts = ( 146 146 )
cd ./afni_pro_glm/sub-09.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-09.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 37553 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub-09.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 37441 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 127
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 01
set minouttr = 127
echo min outlier: run 01, TR 127
tee out.min_outlier.txt
min outlier: run 01, TR 127
3dbucket -prefix vr_base_min_outlier pb00.sub-09.r01.tcat+orig[127]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
align_epi_anat.py -anat2epi -anat sub-09_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-09_T1w+orig
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-09_T1w*
#Script is running (command trimmed):
3dcopy ./sub-09_T1w+orig ./__tt_sub-09_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-09_T1w+orig -prefix ./__tt_sub-09_T1w_ns
#Script is running (command trimmed):
3dinfo ./__tt_sub-08_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/__tt_sub-08_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 345.050415
+ Used gradual clip level = 330.013794 .. 365.519012
+ Number voxels above clip level = 39471
+ Clustering voxels ...
+ Largest cluster has 38949 voxels
+ Clustering voxels ...
+ Largest cluster has 38549 voxels
+ Filled 354 voxels in small holes; now have 38903 voxels
+ Filled 3 voxels in large holes; now have 38906 voxels
+ Clustering voxels ...
+ Largest cluster has 38906 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 124934 voxels
+ Mask now has 38906 voxels
++ 38906 voxels in the mask [out of 163840: 23.75%]
++ first 11 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 0 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 3 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 819.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/819.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-08_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-08_T1w_ns+orig -prefix ./sub-08_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-08_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
#Script is running (command trimmed):
3dinfo ./__tt_sub-07_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/__tt_sub-07_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 355.067200
+ Used gradual clip level = 342.547211 .. 370.037811
+ Number voxels above clip level = 41073
+ Clustering voxels ...
+ Largest cluster has 40053 voxels
+ Clustering voxels ...
+ Largest cluster has 39582 voxels
+ Filled 250 voxels in small holes; now have 39832 voxels
+ Clustering voxels ...
+ Largest cluster has 39832 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 124008 voxels
+ Mask now has 39832 voxels
++ 39832 voxels in the mask [out of 163840: 24.31%]
++ first 10 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 2 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 3 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 845.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/845.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-07_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-07_T1w_ns+orig -prefix ./sub-07_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-07_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-07_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 1583872 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=8 ytop=3
++ Zero-pad: zbot=7 ztop=5
++ 38906 voxels [15.6%] in weight mask
++ Output dataset ./__tt_sub-08_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 38906
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.963 35.048 27.741 (index)
+ source center of mass = 84.800 114.693 116.298 (index)
+ source-target CM = 86.190 -3.441 -1.709 (xyz)
+ estimated center of mass shifts = 86.190 -3.441 -1.709
++ shift param auto-range: 25.5..146.9 -74.7..67.8 -67.2..63.8
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.190132 .. 96.190132 center = 86.190132
+ Range param#2 [y-shift] = -13.440697 .. 6.559303 center = -3.440697
+ Range param#3 [z-shift] = -11.708786 .. 8.291214 center = -1.708786
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.190132 .. 126.190132 center = 86.190132
+ Range param#2 [y-shift] = -43.440697 .. 36.559303 center = -3.440697
+ Range param#3 [z-shift] = -41.708786 .. 38.291214 center = -1.708786
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000088
++ Final parameter search ranges:
+ x-shift = 46.190 .. 126.190
+ y-shift = -43.441 .. 36.559
+ z-shift = -41.709 .. 38.291
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1583872 [out of 11534336] voxels
+ base mask has 52253 [out of 249600] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=64 usiz=159.5
+ - copying weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 32391 total points stored in 63 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
++ 1554288 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=6 ytop=4
++ Zero-pad: zbot=8 ztop=5
++ 39832 voxels [15.9%] in weight mask
++ Output dataset ./__tt_sub-07_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 39832
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.328 33.595 27.760 (index)
+ source center of mass = 86.770 111.660 125.629 (index)
+ source-target CM = 86.255 -1.657 -0.020 (xyz)
+ estimated center of mass shifts = 86.255 -1.657 -0.020
++ shift param auto-range: 25.6..146.9 -72.0..68.6 -66.8..66.7
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.254662 .. 96.254662 center = 86.254662
+ Range param#2 [y-shift] = -11.657471 .. 8.342529 center = -1.657471
+ Range param#3 [z-shift] = -10.019867 .. 9.980133 center = -0.019867
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.254662 .. 126.254662 center = 86.254662
+ Range param#2 [y-shift] = -41.657471 .. 38.342529 center = -1.657471
+ Range param#3 [z-shift] = -40.019867 .. 39.980133 center = -0.019867
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000087
++ Final parameter search ranges:
+ x-shift = 46.255 .. 126.255
+ y-shift = -41.657 .. 38.343
+ z-shift = -40.020 .. 39.980
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1554288 [out of 11534336] voxels
+ base mask has 52970 [out of 251008] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=52 usiz=160
+ - copying weight image
+ - using 39832 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 32246 total points stored in 62 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o-.+.o--.--+oo-.++-.o-+-.+o+o---+.--.oo.+++o+$+.--.+..o.o + - best 88 costs found:
o= 0 v=-0.078395: 86.19 -3.44 -1.71 0.00 0.00 0.00 [grid]
o= 1 v=-0.024170: 91.62 4.15 28.38 -38.19 22.74 -41.67 [rand]
o= 2 v=-0.022830: 100.19 -18.97 33.97 -14.61 -39.00 -28.71 [rand]
o= 3 v=-0.022358: 92.86 -30.11 24.96 7.50 -30.00 -30.00 [grid]
o= 4 v=-0.022101: 121.80 -7.85 -38.58 -37.76 24.22 3.86 [rand]
o= 5 v=-0.021851: 97.68 -19.75 -35.46 -19.17 13.96 2.56 [rand]
o= 6 v=-0.021730: 122.84 -20.67 20.55 5.26 -41.39 -40.03 [rand]
o= 7 v=-0.021515: 101.83 -12.20 -35.54 -23.16 17.17 -3.59 [rand]
o= 8 v=-0.021205: 91.12 -0.47 27.39 -40.99 18.21 -33.12 [rand]
o= 9 v=-0.020806: 121.80 -7.85 -38.58 -37.76 24.22 -3.86 [rand]
o=10 v=-0.020703: 97.68 -19.75 -35.46 -19.17 13.96 -2.56 [rand]
o=11 v=-0.020229: 121.80 0.97 35.16 37.76 -24.22 -3.86 [rand]
o=12 v=-0.020075: 90.34 20.37 30.18 31.52 -11.26 -33.63 [rand]
o=13 v=-0.019908: 100.19 -18.97 33.97 14.61 -39.00 -28.71 [rand]
o=14 v=-0.019694: 120.99 -17.22 -35.72 10.62 -7.10 -10.42 [rand]
o=15 v=-0.019540: 119.22 1.58 -38.52 -11.98 4.28 -19.20 [rand]
o=16 v=-0.019170: 88.48 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=17 v=-0.018913: 121.80 0.97 -38.58 -37.76 24.22 3.86 [rand]
o=18 v=-0.018894: 96.65 -30.71 19.79 -34.95 -5.03 -42.69 [rand]
o=19 v=-0.018891: 115.81 31.17 35.11 24.31 -11.89 -2.84 [rand]
o=20 v=-0.018703: 92.86 -30.11 24.96 -7.50 -30.00 -30.00 [grid]
o=21 v=-0.018636: 115.81 31.17 35.11 24.31 -11.89 2.84 [rand]
o=22 v=-0.018271: 74.70 12.87 -35.46 19.17 13.96 2.56 [rand]
o=23 v=-0.018249: 83.90 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=24 v=-0.018182: 119.22 -8.46 -38.52 -11.98 4.28 -19.20 [rand]
o=25 v=-0.018157: 112.86 3.23 -28.38 -30.00 7.50 7.50 [grid]
o=26 v=-0.018092: 113.03 -16.54 -34.29 -39.07 28.37 8.01 [rand]
o=27 v=-0.018024: 121.80 -7.85 35.16 37.76 -24.22 -3.86 [rand]
o=28 v=-0.017957: 91.12 -6.41 27.39 -40.99 18.21 -33.12 [rand]
o=29 v=-0.017956: 101.83 -12.20 -35.54 -23.16 17.17 3.59 [rand]
o=30 v=-0.017508: 120.99 -17.22 -35.72 -10.62 -7.10 -10.42 [rand]
o=31 v=-0.017362: 74.70 12.87 -35.46 19.17 13.96 -2.56 [rand]
o=32 v=-0.017349: 120.99 -17.22 -35.72 -10.62 7.10 -10.42 [rand]
o=33 v=-0.016693: 105.31 19.97 -35.01 -37.88 13.88 18.42 [rand]
o=34 v=-0.016675: 102.99 15.02 30.65 10.10 -16.32 -39.31 [rand]
o=35 v=-0.016669: 105.31 19.97 -35.01 -37.88 13.88 -18.42 [rand]
o=36 v=-0.016632: 120.99 -17.22 -35.72 10.62 7.10 -10.42 [rand]
o=37 v=-0.016450: 99.53 -27.69 29.08 6.84 -32.76 -20.90 [rand]
o=38 v=-0.016261: 79.52 -30.11 24.96 -30.00 -7.50 -30.00 [grid]
o=39 v=-0.016134: 99.35 34.44 35.49 14.03 -8.39 -25.20 [rand]
o=40 v=-0.016094: 79.78 30.50 -38.28 -19.28 27.13 15.13 [rand]
o=41 v=-0.015858: 95.39 -15.21 -23.44 -6.82 34.51 24.39 [rand]
o=42 v=-0.015801: 120.99 -17.22 -35.72 10.62 -7.10 10.42 [rand]
o=43 v=-0.015734: 70.55 5.32 -35.54 23.16 17.17 -3.59 [rand]
o=44 v=-0.015723: 67.07 19.97 -35.01 37.88 13.88 -18.42 [rand]
o=45 v=-0.015700: 92.86 23.23 24.96 30.00 -7.50 -30.00 [grid]
o=46 v=-0.015623: 113.12 -31.58 -39.65 15.05 4.71 -12.78 [rand]
o=47 v=-0.015567: 92.86 -10.11 -28.38 -7.50 7.50 30.00 [grid]
o=48 v=-0.015330: 88.48 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=49 v=-0.015192: 92.03 -32.53 20.97 30.43 -41.57 -36.66 [rand]
o=50 v=-0.015111: 103.24 -26.13 22.54 -12.15 -12.44 -39.75 [rand]
o=51 v=-0.015075: 122.78 -30.74 29.53 -25.97 -37.53 -40.21 [rand]
o=52 v=-0.014925: 92.86 -10.11 -28.38 -7.50 30.00 30.00 [grid]
o=53 v=-0.014904: 121.80 0.97 -38.58 -37.76 24.22 -3.86 [rand]
o=54 v=-0.014835: 102.99 -21.90 30.65 -10.10 -16.32 -39.31 [rand]
o=55 v=-0.014711: 102.99 15.02 -34.07 -10.10 -16.32 39.31 [rand]
o=56 v=-0.014651: 119.22 -8.46 -38.52 -11.98 -4.28 -19.20 [rand]
o=57 v=-0.014527: 99.35 -41.33 -38.90 14.03 8.39 -25.20 [rand]
o=58 v=-0.014469: 70.55 -12.20 -35.54 23.16 17.17 3.59 [rand]
o=59 v=-0.014214: 121.80 -7.85 35.16 37.76 -24.22 3.86 [rand]
o=60 v=-0.014171: 70.55 -12.20 -35.54 23.16 17.17 -3.59 [rand]
o=61 v=-0.014132: 94.85 -27.60 34.86 27.41 -42.10 -24.35 [rand]
o=62 v=-0.014108: 92.86 -10.11 -28.38 30.00 7.50 7.50 [grid]
o=63 v=-0.013985: 92.61 -37.38 -38.28 19.28 27.13 -15.13 [rand]
o=64 v=-0.013771: 56.91 -19.20 28.15 13.45 -19.96 14.96 [rand]
o=65 v=-0.013738: 104.93 -39.84 -34.76 24.90 4.74 -17.23 [rand]
o=66 v=-0.013600: 92.86 3.23 -28.38 -7.50 30.00 30.00 [grid]
o=67 v=-0.013545: 102.99 15.02 -34.07 10.10 -16.32 39.31 [rand]
o=68 v=-0.013506: 97.68 12.87 -35.46 -19.17 13.96 -2.56 [rand]
o=69 v=-0.013420: 92.86 3.23 24.96 30.00 -30.00 -30.00 [grid]
o=70 v=-0.013384: 83.90 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=71 v=-0.013341: 115.81 -38.05 -38.52 24.31 -11.89 -2.84 [rand]
o=72 v=-0.013295: 59.26 -31.58 36.24 15.05 -4.71 12.78 [rand]
o=73 v=-0.013221: 79.78 30.50 -38.28 19.28 27.13 15.13 [rand]
o=74 v=-0.013191: 59.35 9.66 -34.29 39.07 28.37 -8.01 [rand]
o=75 v=-0.013149: 118.40 -32.49 34.43 -40.41 -19.09 -40.75 [rand]
o=76 v=-0.013072: 70.55 5.32 -35.54 23.16 17.17 3.59 [rand]
o=77 v=-0.013043: 65.94 23.84 -36.19 37.90 5.03 3.08 [rand]
o=78 v=-0.012929: 92.86 -10.11 -28.38 7.50 7.50 7.50 [grid]
o=79 v=-0.012857: 60.67 -34.78 32.57 42.46 -41.07 35.63 [rand]
o=80 v=-0.012789: 99.35 -41.33 35.49 14.03 -8.39 -25.20 [rand]
o=81 v=-0.012665: 97.68 12.87 -35.46 -19.17 13.96 2.56 [rand]
o=82 v=-0.012652: 112.86 -30.11 -28.38 30.00 7.50 7.50 [grid]
o=83 v=-0.012596: 104.93 -39.84 31.34 -24.90 -4.74 -17.23 [rand]
o=84 v=-0.012570: 83.90 -10.62 -33.40 10.97 18.37 -5.04 [rand]
o=85 v=-0.012553: 92.86 -10.11 -28.38 -7.50 30.00 7.50 [grid]
o=86 v=-0.012529: 92.86 -10.11 24.96 -7.50 -7.50 -30.00 [grid]
o=87 v=-0.012519: 80.37 10.75 27.93 38.21 -15.41 -13.51 [rand]
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o-.+o--o.-o.+-+o++o-..--o++.+.-o---+oo-o.oo$-..+o+oo+++ + - best 88 costs found:
o= 0 v=-0.079318: 86.25 -1.66 -0.02 0.00 0.00 0.00 [grid]
o= 1 v=-0.022845: 101.90 -10.42 -33.85 -23.16 17.17 -3.59 [rand]
o= 2 v=-0.022051: 113.10 -14.75 -32.60 -39.07 28.37 8.01 [rand]
o= 3 v=-0.021369: 67.14 21.76 -33.32 37.88 13.88 -18.42 [rand]
o= 4 v=-0.020802: 60.88 25.60 -37.53 41.23 -14.90 -41.72 [rand]
o= 5 v=-0.020670: 101.90 -10.42 -33.85 -23.16 17.17 3.59 [rand]
o= 6 v=-0.020284: 88.55 -8.84 -31.71 -10.97 18.37 -5.04 [rand]
o= 7 v=-0.019795: 60.73 29.68 -34.30 42.46 -41.07 -35.63 [rand]
o= 8 v=-0.019656: 83.96 -8.84 -31.71 -10.97 18.37 -5.04 [rand]
o= 9 v=-0.019637: 79.59 -8.32 -26.69 7.50 30.00 -30.00 [grid]
o=10 v=-0.018782: 83.96 -8.84 -31.71 -10.97 18.37 5.04 [rand]
o=11 v=-0.018313: 92.92 -8.32 -26.69 -7.50 30.00 30.00 [grid]
o=12 v=-0.017438: 95.46 -13.43 -21.75 -6.82 34.51 24.39 [rand]
o=13 v=-0.017427: 61.29 27.39 -35.47 17.69 -14.58 -34.92 [rand]
o=14 v=-0.017152: 88.55 -8.84 -31.71 -10.97 18.37 5.04 [rand]
o=15 v=-0.016563: 121.05 -15.44 -34.03 -10.62 7.10 -10.42 [rand]
o=16 v=-0.015806: 121.86 -6.07 -36.89 -37.76 24.22 3.86 [rand]
o=17 v=-0.015692: 77.05 -13.43 -21.75 6.82 34.51 -24.39 [rand]
o=18 v=-0.015436: 92.35 -8.01 -16.44 -16.81 28.15 41.40 [rand]
o=19 v=-0.015331: 113.10 -14.75 -32.60 -39.07 28.37 -8.01 [rand]
o=20 v=-0.015324: 121.05 -15.44 -34.03 10.62 -7.10 -10.42 [rand]
o=21 v=-0.014957: 79.59 -8.32 -26.69 -7.50 30.00 -7.50 [grid]
o=22 v=-0.014761: 105.37 21.76 -33.32 -37.88 13.88 -18.42 [rand]
o=23 v=-0.014677: 79.84 32.29 -36.59 -19.28 27.13 15.13 [rand]
o=24 v=-0.014628: 92.92 -8.32 -26.69 -7.50 30.00 7.50 [grid]
o=25 v=-0.014614: 105.37 21.76 -33.32 -37.88 13.88 18.42 [rand]
o=26 v=-0.014514: 59.59 5.01 -26.69 30.00 7.50 -7.50 [grid]
o=27 v=-0.014494: 111.63 25.60 -37.53 -41.23 -14.90 41.72 [rand]
o=28 v=-0.014400: 92.92 5.01 -26.69 -7.50 7.50 30.00 [grid]
o=29 v=-0.014180: 79.59 5.01 -26.69 7.50 30.00 -30.00 [grid]
o=30 v=-0.013896: 67.14 21.76 -33.32 37.88 13.88 18.42 [rand]
o=31 v=-0.013699: 119.28 -6.68 -36.84 -11.98 4.28 -19.20 [rand]
o=32 v=-0.013602: 83.96 5.52 -31.71 -10.97 18.37 5.04 [rand]
o=33 v=-0.013456: 79.20 18.16 -35.77 25.15 27.05 -27.90 [rand]
o=34 v=-0.013391: 92.92 5.01 -26.69 -7.50 30.00 30.00 [grid]
o=35 v=-0.013168: 70.61 7.10 -33.85 23.16 17.17 -3.59 [rand]
o=36 v=-0.013003: 122.85 -28.96 -31.26 25.97 37.53 -40.21 [rand]
o=37 v=-0.012994: 79.59 5.01 -26.69 -7.50 30.00 -7.50 [grid]
o=38 v=-0.012926: 70.04 12.45 -35.15 15.74 34.09 -7.78 [rand]
o=39 v=-0.012804: 100.25 13.88 -35.70 -14.61 39.00 28.71 [rand]
o=40 v=-0.012767: 119.28 -6.68 -36.84 -11.98 -4.28 -19.20 [rand]
o=41 v=-0.012719: 111.77 29.68 -34.30 -42.46 -41.07 35.63 [rand]
o=42 v=-0.012626: 79.59 5.01 -26.69 -7.50 7.50 -7.50 [grid]
o=43 v=-0.012601: 115.27 -28.80 -33.63 7.44 27.42 -25.63 [rand]
o=44 v=-0.012492: 112.92 -8.32 -26.69 -30.00 7.50 7.50 [grid]
o=45 v=-0.012466: 106.51 25.62 -34.50 -37.90 5.03 -3.08 [rand]
o=46 v=-0.012425: 97.74 -17.97 -33.77 -19.17 13.96 -2.56 [rand]
o=47 v=-0.012307: 119.28 3.36 -36.84 -11.98 4.28 -19.20 [rand]
o=48 v=-0.012203: 119.28 3.36 -36.84 -11.98 -4.28 -19.20 [rand]
o=49 v=-0.012176: 83.96 5.52 -31.71 -10.97 18.37 -5.04 [rand]
o=50 v=-0.012048: 97.74 -17.97 -33.77 -19.17 13.96 2.56 [rand]
o=51 v=-0.012038: 121.05 -15.44 -34.03 10.62 7.10 -10.42 [rand]
o=52 v=-0.011970: 59.59 5.01 -26.69 30.00 7.50 -30.00 [grid]
o=53 v=-0.011925: 88.55 5.52 -31.71 -10.97 18.37 -5.04 [rand]
o=54 v=-0.011875: 92.92 5.01 -26.69 7.50 7.50 7.50 [grid]
o=55 v=-0.011744: 69.46 16.81 -32.38 10.10 -16.32 -39.31 [rand]
o=56 v=-0.011718: 59.59 5.01 -26.69 30.00 -7.50 -30.00 [grid]
o=57 v=-0.011552: 70.61 -10.42 -33.85 23.16 17.17 3.59 [rand]
o=58 v=-0.011551: 92.92 -8.32 -26.69 -30.00 30.00 30.00 [grid]
o=59 v=-0.011533: 70.61 -10.42 -33.85 23.16 17.17 -3.59 [rand]
o=60 v=-0.011498: 79.59 -28.32 -26.69 -7.50 30.00 -7.50 [grid]
o=61 v=-0.011465: 60.93 27.01 -24.01 42.33 -22.93 -32.02 [rand]
o=62 v=-0.011374: 112.92 -8.32 -26.69 -30.00 30.00 -7.50 [grid]
o=63 v=-0.011330: 79.59 -8.32 -26.69 -7.50 7.50 -30.00 [grid]
o=64 v=-0.011324: 121.05 12.12 -34.03 -10.62 7.10 -10.42 [rand]
o=65 v=-0.011185: 112.92 5.01 -26.69 -30.00 7.50 7.50 [grid]
o=66 v=-0.010996: 84.13 4.85 -30.12 6.85 38.25 -36.13 [rand]
o=67 v=-0.010904: 66.00 25.62 -34.50 37.90 5.03 3.08 [rand]
o=68 v=-0.010885: 74.76 -17.97 -33.77 19.17 13.96 2.56 [rand]
o=69 v=-0.010833: 112.92 5.01 -26.69 -30.00 -7.50 30.00 [grid]
o=70 v=-0.010831: 59.59 5.01 -26.69 7.50 30.00 7.50 [grid]
o=71 v=-0.010786: 113.10 11.44 -32.60 -39.07 28.37 -8.01 [rand]
o=72 v=-0.010784: 92.92 -8.32 -26.69 -7.50 7.50 30.00 [grid]
o=73 v=-0.010771: 77.05 -13.43 -21.75 -6.82 34.51 -24.39 [rand]
o=74 v=-0.010745: 121.05 -15.44 -34.03 -10.62 -7.10 -10.42 [rand]
o=75 v=-0.010632: 112.92 25.01 -26.69 -30.00 -7.50 30.00 [grid]
o=76 v=-0.010449: 113.10 11.44 -32.60 -39.07 28.37 8.01 [rand]
o=77 v=-0.010322: 74.76 -17.97 -33.77 19.17 13.96 -2.56 [rand]
o=78 v=-0.010260: 103.05 16.81 -32.38 -10.10 -16.32 39.31 [rand]
o=79 v=-0.010243: 101.90 7.10 -33.85 -23.16 17.17 -3.59 [rand]
o=80 v=-0.010201: 73.09 36.23 -37.22 -14.03 -8.39 -25.20 [rand]
o=81 v=-0.010186: 82.67 -37.86 -16.60 -36.12 41.59 17.14 [rand]
o=82 v=-0.010179: 94.92 -25.81 -36.59 -27.41 42.10 -24.35 [rand]
o=83 v=-0.010176: 79.59 -8.32 -26.69 -30.00 7.50 -7.50 [grid]
o=84 v=-0.010095: 59.59 25.01 -26.69 30.00 30.00 -7.50 [grid]
o=85 v=-0.010072: 121.05 -15.44 -34.03 10.62 -7.10 10.42 [rand]
o=86 v=-0.009973: 79.59 -8.32 -26.69 -7.50 7.50 -7.50 [grid]
o=87 v=-0.009966: 59.59 25.01 -26.69 30.00 -7.50 -30.00 [grid]
+ - A little optimization:*[#16335=-0.0833147] *[#16338=-0.0996876] *[#16339=-0.122258] *[#16342=-0.125257] *[#16344=-0.126031] *[#16346=-0.127847] *[#16349=-0.129014] *[#16352=-0.129155] *[#16355=-0.129328] *[#16356=-0.129367] *[#16360=-0.12939] *[#16362=-0.129425] *[#16363=-0.129437] *[#16366=-0.129478] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.129478: 86.56 -2.92 0.24 -1.17 0.39 0.22 [grid] [f=50]
o= 1 v=-0.025208: 91.85 3.62 28.37 -37.98 22.65 -41.84 [rand] [f=41]
o= 2 v=-0.037638: 97.07 -22.70 36.86 -10.25 -41.88 -25.72 [rand] [f=62]
o= 3 v=-0.026228: 94.01 -30.11 26.00 6.89 -35.26 -29.95 [grid] [f=67]
o= 4 v=-0.025109: 122.87 -3.88 -38.32 -37.64 24.31 3.06 [rand] [f=61]
o= 5 v=-0.023467: 97.59 -19.20 -35.11 -18.55 14.45 2.78 [rand] [f=47]
o= 6 v=-0.032558: 124.34 -28.56 20.34 5.63 -41.94 -44.51 [rand] [f=52]
o= 7 v=-0.024558: 103.87 -12.91 -36.21 -26.12 14.64 -5.56 [rand] [f=68]
o= 8 v=-0.026595: 92.49 0.17 27.38 -37.57 16.88 -34.60 [rand] [f=47]
o= 9 v=-0.025748: 122.87 -7.62 -36.96 -38.73 25.59 5.74 [rand] [f=37]
o=10 v=-0.023050: 97.81 -19.65 -34.79 -19.15 13.89 2.05 [rand] [f=36]
o=11 v=-0.034817: 125.56 3.51 37.77 32.42 -27.27 -8.86 [rand] [f=47]
o=12 v=-0.026636: 90.62 19.43 30.46 30.75 -11.47 -27.04 [rand] [f=54]
o=13 v=-0.029615: 95.65 -16.42 33.99 15.06 -38.94 -27.36 [rand] [f=44]
o=14 v=-0.025724: 121.05 -17.94 -34.73 10.44 -8.00 -4.28 [rand] [f=40]
o=15 v=-0.027042: 119.10 0.10 -39.89 -13.81 11.33 -14.01 [rand] [f=42]
o=16 v=-0.029803: 88.38 -15.30 -33.36 -10.38 12.73 4.37 [rand] [f=58]
o=17 v=-0.025471: 120.77 -7.35 -37.30 -34.55 25.97 6.34 [rand] [f=51]
o=18 v=-0.021813: 96.06 -31.35 20.30 -37.16 -7.69 -44.56 [rand] [f=37]
o=19 v=-0.039610: 120.38 31.62 37.96 31.21 -11.89 -1.49 [rand] [f=60]
o=20 v=-0.030208: 99.98 -30.12 26.92 -4.50 -28.25 -34.17 [grid] [f=62]
o=21 v=-0.033973: 115.95 30.51 37.09 27.08 -7.23 4.45 [rand] [f=45]
o=22 v=-0.024448: 68.66 12.44 -38.44 19.11 14.37 2.63 [rand] [f=52]
o=23 v=-0.030816: 88.36 -12.05 -34.28 -12.31 11.56 5.36 [rand] [f=57]
o=24 v=-0.026831: 119.70 -7.09 -37.52 -10.99 4.50 -13.47 [rand] [f=43]
o=25 v=-0.021834: 110.54 4.11 -28.89 -31.19 6.72 7.75 [grid] [f=46]
o=26 v=-0.029659: 112.69 -13.96 -35.31 -28.09 23.99 4.39 [rand] [f=64]
o=27 v=-0.036380: 124.25 -11.62 38.17 33.28 -31.14 -4.50 [rand] [f=57]
o=28 v=-0.026330: 92.18 1.00 27.77 -39.33 18.23 -32.61 [rand] [f=51]
o=29 v=-0.022613: 102.63 -11.90 -37.10 -25.08 13.86 -0.27 [rand] [f=35]
o=30 v=-0.023502: 119.40 -16.34 -38.41 -9.52 -2.70 -13.24 [rand] [f=49]
o=31 v=-0.024601: 69.46 14.02 -37.34 19.44 12.69 -2.24 [rand] [f=44]
o=32 v=-0.025824: 119.43 -8.11 -37.98 -7.01 10.78 -13.52 [rand] [f=83]
o=33 v=-0.028485: 111.32 17.78 -33.51 -44.98 10.27 21.60 [rand] [f=50]
o=34 v=-0.021606: 103.05 11.34 30.91 10.44 -16.63 -39.34 [rand] [f=54]
o=35 v=-0.025812: 105.92 18.80 -34.03 -38.48 13.27 -12.88 [rand] [f=49]
o=36 v=-0.023639: 120.56 -21.02 -38.02 5.46 4.63 -15.69 [rand] [f=51]
o=37 v=-0.022421: 96.59 -28.00 29.09 6.71 -31.63 -25.55 [rand] [f=39]
o=38 v=-0.020190: 79.59 -26.31 24.88 -29.92 -8.68 -28.78 [grid] [f=49]
o=39 v=-0.021749: 96.94 33.57 35.86 11.35 -6.94 -19.87 [rand] [f=66]
o=40 v=-0.019895: 82.95 28.96 -37.93 -17.75 28.76 13.02 [rand] [f=37]
o=41 v=-0.021356: 91.66 -14.96 -22.76 -7.10 34.58 24.82 [rand] [f=46]
o=42 v=-0.027107: 124.49 -20.18 -32.05 10.69 -7.44 9.91 [rand] [f=45]
o=43 v=-0.026319: 68.91 11.17 -38.05 20.29 19.40 -6.43 [rand] [f=56]
o=44 v=-0.024913: 67.84 15.42 -35.49 37.39 14.37 -18.96 [rand] [f=37]
o=45 v=-0.023354: 93.76 23.30 27.77 29.47 -7.75 -31.27 [grid] [f=40]
o=46 v=-0.019407: 109.48 -31.82 -39.14 15.51 4.62 -13.43 [rand] [f=46]
o=47 v=-0.022879: 92.79 -14.50 -30.57 -6.67 8.67 35.86 [grid] [f=59]
o=48 v=-0.028698: 88.61 -13.59 -36.48 -10.26 12.62 0.84 [rand] [f=53]
o=49 v=-0.019705: 92.37 -28.32 21.44 30.84 -39.72 -37.84 [rand] [f=65]
o=50 v=-0.022741: 99.40 -24.95 21.80 -13.00 -13.24 -40.83 [rand] [f=46]
o=51 v=-0.025023: 125.97 -26.96 37.93 -27.17 -36.78 -38.74 [rand] [f=57]
o=52 v=-0.018040: 92.91 -9.23 -27.61 -8.65 30.90 29.96 [grid] [f=65]
o=53 v=-0.026083: 119.13 -8.06 -39.09 -31.95 24.41 -0.92 [rand] [f=47]
o=54 v=-0.021481: 97.21 -15.76 28.19 -9.68 -14.20 -39.20 [rand] [f=52]
o=55 v=-0.029622: 102.22 23.54 -40.40 -8.51 -15.66 40.94 [rand] [f=57]
o=56 v=-0.024318: 119.26 -12.01 -40.99 -6.33 -3.92 -15.68 [rand] [f=67]
o=57 v=-0.016309: 99.90 -40.50 -41.08 13.83 7.03 -28.14 [rand] [f=41]
o=58 v=-0.020067: 70.82 -7.68 -36.96 23.60 18.14 3.28 [rand] [f=53]
o=59 v=-0.029083: 122.02 -8.54 38.29 35.74 -24.75 2.19 [rand] [f=40]
o=60 v=-0.018760: 69.81 -16.27 -37.23 22.97 16.08 -3.59 [rand] [f=33]
o=61 v=-0.027637: 95.70 -14.89 33.59 24.68 -37.62 -25.78 [rand] [f=55]
o=62 v=-0.021869: 90.87 -7.14 -30.34 27.19 5.03 7.77 [grid] [f=44]
o=63 v=-0.016562: 93.04 -33.28 -38.26 19.24 27.71 -15.44 [rand] [f=35]
o=64 v=-0.017593: 53.52 -19.20 30.34 13.28 -18.83 16.03 [rand] [f=60]
o=65 v=-0.020603: 105.13 -39.25 -35.37 22.69 13.32 -21.57 [rand] [f=50]
o=66 v=-0.022279: 93.18 9.76 -30.87 -8.28 32.53 27.85 [grid] [f=76]
o=67 v=-0.023893: 102.80 20.21 -36.92 8.62 -14.85 40.53 [rand] [f=50]
o=68 v=-0.030230: 94.78 11.44 -41.70 -27.01 18.02 -15.08 [rand] [f=53]
o=69 v=-0.025604: 92.34 2.49 30.25 29.16 -28.24 -27.67 [grid] [f=63]
o=70 v=-0.028099: 84.44 -14.51 -37.03 -5.85 10.74 5.83 [rand] [f=62]
o=71 v=-0.024499: 117.76 -30.97 -33.13 30.69 -12.85 -3.72 [rand] [f=61]
o=72 v=-0.017104: 59.48 -35.24 36.02 15.19 -4.00 12.18 [rand] [f=44]
o=73 v=-0.020121: 78.49 34.05 -38.37 19.48 26.09 14.47 [rand] [f=43]
o=74 v=-0.027725: 66.37 14.95 -34.10 33.91 20.53 -10.37 [rand] [f=70]
o=75 v=-0.021457: 118.15 -34.24 32.27 -35.98 -26.19 -42.12 [rand] [f=53]
o=76 v=-0.033583: 84.74 5.08 -40.21 22.43 10.74 3.93 [rand] [f=82]
o=77 v=-0.022471: 66.16 19.67 -38.88 38.58 4.87 -0.04 [rand] [f=62]
o=78 v=-0.022165: 93.72 -7.71 -26.28 17.15 7.89 8.13 [grid] [f=39]
o=79 v=-0.019392: 56.80 -35.73 32.75 42.93 -41.01 35.05 [rand] [f=40]
o=80 v=-0.024286: 99.09 -41.34 36.23 14.84 -11.57 -20.31 [rand] [f=40]
o=81 v=-0.027989: 100.32 13.71 -36.07 -26.64 6.16 -4.67 [rand] [f=61]
o=82 v=-0.018578: 114.04 -25.07 -27.78 31.22 5.83 3.98 [grid] [f=41]
o=83 v=-0.020005: 108.98 -40.26 31.30 -22.78 -4.31 -16.42 [rand] [f=52]
o=84 v=-0.022922: 85.64 -10.65 -38.37 5.12 14.67 -7.14 [rand] [f=60]
o=85 v=-0.022479: 89.13 -11.37 -28.03 -5.87 37.76 7.84 [grid] [f=59]
o=86 v=-0.019822: 93.58 -9.73 28.66 -9.65 -11.27 -31.86 [grid] [f=57]
o=87 v=-0.031659: 80.56 10.85 31.85 39.61 -23.66 -12.54 [rand] [f=68]
+ - saving # 0 for use with twobest
+ - saving #19 for use with twobest
+ - saving # 2 for use with twobest
+ - saving #27 for use with twobest
+ - saving #11 for use with twobest
+ - saving #21 for use with twobest
+ - saving #76 for use with twobest
+ - saving # 6 for use with twobest
+ - saving #87 for use with twobest
+ - saving #23 for use with twobest
+ - saving #68 for use with twobest
+ - saving #20 for use with twobest
+ - skip #16 for twobest: too close to set #23
+ - saving #26 for use with twobest
+ - saving #55 for use with twobest
+ - saving #13 for use with twobest
+ - saving #59 for use with twobest
+ - skip #48 for twobest: too close to set #16
+ - saving #33 for use with twobest
+ - saving #70 for use with twobest
+ - saving #81 for use with twobest
+ - saving #74 for use with twobest
+ - saving #61 for use with twobest
+ - saving #42 for use with twobest
+ - saving #15 for use with twobest
+ - saving #24 for use with twobest
+ - saving #12 for use with twobest
+ - saving # 8 for use with twobest
+ - skip #28 for twobest: too close to set # 8
+ - saving #43 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #53 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31909 total points stored in 63 'TOHD(17.3748)' bloks (0 duplicates)
*[#20978=-0.169331] *[#20986=-0.170313] *[#21000=-0.170526] *[#21001=-0.172023] *[#21005=-0.172274] *[#21008=-0.173681] *[#21009=-0.1739] *[#21013=-0.174339] *[#21016=-0.175108] *[#21017=-0.176175] *[#21018=-0.176452] *[#21019=-0.176809] *[#21021=-0.177261] *[#21024=-0.17761] *[#21026=-0.178186] *[#21027=-0.178833] *[#21028=-0.179343] *[#21031=-0.179472] *[#21040=-0.180395] *[#21043=-0.180428] *[#21045=-0.180429] *[#21047=-0.180514] *[#21048=-0.180599] *[#21049=-0.180672] *[#21051=-0.180752] *[#21052=-0.180786] *[#21053=-0.180808] *[#21054=-0.18082] *[#21055=-0.180832] *[#21056=-0.180858] *[#21057=-0.180864] *[#21059=-0.180909] *[#21060=-0.180915] *[#21061=-0.180934] *[#21062=-0.181065] *[#21063=-0.181087] *[#21064=-0.181114] *[#21065=-0.181253] *[#21068=-0.181324] *[#21069=-0.181432] *[#21070=-0.18147] *[#21073=-0.181504] *[#21074=-0.18155] *[#21075=-0.181565]
+ - param set #1 has cost=-0.181565 [o=0 t=0]
+ -- Parameters = 86.2417 -3.0340 0.1839 -0.7927 0.0198 0.1572 0.9849 1.0186 0.9937 -0.0067 -0.0009 -0.0030
+ - param set #2 has cost=-0.029599 [o=19 t=1]
+ -- Parameters = 120.3747 31.4269 37.8606 31.0260 -11.8449 -1.1094 1.0067 0.9979 0.9877 -0.0009 0.0021 -0.0001
+ - param set #3 has cost=-0.032947 [o=2 t=2]
+ -- Parameters = 97.1422 -22.9068 36.7125 -10.4271 -42.0244 -25.8414 0.9953 0.9994 0.9958 -0.0016 0.0013 -0.0002
+ - param set #4 has cost=-0.025913 [o=27 t=3]
+ -- Parameters = 124.2213 -11.7386 37.9231 33.3757 -31.0928 -4.5253 1.0015 1.0012 0.9970 0.0002 0.0015 0.0004
+ - param set #5 has cost=-0.028112 [o=11 t=4]
+ -- Parameters = 122.3987 3.2658 37.8554 32.0364 -27.2812 -8.8197 0.9993 1.0000 0.9989 -0.0002 -0.0001 -0.0002
+ - param set #6 has cost=-0.027098 [o=21 t=5]
+ -- Parameters = 115.6655 30.6513 37.9152 27.7891 -10.1989 1.4674 0.9984 0.9912 0.9884 0.0004 -0.0012 0.0008
+ - param set #7 has cost=-0.025330 [o=76 t=6]
+ -- Parameters = 84.5296 4.8670 -40.1157 22.4018 10.4986 3.9660 1.0038 0.9999 1.0018 -0.0005 -0.0003 0.0016
+ - param set #8 has cost=-0.027671 [o=6 t=7]
+ -- Parameters = 124.4184 -28.3909 20.3705 5.2792 -41.8702 -44.1177 1.0047 0.9790 1.0062 -0.0035 -0.0002 -0.0054
+ - param set #9 has cost=-0.026880 [o=87 t=8]
+ -- Parameters = 80.6211 10.7956 31.8627 39.3826 -23.6462 -12.7049 0.9977 1.0051 0.9899 -0.0002 0.0006 -0.0006
+ - param set #10 has cost=-0.025438 [o=23 t=9]
+ -- Parameters = 88.5119 -12.3057 -34.5255 -12.2625 11.3388 4.8335 1.0036 0.9951 0.9911 0.0147 -0.0003 -0.0010
+ - param set #11 has cost=-0.025056 [o=68 t=10]
+ -- Parameters = 95.0321 11.2848 -41.6982 -27.1384 18.0539 -14.7625 0.9967 0.9975 1.0046 0.0010 -0.0017 -0.0002
+ - param set #12 has cost=-0.026301 [o=20 t=11]
+ -- Parameters = 99.7295 -29.7606 26.8107 -4.8322 -28.5727 -34.7626 1.0025 0.9939 1.0000 0.0001 -0.0010 0.0011
+ - param set #13 has cost=-0.024785 [o=26 t=12]
+ -- Parameters = 112.7484 -13.9672 -35.1899 -28.0512 24.1536 4.0959 0.9974 1.0021 1.0031 -0.0009 0.0004 -0.0004
+ - param set #14 has cost=-0.023818 [o=55 t=13]
+ -- Parameters = 102.3059 23.5118 -40.3609 -8.3749 -15.9182 41.1731 0.9987 1.0004 1.0059 0.0001 -0.0012 -0.0005
+ - param set #15 has cost=-0.025346 [o=13 t=14]
+ -- Parameters = 95.5049 -16.5664 34.3042 14.7561 -38.9196 -27.3528 1.0042 1.0061 0.9993 0.0005 -0.0002 -0.0014
+ - param set #16 has cost=-0.028890 [o=59 t=15]
+ -- Parameters = 121.0495 -8.3479 38.2577 35.4740 -28.4600 -2.1114 0.9475 0.9673 0.9742 0.0001 -0.0099 0.0125
+ - param set #17 has cost=-0.022857 [o=33 t=16]
+ -- Parameters = 111.2125 17.7604 -33.8452 -44.8026 10.9680 21.5318 0.9972 1.0001 1.0062 -0.0000 0.0110 -0.0001
+ - param set #18 has cost=-0.024121 [o=70 t=17]
+ -- Parameters = 84.6139 -14.8654 -35.5807 -6.2385 9.4374 4.5597 0.9971 1.0019 0.9957 -0.0008 0.0001 -0.0020
+ - param set #19 has cost=-0.026862 [o=81 t=18]
+ -- Parameters = 100.4019 15.0228 -35.6483 -31.5537 6.5158 -4.5506 0.9870 0.9952 0.9971 0.0007 0.0006 0.0008
+ - param set #20 has cost=-0.019718 [o=74 t=19]
+ -- Parameters = 66.3511 14.9204 -34.0041 33.9302 20.6377 -10.4785 0.9995 1.0003 0.9990 0.0002 -0.0001 -0.0002
+ - param set #21 has cost=-0.024596 [o=61 t=20]
+ -- Parameters = 94.9530 -15.7971 34.3648 21.1914 -38.2038 -26.5993 0.9967 1.0153 0.9925 0.0030 0.0004 -0.0029
+ - param set #22 has cost=-0.030211 [o=42 t=21]
+ -- Parameters = 125.4314 -20.5678 -30.0616 11.1232 -9.1532 7.8234 0.9842 0.9823 0.9478 -0.0144 -0.0209 0.0117
+ - param set #23 has cost=-0.022798 [o=15 t=22]
+ -- Parameters = 119.2795 0.0863 -39.9379 -13.8465 11.2937 -14.2121 0.9972 0.9982 1.0023 0.0015 -0.0005 -0.0005
+ - param set #24 has cost=-0.022617 [o=24 t=23]
+ -- Parameters = 119.5177 -7.0698 -37.4677 -10.8037 4.7487 -13.3916 1.0004 1.0044 0.9954 0.0003 -0.0039 -0.0012
+ - param set #25 has cost=-0.025054 [o=12 t=24]
+ -- Parameters = 90.2415 19.2854 31.2812 30.8214 -12.2280 -26.6292 0.9929 0.9856 0.9532 -0.0054 0.0058 -0.0182
+ - param set #26 has cost=-0.022589 [o=8 t=25]
+ -- Parameters = 92.2565 -0.3541 27.7222 -37.8739 16.7082 -34.1457 1.0015 0.9972 0.9965 0.0001 0.0001 -0.0009
+ - param set #27 has cost=-0.023658 [o=43 t=26]
+ -- Parameters = 67.8001 10.6026 -38.1869 20.2024 17.6918 -6.5006 0.9989 0.9962 0.9715 0.0005 -0.0034 0.0218
+ - param set #28 has cost=-0.023512 [o=3 t=27]
+ -- Parameters = 93.6271 -30.3348 25.2672 6.4093 -34.2989 -30.7456 1.0028 0.9979 1.0102 0.0018 0.0004 -0.0028
+ - param set #29 has cost=-0.022179 [o=53 t=28]
+ -- Parameters = 119.1053 -8.1075 -39.0073 -32.2735 24.1427 -0.5844 0.9994 1.0016 1.0011 -0.0012 0.0000 0.0001
*[#23803=-0.181994] *[#23804=-0.182059] *[#23808=-0.182443] *[#23813=-0.182802] *[#23817=-0.182824] *[#23821=-0.183125]
+ - param set #30 has cost=-0.183125 [o=-1 t=-1]
+ -- Parameters = 86.2790 -2.9386 0.2322 -0.5467 -0.0139 0.1276 0.9790 1.0183 0.9889 0.0075 0.0058 -0.0075
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0636
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.489
+ --- dist(#5,#1) = 0.47
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.47
+ --- dist(#8,#1) = 0.492
+ --- dist(#9,#1) = 0.471
+ --- dist(#10,#1) = 0.444
+ --- dist(#11,#1) = 0.449
+ --- dist(#12,#1) = 0.388
+ --- dist(#13,#1) = 0.474
+ --- dist(#14,#1) = 0.434
+ --- dist(#15,#1) = 0.432
+ --- dist(#16,#1) = 0.504
+ --- dist(#17,#1) = 0.524
+ --- dist(#18,#1) = 0.388
+ --- dist(#19,#1) = 0.443
+ --- dist(#20,#1) = 0.427
+ --- dist(#21,#1) = 0.448
+ --- dist(#22,#1) = 0.507
+ --- dist(#23,#1) = 0.48
+ --- dist(#24,#1) = 0.381
+ --- dist(#25,#1) = 0.492
+ --- dist(#26,#1) = 0.502
+ --- dist(#27,#1) = 0.471
+ --- dist(#28,#1) = 0.415
+ --- dist(#29,#1) = 0.49
+ --- dist(#30,#1) = 0.428
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ - A little optimization:*[#16335=-0.0816929] *[#16336=-0.0937804] *[#16337=-0.100779] *[#16340=-0.104372] *[#16343=-0.105499] *[#16345=-0.105729] *[#16347=-0.107023] *[#16348=-0.107067] *[#16351=-0.108173] *[#16355=-0.108191] *[#16356=-0.10844] *[#16357=-0.108954] *[#16358=-0.109063] *[#16359=-0.109144] *[#16361=-0.10921] *[#16362=-0.109244] *[#16363=-0.109332] *[#16365=-0.109347] *[#16366=-0.109407] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.109407: 86.86 -1.89 1.09 -1.21 -0.18 -0.85 [grid] [f=51]
o= 1 v=-0.034862: 108.02 -10.74 -33.15 -29.67 18.42 -2.79 [rand] [f=57]
o= 2 v=-0.035313: 111.56 -14.54 -33.80 -34.82 23.54 7.76 [rand] [f=51]
o= 3 v=-0.028460: 67.97 17.82 -33.53 36.08 14.99 -18.39 [rand] [f=61]
o= 4 v=-0.024921: 60.22 24.25 -38.05 44.77 -14.40 -41.36 [rand] [f=35]
o= 5 v=-0.032166: 105.09 -10.03 -33.18 -24.90 14.47 -0.83 [rand] [f=83]
o= 6 v=-0.021639: 88.50 -12.74 -31.67 -10.99 18.37 -4.92 [rand] [f=31]
o= 7 v=-0.025210: 61.33 30.37 -33.56 43.86 -40.46 -29.30 [rand] [f=52]
o= 8 v=-0.026893: 84.19 -4.67 -32.25 -11.40 18.18 -4.92 [rand] [f=51]
o= 9 v=-0.030644: 80.30 -11.95 -24.63 7.91 27.66 -23.83 [grid] [f=61]
o=10 v=-0.029696: 84.85 -5.08 -33.93 -14.08 11.91 -1.84 [rand] [f=52]
o=11 v=-0.023979: 94.40 -6.65 -23.26 -13.25 27.33 23.53 [grid] [f=45]
o=12 v=-0.023858: 93.67 -7.63 -23.39 -5.97 31.16 23.44 [rand] [f=50]
o=13 v=-0.030048: 64.54 24.23 -32.63 19.40 -12.92 -33.55 [rand] [f=44]
o=14 v=-0.019898: 84.68 -8.29 -30.52 -10.79 18.74 4.28 [rand] [f=41]
o=15 v=-0.028233: 118.13 -6.82 -33.79 -8.27 9.09 -10.24 [rand] [f=56]
o=16 v=-0.021104: 117.31 -7.60 -35.35 -38.08 23.85 5.02 [rand] [f=52]
o=17 v=-0.030758: 83.71 -14.33 -22.05 6.86 31.23 -22.47 [rand] [f=83]
o=18 v=-0.020099: 91.20 -8.78 -17.65 -15.91 24.92 38.81 [rand] [f=64]
o=19 v=-0.038099: 107.98 -14.44 -36.24 -32.52 20.88 -4.98 [rand] [f=79]
o=20 v=-0.018339: 121.46 -15.76 -36.90 8.85 -6.47 -6.83 [rand] [f=40]
o=21 v=-0.023340: 84.38 -8.36 -26.84 -6.84 30.08 -6.30 [grid] [f=41]
o=22 v=-0.040221: 106.58 19.48 -35.57 -44.65 12.32 -8.75 [rand] [f=83]
o=23 v=-0.023412: 85.73 33.03 -39.22 -20.58 25.67 12.15 [rand] [f=63]
o=24 v=-0.025449: 89.32 -9.76 -27.82 -7.23 30.67 7.44 [grid] [f=44]
o=25 v=-0.020872: 103.77 21.91 -36.69 -38.76 10.44 26.75 [rand] [f=47]
o=26 v=-0.023974: 61.01 6.61 -29.84 27.21 14.98 -5.36 [grid] [f=45]
o=27 v=-0.019121: 110.66 21.80 -37.29 -41.16 -14.74 41.62 [rand] [f=33]
o=28 v=-0.028267: 94.43 -1.48 -29.93 -5.34 7.88 27.30 [grid] [f=48]
o=29 v=-0.023293: 82.03 -0.74 -27.70 2.01 31.05 -26.66 [grid] [f=67]
o=30 v=-0.021475: 67.91 22.52 -35.94 40.31 18.49 16.84 [rand] [f=57]
o=31 v=-0.029074: 117.92 -8.84 -33.92 -7.06 7.03 -8.88 [rand] [f=51]
o=32 v=-0.023377: 83.29 3.82 -35.07 -13.03 16.37 0.49 [rand] [f=48]
o=33 v=-0.018506: 76.66 16.54 -35.02 25.64 27.49 -27.33 [rand] [f=31]
o=34 v=-0.020653: 94.34 -3.94 -27.74 -3.11 26.49 26.85 [grid] [f=55]
o=35 v=-0.023464: 62.50 7.78 -35.30 19.72 17.56 -2.43 [rand] [f=48]
o=36 v=-0.021209: 120.52 -23.44 -35.29 18.73 37.22 -39.30 [rand] [f=55]
o=37 v=-0.028429: 82.37 -2.01 -27.46 -1.06 24.73 -19.37 [grid] [f=73]
o=38 v=-0.018827: 68.99 11.67 -35.53 20.36 34.07 -8.17 [rand] [f=51]
o=39 v=-0.022223: 98.96 15.00 -40.02 -14.58 37.35 25.46 [rand] [f=64]
o=40 v=-0.029399: 118.11 -6.16 -36.48 -0.16 -0.80 -14.21 [rand] [f=63]
o=41 v=-0.023792: 113.22 28.90 -39.83 -44.19 -44.75 30.99 [rand] [f=54]
o=42 v=-0.018888: 80.65 2.98 -26.00 -8.01 13.43 -6.87 [grid] [f=40]
o=43 v=-0.028798: 119.77 -33.46 -39.42 6.11 33.12 -19.91 [rand] [f=61]
o=44 v=-0.021498: 110.06 -7.68 -30.60 -31.51 7.02 6.67 [grid] [f=43]
o=45 v=-0.038146: 106.31 18.77 -34.57 -44.23 10.12 -6.49 [rand] [f=69]
o=46 v=-0.033154: 104.43 -9.48 -34.16 -25.43 13.36 -2.47 [rand] [f=82]
o=47 v=-0.019804: 121.14 2.59 -35.78 -13.30 5.43 -13.40 [rand] [f=64]
o=48 v=-0.015594: 119.42 3.14 -37.43 -12.07 0.47 -19.09 [rand] [f=36]
o=49 v=-0.030101: 85.13 -1.40 -39.08 -9.42 14.33 -8.42 [rand] [f=50]
o=50 v=-0.022933: 97.93 -11.78 -33.54 -27.92 13.34 6.25 [rand] [f=57]
o=51 v=-0.019711: 121.11 -15.42 -36.53 5.19 10.37 -11.43 [rand] [f=46]
o=52 v=-0.027777: 60.78 6.39 -27.40 39.11 1.63 -31.51 [grid] [f=40]
o=53 v=-0.026603: 87.31 -1.13 -35.86 -9.63 17.29 -4.75 [rand] [f=55]
o=54 v=-0.016537: 93.14 5.70 -22.80 7.52 7.83 7.18 [grid] [f=54]
o=55 v=-0.021611: 65.67 17.06 -33.54 11.10 -16.11 -38.46 [rand] [f=46]
o=56 v=-0.028445: 59.65 7.98 -29.86 40.17 -7.42 -30.74 [grid] [f=56]
o=57 v=-0.017651: 70.10 -15.07 -34.11 22.38 15.71 3.62 [rand] [f=46]
o=58 v=-0.021543: 92.74 -8.13 -25.76 -20.16 30.60 28.56 [grid] [f=51]
o=59 v=-0.018194: 69.27 -11.06 -29.01 20.58 16.83 -7.33 [rand] [f=51]
o=60 v=-0.021164: 81.19 -19.80 -28.46 -7.85 29.52 -8.04 [grid] [f=45]
o=61 v=-0.024678: 59.44 27.66 -32.23 44.10 -26.75 -36.21 [rand] [f=68]
o=62 v=-0.023460: 108.32 -9.13 -30.72 -29.95 29.95 -8.15 [grid] [f=42]
o=63 v=-0.026382: 80.47 -7.99 -32.80 -1.37 14.36 -18.68 [grid] [f=60]
o=64 v=-0.018561: 119.87 7.67 -32.72 -11.56 9.09 -10.59 [rand] [f=68]
o=65 v=-0.027691: 110.36 12.88 -28.00 -33.60 9.04 10.12 [grid] [f=51]
o=66 v=-0.027360: 80.72 9.91 -35.91 3.45 31.97 -36.63 [rand] [f=46]
o=67 v=-0.027508: 69.46 20.80 -33.58 37.02 7.76 5.04 [rand] [f=83]
o=68 v=-0.017801: 72.41 -12.46 -35.13 18.36 12.81 3.66 [rand] [f=56]
o=69 v=-0.017681: 113.39 5.80 -30.10 -32.68 -3.52 31.12 [grid] [f=55]
o=70 v=-0.025446: 63.53 8.43 -30.78 10.07 25.81 9.38 [grid] [f=68]
o=71 v=-0.029240: 108.01 20.23 -32.51 -41.87 19.48 -5.16 [rand] [f=62]
o=72 v=-0.033764: 93.93 -4.88 -31.32 -3.60 8.49 23.03 [grid] [f=51]
o=73 v=-0.028228: 80.87 -6.78 -28.62 -1.08 31.86 -22.86 [rand] [f=77]
o=74 v=-0.028612: 118.38 -8.51 -33.02 -8.36 5.97 -8.81 [rand] [f=65]
o=75 v=-0.017643: 111.81 22.13 -29.68 -29.19 -7.94 28.65 [grid] [f=54]
o=76 v=-0.030698: 112.72 18.99 -29.68 -36.49 19.10 11.73 [rand] [f=61]
o=77 v=-0.016922: 71.64 -15.59 -34.41 20.86 16.69 5.99 [rand] [f=41]
o=78 v=-0.025328: 101.73 31.59 -39.46 -7.64 -13.47 44.63 [rand] [f=49]
o=79 v=-0.019548: 103.09 1.06 -36.59 -24.83 9.87 -7.25 [rand] [f=73]
o=80 v=-0.019880: 73.18 38.10 -39.99 -8.21 -13.26 -25.63 [rand] [f=61]
o=81 v=-0.016657: 84.51 -39.87 -19.64 -34.49 44.98 22.60 [rand] [f=60]
o=82 v=-0.018929: 94.51 -30.68 -40.00 -29.29 42.05 -23.62 [rand] [f=32]
o=83 v=-0.028125: 82.82 -5.90 -31.69 -9.77 13.97 -6.67 [grid] [f=83]
o=84 v=-0.016672: 60.21 24.44 -26.19 36.58 29.78 -6.93 [grid] [f=70]
o=85 v=-0.014832: 123.61 -14.65 -36.60 10.55 -7.90 7.83 [rand] [f=45]
o=86 v=-0.027259: 80.26 -4.53 -30.51 -12.78 12.90 -6.40 [grid] [f=48]
o=87 v=-0.018895: 61.95 17.71 -30.90 30.81 -7.11 -33.34 [grid] [f=54]
+ - saving # 0 for use with twobest
+ - saving #22 for use with twobest
+ - skip #45 for twobest: too close to set #22
+ - saving #19 for use with twobest
+ - saving # 2 for use with twobest
+ !source mask fill: ubot=64 usiz=159.5
+ - skip # 1 for twobest: too close to set #19
+ - saving #72 for use with twobest
+ - saving #46 for use with twobest
+ - skip # 5 for twobest: too close to set #46
+ - saving #17 for use with twobest
+ - saving #76 for use with twobest
+ - skip # 9 for twobest: too close to set #17
+ - saving #49 for use with twobest
+ - saving #13 for use with twobest
+ - saving #10 for use with twobest
+ - saving #40 for use with twobest
+ - saving #71 for use with twobest
+ - saving #31 for use with twobest
+ - saving #43 for use with twobest
+ - skip #74 for twobest: too close to set #31
+ - saving # 3 for use with twobest
+ - saving #56 for use with twobest
+ - saving #37 for use with twobest
+ - skip #28 for twobest: too close to set #72
+ - skip #15 for twobest: too close to set #31
+ - saving #73 for use with twobest
+ - saving #83 for use with twobest
+ - saving #52 for use with twobest
+ - saving #65 for use with twobest
+ - saving #67 for use with twobest
+ - saving #66 for use with twobest
+ - skip #86 for twobest: too close to set #83
+ - skip # 8 for twobest: too close to set #83
+ - skip #53 for twobest: too close to set #49
+ - saving #63 for use with twobest
+ - saving #24 for use with twobest
+ - saving #70 for use with twobest
+ - saving #78 for use with twobest
+ - saving # 7 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=52 usiz=160
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31999 total points stored in 64 'TOHD(17.2644)' bloks (0 duplicates)
*[#23823=-0.208583] *[#23853=-0.209083] *[#23856=-0.209692] *[#23857=-0.21004] *[#23858=-0.210473] *[#23859=-0.210652] *[#23861=-0.210912] *[#23862=-0.21108] *[#23863=-0.211096] *[#23864=-0.211318] *[#23865=-0.211465] *[#23868=-0.211558] *[#23871=-0.211598] *[#23874=-0.211605] *[#23876=-0.211804] *[#23880=-0.211892] *[#23883=-0.212487] *[#23884=-0.212638] *[#23887=-0.212807] *[#23889=-0.212858] *[#23890=-0.212946] *[#23891=-0.213045] *[#23892=-0.213054] *[#23893=-0.213085] *[#23896=-0.213618] *[#23897=-0.214048] *[#23898=-0.2143] *[#23899=-0.214697] *[#23902=-0.214782] *[#23904=-0.215131] *[#23907=-0.215508] *[#23908=-0.21602] *[#23909=-0.21655] *[#23910=-0.216938] *[#23911=-0.21729] *[#23912=-0.217316] *[#23913=-0.217499] *[#23914=-0.217762] *[#23915=-0.217858] *[#23920=-0.218401] *[#23921=-0.218462] *[#23922=-0.218657] *[#23925=-0.219463] *[#23926=-0.219544] *[#23927=-0.219959] *[#23928=-0.220151] *[#23931=-0.220227]
+ - param set #1 has cost=-0.220227 [o=-1 t=-1]
+ -- Parameters = 86.5690 -3.1646 -0.1381 -0.8542 -1.9133 -0.7763 0.9848 1.0171 0.9535 -0.0108 0.0209 -0.0382
+ - param set #2 has cost=-0.216445 [o=0 t=0]
+ - retaining old weight image
+ -- Parameters = 86.4148 -3.1639 -0.1179 -0.8652 -0.7590 -0.0405 0.9867 1.0242 0.9680 -0.0151 0.0021 -0.0159
+ - using 39832 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32296 total points stored in 64 'TOHD(17.3748)' bloks (0 duplicates)
+ - param set #3 has cost=-0.027810 [o=2 t=2]
+ -- Parameters = 97.2032 -22.9052 36.6921 -10.4554 -42.1013 -25.7685 0.9914 0.9972 0.9931 -0.0021 0.0004 0.0019
+ - param set #4 has cost=-0.030534 [o=42 t=21]
*[#21244=-0.174927] *[#21255=-0.174992] *[#21265=-0.17571] *[#21272=-0.176379] *[#21276=-0.178711] *[#21277=-0.179551] *[#21280=-0.179669] *[#21281=-0.17992] *[#21283=-0.180833] *[#21290=-0.182015] *[#21291=-0.1826] *[#21292=-0.182933] *[#21293=-0.183073] *[#21296=-0.184306] *[#21298=-0.184721] *[#21301=-0.185484] *[#21302=-0.186345] *[#21305=-0.187523] *[#21306=-0.189594] *[#21319=-0.189868] *[#21321=-0.190775] *[#21325=-0.190896] *[#21327=-0.190906] *[#21329=-0.190943] *[#21330=-0.191079] *[#21331=-0.191191] *[#21332=-0.191252] *[#21335=-0.191289] *[#21336=-0.191308] *[#21339=-0.191342] *[#21340=-0.191384] *[#21341=-0.191469] *[#21342=-0.191674]
+ -- Parameters = 124.9320 -22.0031 -26.3056 8.7231 -2.9528 10.9929 0.9078 0.9429 0.9127 -0.0312 -0.0436 -0.0116
+ - param set #1 has cost=-0.191674 [o=0 t=0]
+ - param set #5 has cost=-0.024661 [o=19 t=1]
+ -- Parameters = 120.2512 31.5807 37.8833 31.4755 -11.7813 -0.9877 1.0280 0.9922 0.9858 -0.0015 0.0013 0.0011
+ -- Parameters = 86.6206 -1.9079 0.7240 -0.5697 -0.6829 -0.8597 0.9774 1.0125 0.9617 -0.0068 0.0002 -0.0021
+ - param set #6 has cost=-0.022586 [o=59 t=15]
+ -- Parameters = 121.0138 -8.5765 38.2813 35.5698 -28.5095 -2.1222 0.9444 0.9668 0.9787 0.0009 -0.0099 0.0127
+ - param set #2 has cost=-0.034591 [o=22 t=1]
+ -- Parameters = 106.4086 19.6051 -35.8951 -44.8734 12.4997 -8.7055 0.9967 1.0003 0.9971 -0.0016 0.0016 0.0138
+ - param set #7 has cost=-0.024438 [o=11 t=4]
+ - param set #3 has cost=-0.027945 [o=19 t=2]
+ -- Parameters = 107.9776 -14.7330 -35.9494 -31.8773 21.0582 -5.1372 1.0041 0.9907 1.0010 0.0005 0.0041 0.0001
+ -- Parameters = 122.3828 3.2625 37.8905 32.0936 -27.2969 -8.9957 0.9996 1.0008 0.9996 -0.0003 -0.0004 0.0002
+ - param set #4 has cost=-0.029509 [o=2 t=3]
+ - param set #8 has cost=-0.022973 [o=6 t=7]
+ -- Parameters = 111.6435 -15.3075 -34.4049 -34.4691 22.2932 3.5428 0.9911 0.9799 0.9934 -0.0012 -0.0004 -0.0004
+ -- Parameters = 124.5990 -28.5221 20.3574 5.1992 -41.4910 -44.5459 1.0133 0.9665 0.9986 -0.0044 -0.0021 -0.0082
+ - param set #9 has cost=-0.023141 [o=21 t=5]
+ -- Parameters = 115.3082 31.2509 37.9653 28.2268 -10.2700 2.1299 0.9881 0.9725 1.0137 0.0031 -0.0002 -0.0013
+ - param set #5 has cost=-0.023428 [o=72 t=4]
+ -- Parameters = 94.2680 -5.1911 -31.1941 -3.6464 8.3348 23.0201 0.9964 1.0033 1.0028 0.0001 0.0114 -0.0005
+ - param set #10 has cost=-0.020712 [o=87 t=8]
+ - param set #6 has cost=-0.026077 [o=46 t=5]
+ -- Parameters = 80.6871 10.7820 31.8608 39.2432 -23.8256 -12.8107 0.9960 1.0010 0.9862 0.0001 -0.0012 -0.0004
+ -- Parameters = 106.0056 -10.7151 -34.6467 -25.2732 13.0701 -4.3751 0.9904 1.0041 0.9984 -0.0011 -0.0005 -0.0003
+ - param set #11 has cost=-0.023867 [o=81 t=18]
+ -- Parameters = 100.9144 15.5630 -35.5687 -30.9887 7.2817 -4.8546 0.9920 1.0234 0.9953 -0.0013 0.0007 -0.0033
+ - param set #7 has cost=-0.028354 [o=17 t=6]
+ -- Parameters = 82.7524 -14.3957 -22.6778 6.3373 28.2071 -26.1051 0.9919 0.9914 0.9914 -0.0009 -0.0022 -0.0015
+ - param set #12 has cost=-0.022773 [o=20 t=11]
+ -- Parameters = 99.3746 -29.8610 26.9858 -5.3618 -28.5046 -34.3145 1.0066 0.9859 1.0136 -0.0014 0.0000 0.0010
+ - param set #8 has cost=-0.024084 [o=76 t=7]
+ -- Parameters = 112.5381 18.8757 -29.7618 -36.1762 19.0630 11.7461 0.9956 0.9872 0.9943 0.0006 0.0019 0.0003
+ - param set #13 has cost=-0.021875 [o=27 t=3]
+ -- Parameters = 124.5604 -15.0908 37.9811 33.1165 -31.3178 -4.2711 1.0069 1.0029 0.9948 0.0004 0.0007 0.0018
+ - param set #14 has cost=-0.018972 [o=23 t=9]
+ - param set #9 has cost=-0.024765 [o=49 t=8]
+ -- Parameters = 85.1655 -1.2498 -38.8962 -9.4981 14.4006 -8.2666 1.0023 1.0007 1.0016 0.0013 0.0002 -0.0004
+ -- Parameters = 88.7944 -12.1116 -34.5232 -12.0411 11.5798 4.8169 1.0017 0.9955 0.9899 0.0150 0.0007 -0.0000
+ - param set #10 has cost=-0.025076 [o=13 t=9]
+ -- Parameters = 64.3032 24.1293 -32.0954 19.7469 -12.5877 -33.4535 1.0046 0.9996 1.0042 0.0010 0.0002 0.0009
+ - param set #15 has cost=-0.022093 [o=13 t=14]
+ -- Parameters = 94.6722 -16.0639 34.9181 15.7936 -38.3548 -28.1825 0.9908 0.9977 0.9904 -0.0019 -0.0004 -0.0028
+ - param set #11 has cost=-0.024333 [o=10 t=10]
+ -- Parameters = 84.8235 -5.0371 -33.9793 -13.8857 11.8424 -2.1003 0.9989 0.9995 0.9994 0.0000 -0.0013 0.0001
+ - param set #16 has cost=-0.019604 [o=76 t=6]
+ -- Parameters = 84.2874 4.5998 -40.1267 21.8844 10.7009 4.2715 1.0055 0.9996 1.0026 -0.0011 -0.0010 0.0040
+ - param set #12 has cost=-0.020437 [o=40 t=11]
+ -- Parameters = 118.3890 -6.4954 -36.4349 -0.5499 -1.1358 -14.2508 0.9976 1.0032 1.0022 -0.0002 0.0002 -0.0009
+ - param set #13 has cost=-0.030678 [o=71 t=12]
+ -- Parameters = 106.1421 20.5427 -32.1699 -43.3681 13.2971 -5.8962 1.0036 0.9541 1.0455 0.0085 0.0152 0.0216
+ - param set #17 has cost=-0.018545 [o=68 t=10]
+ -- Parameters = 94.9844 11.4663 -41.6971 -27.1688 18.1245 -14.7945 0.9941 1.0035 1.0045 0.0090 -0.0015 -0.0008
+ - param set #18 has cost=-0.022035 [o=12 t=24]
+ - param set #14 has cost=-0.020999 [o=31 t=13]
+ -- Parameters = 118.0563 -9.0610 -33.3186 -7.2873 7.1884 -8.7947 1.0000 1.0056 1.0027 0.0003 0.0004 -0.0017
+ -- Parameters = 90.4553 19.1335 31.0461 30.9460 -12.3561 -27.0333 0.9931 0.9841 0.9491 -0.0063 0.0070 -0.0183
+ - param set #15 has cost=-0.021676 [o=43 t=14]
+ -- Parameters = 119.8119 -33.5127 -39.4352 6.0758 33.1327 -19.9405 1.0004 0.9998 0.9991 0.0012 -0.0004 -0.0001
+ - param set #19 has cost=-0.017809 [o=26 t=12]
+ -- Parameters = 112.7930 -14.0675 -34.9825 -28.0190 24.1066 3.9824 0.9955 1.0005 1.0055 -0.0008 0.0082 -0.0007
+ - param set #16 has cost=-0.023472 [o=3 t=15]
+ -- Parameters = 68.3286 17.2570 -33.6052 35.6049 15.0826 -17.9640 0.9983 0.9972 0.9897 -0.0038 -0.0016 0.0029
+ - param set #20 has cost=-0.022233 [o=61 t=20]
+ -- Parameters = 94.9835 -15.7922 34.2528 21.3591 -38.3023 -26.6743 0.9942 1.0155 0.9949 0.0025 0.0007 -0.0032
+ - param set #17 has cost=-0.023446 [o=56 t=16]
+ -- Parameters = 59.8106 7.8252 -30.3034 40.2585 -7.2001 -30.9429 0.9982 1.0077 0.9838 0.0012 -0.0004 0.0020
+ - param set #21 has cost=-0.021028 [o=70 t=17]
+ -- Parameters = 86.4718 -14.4547 -34.6323 -6.2583 9.6649 3.7010 0.9942 0.9844 0.9884 0.0090 0.0005 -0.0028
+ - param set #22 has cost=-0.017872 [o=55 t=13]
+ -- Parameters = 102.4708 23.5267 -40.2705 -8.5359 -16.0124 41.0981 1.0009 0.9972 1.0084 -0.0001 -0.0017 -0.0010
+ - param set #23 has cost=-0.017721 [o=43 t=26]
+ -- Parameters = 67.4275 10.6734 -38.2748 20.0915 17.9131 -6.3259 0.9971 0.9935 0.9725 0.0008 0.0052 0.0220
+ - param set #24 has cost=-0.020349 [o=3 t=27]
+ -- Parameters = 92.5580 -30.5120 25.0446 5.9667 -33.9224 -32.3783 0.9901 0.9827 0.9962 0.0134 0.0054 -0.0044
+ - param set #18 has cost=-0.026531 [o=37 t=17]
+ -- Parameters = 82.3461 -1.9520 -27.2448 -0.9430 24.6948 -19.3807 1.0055 0.9977 1.0060 -0.0009 -0.0010 -0.0006
+ - param set #25 has cost=-0.018272 [o=33 t=16]
+ -- Parameters = 110.7879 17.5217 -34.1652 -44.9186 11.3923 22.6426 0.9815 0.9941 0.9763 -0.0011 0.0224 -0.0024
+ - param set #19 has cost=-0.024189 [o=73 t=18]
+ -- Parameters = 80.3910 -6.1155 -27.3355 0.9511 31.3036 -21.5901 1.0369 1.0074 0.9993 0.0038 0.0014 0.0012
+ - param set #26 has cost=-0.016165 [o=15 t=22]
+ -- Parameters = 119.3371 0.1366 -40.0015 -13.9032 11.2916 -14.2405 0.9960 0.9969 1.0033 0.0023 -0.0007 -0.0005
+ - param set #27 has cost=-0.017790 [o=24 t=23]
+ -- Parameters = 119.5484 -7.0770 -37.5706 -10.9167 4.8044 -13.1528 1.0001 1.0034 0.9886 -0.0026 0.0023 -0.0026
+ - param set #20 has cost=-0.025492 [o=83 t=19]
+ -- Parameters = 82.3044 -5.9176 -31.5894 -9.7663 14.0681 -6.1728 1.0393 0.9955 0.9965 0.0011 0.0009 -0.0004
+ - param set #28 has cost=-0.021054 [o=8 t=25]
+ -- Parameters = 92.1249 -0.7941 28.1167 -37.9315 16.4735 -34.0354 0.9818 0.9814 0.9791 -0.0018 -0.0013 -0.0009
+ - param set #21 has cost=-0.022530 [o=52 t=20]
+ - param set #29 has cost=-0.015293 [o=53 t=28]
+ -- Parameters = 60.5250 6.1226 -27.2982 39.1416 1.6479 -31.1314 1.0017 1.0062 0.9920 0.0004 0.0031 -0.0025
+ -- Parameters = 119.0922 -8.1040 -38.9836 -32.2314 24.1018 -0.5725 1.0000 1.0027 1.0031 -0.0012 0.0001 0.0002
+ - param set #30 has cost=-0.014662 [o=74 t=19]
+ -- Parameters = 66.4003 14.9545 -34.0002 33.9259 20.6687 -10.5010 0.9998 1.0006 0.9992 0.0001 -0.0000 0.0007
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.1
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.46
+ --- dist(#5,#1) = 0.475
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.443
+ --- dist(#8,#1) = 0.476
+ --- dist(#9,#1) = 0.486
+ --- dist(#10,#1) = 0.373
+ --- dist(#11,#1) = 0.48
+ --- dist(#12,#1) = 0.43
+ --- dist(#13,#1) = 0.438
+ --- dist(#14,#1) = 0.39
+ --- dist(#15,#1) = 0.476
+ --- dist(#16,#1) = 0.412
+ --- dist(#17,#1) = 0.431
+ --- dist(#18,#1) = 0.446
+ --- dist(#19,#1) = 0.356
+ --- dist(#20,#1) = 0.5
+ --- dist(#21,#1) = 0.43
+ --- dist(#22,#1) = 0.519
+ --- dist(#23,#1) = 0.49
+ --- dist(#24,#1) = 0.502
+ --- dist(#25,#1) = 0.436
+ - param set #22 has cost=-0.030740 [o=65 t=21]
+ --- dist(#26,#1) = 0.468
+ -- Parameters = 110.3402 17.6552 -28.6320 -32.5642 15.3099 9.3766 0.9529 0.8413 0.8582 -0.0199 0.0242 -0.0248
+ --- dist(#27,#1) = 0.477
+ --- dist(#28,#1) = 0.498
+ --- dist(#29,#1) = 0.486
+ --- dist(#30,#1) = 0.423
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ - param set #23 has cost=-0.021991 [o=67 t=22]
+ -- Parameters = 69.4308 20.8958 -33.5659 37.0987 7.7821 4.9826 1.0034 0.9996 1.0003 -0.0002 0.0001 -0.0006
+ - param set #24 has cost=-0.023728 [o=66 t=23]
+ -- Parameters = 80.7546 9.5263 -35.6999 3.8794 30.9339 -31.3952 1.0011 1.0012 1.0065 -0.0024 0.0016 -0.0002
+ - param set #25 has cost=-0.023562 [o=63 t=24]
+ -- Parameters = 80.6018 -8.5287 -32.6702 -1.4456 14.4821 -18.9136 1.0020 1.0097 1.0053 0.0006 -0.0006 -0.0012
+ !source mask fill: ubot=64 usiz=159.5
+ - param set #26 has cost=-0.020455 [o=24 t=25]
+ -- Parameters = 89.2607 -9.9893 -27.5142 -7.1198 30.7141 7.3076 0.9972 1.0071 1.0025 0.0007 -0.0006 -0.0025
+ - param set #27 has cost=-0.019267 [o=70 t=26]
+ -- Parameters = 63.6922 8.4567 -30.9719 9.9113 26.0475 9.1903 0.9989 1.0017 0.9928 0.0011 0.0014 -0.0012
+ - param set #28 has cost=-0.020618 [o=78 t=27]
+ -- Parameters = 102.3947 30.8560 -39.5328 -7.6875 -13.6336 44.1400 0.9956 0.9977 1.0024 0.0025 0.0139 0.0040
+ - param set #29 has cost=-0.024607 [o=7 t=28]
+ -- Parameters = 61.4804 31.4802 -33.8604 44.4563 -40.5391 -28.8442 1.0027 0.9972 0.9894 -0.0000 -0.0027 0.0147
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32677 total points stored in 67 'TOHD(17.1973)' bloks (0 duplicates)
+ - param set #30 has cost=-0.183324 [o=-1 t=-1]
+ -- Parameters = 86.5364 -1.8144 0.8596 -0.6394 -0.1600 -0.9487 0.9958 1.0078 0.9702 -0.0028 0.0060 0.0098
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0539
+ --- dist(#3,#1) = 0.492
+ --- dist(#4,#1) = 0.367
+ --- dist(#5,#1) = 0.476
+ --- dist(#6,#1) = 0.439
+ --- dist(#7,#1) = 0.321
+ --- dist(#8,#1) = 0.458
+ --- dist(#9,#1) = 0.35
+ --- dist(#10,#1) = 0.442
+ --- dist(#11,#1) = 0.404
+ --- dist(#12,#1) = 0.41
+ --- dist(#13,#1) = 0.495
+ --- dist(#14,#1) = 0.5
+ --- dist(#15,#1) = 0.434
+ --- dist(#16,#1) = 0.355
+ --- dist(#17,#1) = 0.396
+ --- dist(#18,#1) = 0.455
+ --- dist(#19,#1) = 0.417
+ --- dist(#20,#1) = 0.429
+ --- dist(#21,#1) = 0.454
*[#26935=-0.23117] *[#26944=-0.232084] *[#26959=-0.2323] *[#26962=-0.232318] *[#26964=-0.232417] *[#26965=-0.232543] *[#26966=-0.232579] *[#26967=-0.232897] *[#26968=-0.233239] *[#26970=-0.233374] *[#26971=-0.233673] *[#26972=-0.233862] *[#26973=-0.234107] *[#26976=-0.234183] *[#26979=-0.234351] *[#26982=-0.234738] *[#26989=-0.234754] *[#26991=-0.234778] *[#26993=-0.234835] *[#26994=-0.234875] *[#26995=-0.23489] *[#26998=-0.234922] *[#27003=-0.234952] *[#27008=-0.23499] *[#27022=-0.235079] *[#27024=-0.235114] *[#27025=-0.235125] *[#27026=-0.235141] *[#27029=-0.235189] *[#27030=-0.235209] *[#27033=-0.235232] *[#27034=-0.235249] *[#27037=-0.235325] *[#27038=-0.235358] *[#27039=-0.235442] *[#27044=-0.235475] *[#27045=-0.235495] *[#27046=-0.235521] *[#27047=-0.235525] *[#27049=-0.235599] *[#27050=-0.235614] *[#27052=-0.235647] *[#27054=-0.235672]
+ - param set #1 has cost=-0.235672 [o=-1 t=-1]
+ --- dist(#22,#1) = 0.399
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ --- dist(#23,#1) = 0.441
+ --- dist(#24,#1) = 0.429
+ --- dist(#25,#1) = 0.502
+ --- dist(#26,#1) = 0.426
+ --- dist(#27,#1) = 0.503
+ --- dist(#28,#1) = 0.353
+ --- dist(#29,#1) = 0.464
+ --- dist(#30,#1) = 0.396
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ - param set #2 has cost=-0.234666 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - param set #3 has cost=-0.025822 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - param set #4 has cost=-0.024015 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ !source mask fill: ubot=52 usiz=160
+ - param set #5 has cost=-0.020677 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - param set #6 has cost=-0.021109 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - param set #7 has cost=-0.021742 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - param set #8 has cost=-0.019976 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - param set #9 has cost=-0.019055 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - retaining old weight image
+ - using 39832 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32660 total points stored in 66 'TOHD(17.2644)' bloks (0 duplicates)
+ - param set #10 has cost=-0.019574 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - param set #11 has cost=-0.019180 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
*[#24151=-0.210067] *[#24181=-0.210317] *[#24182=-0.210386] *[#24183=-0.210432] *[#24185=-0.2105] *[#24186=-0.210597] *[#24191=-0.210715] *[#24194=-0.210947] *[#24195=-0.211032] *[#24198=-0.211127] *[#24209=-0.211295] *[#24210=-0.211355] *[#24217=-0.211459] *[#24222=-0.211497] *[#24225=-0.211563] *[#24226=-0.211572] *[#24236=-0.211583] *[#24237=-0.211596] *[#24238=-0.21163] *[#24239=-0.211664] *[#24240=-0.211681] *[#24243=-0.211709] *[#24244=-0.211718] *[#24245=-0.211736] *[#24246=-0.211749] *[#24247=-0.211752] *[#24248=-0.211752] *[#24250=-0.211755] *[#24252=-0.211763] *[#24253=-0.211771] *[#24254=-0.211781] *[#24257=-0.211824] *[#24258=-0.21184] *[#24259=-0.21184]
+ - param set #12 has cost=-0.018566 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - param set #1 has cost=-0.211840 [o=0 t=0]
+ -- Parameters = 86.6831 -2.0317 0.8447 -0.6378 -1.0042 -0.8735 0.9855 1.0164 0.9715 -0.0087 0.0016 -0.0091
+ - param set #2 has cost=-0.210204 [o=-1 t=-1]
+ -- Parameters = 86.6147 -2.0135 0.7367 -0.4354 -0.7596 -0.9919 0.9823 1.0169 0.9657 -0.0026 0.0028 -0.0019
+ - param set #3 has cost=-0.027042 [o=22 t=1]
+ -- Parameters = 106.5085 20.1087 -36.0927 -44.8368 12.7504 -8.8824 0.9896 0.9954 0.9858 -0.0027 0.0014 0.0129
+ - param set #13 has cost=-0.018186 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - param set #4 has cost=-0.024127 [o=65 t=21]
+ - param set #14 has cost=-0.018618 [o=12 t=24]
+ -- Parameters = 110.0660 17.6562 -28.3811 -32.4215 15.2348 9.4391 0.9570 0.8389 0.8611 -0.0205 0.0328 -0.0252
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - param set #5 has cost=-0.024092 [o=71 t=12]
+ -- Parameters = 106.0959 20.7701 -31.9103 -44.0888 13.1036 -5.5823 1.0023 0.9548 1.0411 0.0080 0.0172 0.0198
+ - param set #15 has cost=-0.018967 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - param set #6 has cost=-0.020653 [o=2 t=3]
+ -- Parameters = 111.5365 -15.5360 -34.2479 -34.2638 21.9944 3.5282 0.9847 0.9772 0.9959 0.0085 -0.0012 -0.0001
+ - param set #16 has cost=-0.017468 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - param set #7 has cost=-0.022119 [o=17 t=6]
+ -- Parameters = 82.4646 -14.3521 -22.7307 5.9224 28.3798 -25.7230 0.9856 0.9849 0.9908 -0.0041 -0.0044 -0.0019
+ - param set #17 has cost=-0.018075 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - param set #8 has cost=-0.019216 [o=19 t=2]
+ -- Parameters = 108.0066 -14.8867 -35.7909 -31.7852 21.1295 -5.4882 1.0016 0.9936 1.0107 0.0002 0.0033 0.0016
+ - param set #18 has cost=-0.016481 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - param set #9 has cost=-0.024357 [o=37 t=17]
+ -- Parameters = 82.0175 -1.8633 -24.6022 -0.9774 24.9642 -18.4170 1.0084 0.9977 0.9908 -0.0019 -0.0010 -0.0010
+ - param set #19 has cost=-0.017711 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - param set #10 has cost=-0.017902 [o=46 t=5]
+ -- Parameters = 106.0000 -10.6877 -34.5551 -25.3985 13.1112 -4.2491 0.9902 1.0065 1.0003 0.0067 -0.0001 -0.0005
+ - param set #11 has cost=-0.020188 [o=83 t=19]
+ -- Parameters = 82.6283 -5.8731 -31.4461 -9.9141 14.0996 -6.1519 1.0388 0.9947 1.0007 0.0017 0.0009 -0.0010
+ - param set #12 has cost=-0.019722 [o=13 t=9]
+ -- Parameters = 64.3730 24.5243 -31.8189 19.9781 -12.3195 -33.4344 1.0073 0.9958 1.0048 -0.0000 -0.0000 0.0033
+ - param set #20 has cost=-0.021740 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - param set #13 has cost=-0.019431 [o=49 t=8]
+ -- Parameters = 84.4771 -0.6586 -39.0370 -9.5456 14.3587 -7.8427 1.0040 0.9987 1.0039 0.0010 -0.0006 -0.0020
+ - param set #21 has cost=-0.015678 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - param set #14 has cost=-0.019934 [o=7 t=28]
+ -- Parameters = 61.6011 31.3060 -33.9185 44.3119 -40.4583 -29.2035 1.0063 1.0007 0.9835 -0.0011 -0.0035 0.0150
+ - param set #15 has cost=-0.018852 [o=10 t=10]
+ -- Parameters = 84.8373 -5.0092 -33.9915 -13.7302 11.8253 -2.0771 0.9998 0.9993 0.9987 -0.0000 -0.0011 0.0005
+ - param set #22 has cost=-0.016589 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - param set #16 has cost=-0.020329 [o=73 t=18]
+ -- Parameters = 80.3776 -6.1376 -27.4640 0.9473 31.2972 -21.5061 1.0386 1.0084 0.9965 0.0036 -0.0001 0.0010
+ - param set #23 has cost=-0.016828 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - param set #17 has cost=-0.018875 [o=76 t=7]
+ -- Parameters = 111.2448 21.3343 -29.2001 -36.7747 20.7625 12.5130 0.9835 0.9801 0.9838 0.0046 0.0045 0.0050
+ - param set #24 has cost=-0.016103 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - param set #18 has cost=-0.019222 [o=66 t=23]
+ -- Parameters = 80.5526 9.5944 -35.1741 3.9613 30.9366 -30.9931 1.0118 0.9993 1.0188 -0.0053 0.0008 -0.0014
+ - param set #25 has cost=-0.014262 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - param set #19 has cost=-0.021144 [o=63 t=24]
+ -- Parameters = 80.6367 -8.4786 -32.3401 -1.6325 14.2809 -18.7626 1.0124 1.0091 1.0111 0.0021 0.0001 -0.0040
+ - param set #26 has cost=-0.012861 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - param set #20 has cost=-0.018049 [o=3 t=15]
+ -- Parameters = 68.5322 17.2000 -33.6094 35.3192 15.2041 -17.9289 0.9985 0.9922 0.9925 -0.0055 -0.0015 0.0027
+ - param set #27 has cost=-0.017379 [o=43 t=26]
+ - param set #21 has cost=-0.017422 [o=56 t=16]
+ -- Parameters = 59.4743 7.5371 -30.4237 40.2680 -7.2688 -30.9022 1.0033 1.0109 0.9799 0.0019 -0.0003 0.0009
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - param set #28 has cost=-0.012238 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - param set #22 has cost=-0.020247 [o=72 t=4]
+ -- Parameters = 94.7163 -6.2551 -31.3344 -5.6451 7.7706 21.2591 0.9881 1.0134 0.9854 0.0003 0.0115 -0.0033
+ - param set #29 has cost=-0.013300 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - param set #23 has cost=-0.016656 [o=52 t=20]
+ -- Parameters = 60.4674 6.1673 -27.2013 39.1418 1.7522 -31.0917 0.9990 1.0082 0.9930 0.0090 0.0032 -0.0020
+ - param set #30 has cost=-0.012264 [o=74 t=19]
+ - param set #24 has cost=-0.018844 [o=67 t=22]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - sorting parameter sets by cost
+ -- Parameters = 69.8186 20.9295 -34.5265 37.4317 6.7280 4.7808 0.9963 0.9951 0.9870 -0.0010 0.0011 -0.0020
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0383
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.456
+ --- dist(#5,#1) = 0.44
+ --- dist(#6,#1) = 0.495
+ --- dist(#7,#1) = 0.473
+ --- dist(#8,#1) = 0.473
+ --- dist(#9,#1) = 0.474
+ --- dist(#10,#1) = 0.377
+ --- dist(#11,#1) = 0.478
+ --- dist(#12,#1) = 0.491
+ --- dist(#13,#1) = 0.476
+ --- dist(#14,#1) = 0.392
+ --- dist(#15,#1) = 0.428
+ --- dist(#16,#1) = 0.438
+ --- dist(#17,#1) = 0.433
+ --- dist(#18,#1) = 0.36
+ --- dist(#19,#1) = 0.413
+ --- dist(#20,#1) = 0.482
+ --- dist(#21,#1) = 0.49
+ --- dist(#22,#1) = 0.522
+ --- dist(#23,#1) = 0.443
+ --- dist(#24,#1) = 0.507
+ --- dist(#25,#1) = 0.432
+ --- dist(#26,#1) = 0.437
+ --- dist(#27,#1) = 0.486
+ --- dist(#28,#1) = 0.472
+ --- dist(#29,#1) = 0.426
+ --- dist(#30,#1) = 0.501
+ - Total coarse refinement net CPU time = 0.0 s; 9153 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ - param set #25 has cost=-0.017713 [o=43 t=14]
+ -- Parameters = 119.6089 -33.2981 -39.4999 5.1211 32.5009 -21.1545 0.9896 0.9936 0.9998 0.0011 0.0006 0.0028
+ - param set #26 has cost=-0.016425 [o=31 t=13]
+ -- Parameters = 117.9464 -9.3392 -32.9418 -7.3419 6.7233 -8.7240 0.9974 1.0079 1.0161 0.0018 -0.0006 -0.0034
+ - param set #27 has cost=-0.017652 [o=78 t=27]
+ -- Parameters = 102.2726 30.8430 -39.4950 -7.8650 -13.7218 43.9992 1.0007 0.9913 0.9973 0.0023 0.0129 0.0041
+ !source mask fill: ubot=64 usiz=159.5
+ - param set #28 has cost=-0.015924 [o=24 t=25]
+ -- Parameters = 87.8436 -9.5722 -26.0819 -6.3910 31.6894 8.0155 0.9962 1.0145 1.0054 0.0023 0.0087 -0.0033
+ - param set #29 has cost=-0.015863 [o=40 t=11]
+ -- Parameters = 118.3780 -9.4251 -36.5595 -0.7873 -1.1197 -14.1428 0.9925 1.0041 1.0004 -0.0005 0.0001 -0.0020
+ - param set #30 has cost=-0.013065 [o=70 t=26]
+ -- Parameters = 63.8028 8.2389 -30.6017 10.0153 25.9268 8.9025 0.9989 1.0037 0.9917 0.0009 0.0010 -0.0020
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0325
+ --- dist(#3,#1) = 0.491
+ --- dist(#4,#1) = 0.318
+ --- dist(#5,#1) = 0.365
+ --- dist(#6,#1) = 0.483
+ --- dist(#7,#1) = 0.326
+ --- dist(#8,#1) = 0.415
+ --- dist(#9,#1) = 0.439
+ --- dist(#10,#1) = 0.359
+ --- dist(#11,#1) = 0.402
+ --- dist(#12,#1) = 0.404
+ --- dist(#13,#1) = 0.499
+ --- dist(#14,#1) = 0.408
+ --- dist(#15,#1) = 0.499
+ --- dist(#16,#1) = 0.45
+ --- dist(#17,#1) = 0.458
+ --- dist(#18,#1) = 0.402
+ --- dist(#19,#1) = 0.435
+ --- dist(#20,#1) = 0.442
+ --- dist(#21,#1) = 0.431
+ --- dist(#22,#1) = 0.442
+ --- dist(#23,#1) = 0.504
+ --- dist(#24,#1) = 0.504
+ --- dist(#25,#1) = 0.455
+ --- dist(#26,#1) = 0.442
+ --- dist(#27,#1) = 0.422
+ --- dist(#28,#1) = 0.363
+ --- dist(#29,#1) = 0.468
+ --- dist(#30,#1) = 0.393
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 32726 total points stored in 68 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.229302 * [o=-1 t=-1]
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ - cost(#2)=-0.228654 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - cost(#3)=-0.020796 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - cost(#4)=-0.021017 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ - cost(#5)=-0.019426 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - cost(#6)=-0.019844 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - cost(#7)=-0.018617 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - cost(#8)=-0.016587 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - cost(#9)=-0.017151 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - cost(#10)=-0.018519 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - cost(#11)=-0.016436 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
+ - cost(#12)=-0.016901 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - cost(#13)=-0.016877 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - cost(#14)=-0.016763 [o=12 t=24]
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - cost(#15)=-0.016364 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - cost(#16)=-0.015450 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - cost(#17)=-0.016063 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - cost(#18)=-0.015527 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - cost(#19)=-0.015525 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - cost(#20)=-0.016496 [o=43 t=26]
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - cost(#21)=-0.014669 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - cost(#22)=-0.015020 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - cost(#23)=-0.014942 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - cost(#24)=-0.014574 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - cost(#25)=-0.014193 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - cost(#26)=-0.012148 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - cost(#27)=-0.012000 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - cost(#28)=-0.011452 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - cost(#29)=-0.011364 [o=74 t=19]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - cost(#30)=-0.010783 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - cost(#31)=-0.129342 [o=-2 t=-2]
+ -- Parameters = 86.1901 -3.4407 -1.7088 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.229705 * [o=-1 t=-1]
+ -- Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ !source mask fill: ubot=52 usiz=160
+ - cost(#2)=-0.229554 [o=0 t=0]
+ -- Parameters = 86.4108 -3.3549 0.0075 -0.6847 -1.9108 -0.2988 0.9897 1.0254 0.9729 -0.0093 0.0067 -0.0411
+ - cost(#3)=-0.020800 [o=42 t=21]
+ -- Parameters = 124.8687 -21.9275 -26.3385 8.9060 -2.8977 11.2572 0.9066 0.9499 0.9145 -0.0261 -0.0428 -0.0111
+ - cost(#4)=-0.021932 [o=2 t=2]
+ -- Parameters = 96.7132 -21.8547 37.0594 -11.0616 -41.9192 -26.4843 0.9931 0.9931 0.9950 -0.0013 0.0013 0.0069
+ - cost(#5)=-0.019536 [o=81 t=18]
+ -- Parameters = 100.8433 15.8919 -35.1646 -30.9880 7.4855 -4.8544 0.9870 1.0216 0.9993 -0.0015 0.0070 -0.0025
+ - retaining old weight image
+ - using 39832 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - cost(#6)=-0.021827 [o=76 t=6]
+ -- Parameters = 82.3364 4.8772 -39.6003 20.1755 10.4514 3.8632 1.0049 1.0059 1.0260 -0.0085 -0.0025 0.0079
+ 32435 total points stored in 66 'TOHD(17.1973)' bloks (0 duplicates)
+ - cost(#7)=-0.019391 [o=11 t=4]
+ -- Parameters = 122.1885 3.3850 37.7804 31.9993 -26.7764 -9.1048 1.0047 1.0043 1.0237 -0.0004 -0.0009 0.0010
+ - cost(#8)=-0.016887 [o=19 t=1]
+ -- Parameters = 120.3862 31.8744 37.9044 31.2181 -11.7938 -1.2006 1.0284 0.9884 0.9874 -0.0101 0.0025 0.0006
+ - cost(#9)=-0.017390 [o=21 t=5]
+ -- Parameters = 115.3469 31.2859 37.8570 28.2962 -10.3465 2.2030 0.9873 0.9648 1.0135 0.0016 0.0010 -0.0018
+ - cost(#10)=-0.018693 [o=20 t=11]
+ -- Parameters = 99.3262 -29.9447 26.9371 -5.3159 -28.4886 -34.0726 1.0107 0.9849 1.0213 -0.0036 -0.0003 0.0007
+ - cost(#11)=-0.016689 [o=59 t=15]
+ -- Parameters = 120.8994 -8.5654 38.2884 35.5660 -28.3844 -2.2898 0.9449 0.9652 0.9808 0.0065 -0.0097 0.0202
+ - cost(#12)=-0.017087 [o=6 t=7]
*[#27403=-0.232336] *[#27429=-0.232375] *[#27435=-0.232417] *[#27436=-0.232497] *[#27438=-0.232553] *[#27439=-0.232867] *[#27440=-0.232891] *[#27442=-0.233036] *[#27447=-0.233059] *[#27449=-0.233135] *[#27452=-0.233238] *[#27453=-0.233301] *[#27454=-0.233436] *[#27455=-0.233473] *[#27457=-0.233733] *[#27459=-0.233807] *[#27460=-0.233844] *[#27462=-0.233878] *[#27463=-0.233934] *[#27466=-0.234072] *[#27467=-0.234156] *[#27470=-0.23419] *[#27475=-0.234198] *[#27479=-0.234257] *[#27481=-0.234407] *[#27482=-0.234442] *[#27484=-0.234529] *[#27487=-0.234659] *[#27488=-0.234749] *[#27489=-0.234947] *[#27490=-0.235093] *[#27491=-0.235231] *[#27492=-0.235304] *[#27495=-0.235337] *[#27502=-0.235383] *[#27503=-0.23543] *[#27505=-0.235635] *[#27509=-0.235683] *[#27510=-0.23574] *[#27513=-0.23574] *[#27515=-0.235831] *[#27516=-0.235847] *[#27518=-0.235996] *[#27519=-0.236042] *[#27520=-0.23608]
+ -- Parameters = 124.5712 -28.6409 20.3221 5.1349 -41.6487 -44.5124 1.0125 0.9652 0.9978 -0.0043 -0.0015 -0.0151
+ - param set #1 has cost=-0.236080 [o=0 t=0]
+ -- Parameters = 86.7063 -2.0554 0.8724 -0.5856 -1.9323 -1.2648 0.9893 1.0180 0.9736 -0.0076 0.0106 -0.0273
+ - param set #2 has cost=-0.234027 [o=-1 t=-1]
+ -- Parameters = 86.6848 -2.0691 0.8802 -0.5271 -1.3065 -1.3295 0.9893 1.0196 0.9779 -0.0049 0.0117 -0.0145
+ - cost(#13)=-0.018747 [o=27 t=3]
+ -- Parameters = 124.8220 -15.4132 35.7434 32.8513 -32.2128 -4.2913 1.0051 1.0020 0.9925 -0.0014 0.0061 0.0027
+ - cost(#14)=-0.016958 [o=12 t=24]
+ -- Parameters = 90.6424 19.0892 31.4461 30.8611 -12.4391 -26.9602 1.0001 0.9900 0.9213 -0.0096 0.0069 -0.0164
+ - param set #3 has cost=-0.023005 [o=22 t=1]
+ -- Parameters = 106.5914 20.1549 -36.2959 -44.8689 12.8005 -8.9907 0.9892 0.9951 0.9825 -0.0029 0.0007 0.0136
+ - param set #4 has cost=-0.020592 [o=37 t=17]
+ -- Parameters = 81.8583 -1.9729 -24.6700 -0.9716 24.9751 -18.3067 1.0111 1.0015 0.9891 -0.0017 -0.0012 -0.0010
+ - param set #5 has cost=-0.019942 [o=65 t=21]
+ -- Parameters = 110.0321 17.7037 -28.3614 -32.4523 15.1376 9.4693 0.9565 0.8369 0.8625 -0.0210 0.0333 -0.0251
+ - param set #6 has cost=-0.020356 [o=71 t=12]
+ -- Parameters = 106.1047 20.6599 -31.9738 -43.9775 13.0946 -5.6192 1.0004 0.9539 1.0404 0.0078 0.0169 0.0259
+ - cost(#15)=-0.016621 [o=61 t=20]
+ -- Parameters = 95.2265 -16.0288 34.3051 18.8769 -38.6949 -27.3742 0.9949 1.0116 0.9938 0.0095 0.0005 -0.0030
+ - param set #7 has cost=-0.019654 [o=17 t=6]
+ - cost(#16)=-0.015480 [o=13 t=14]
+ -- Parameters = 82.6351 -14.6963 -22.6522 5.8338 28.0412 -25.8251 0.9860 0.9667 0.9910 -0.0068 -0.0030 0.0013
+ -- Parameters = 94.6942 -15.8722 35.0063 16.0018 -38.3548 -28.1013 0.9892 1.0073 0.9938 -0.0018 0.0006 -0.0023
+ - param set #8 has cost=-0.019078 [o=63 t=24]
+ -- Parameters = 80.5173 -8.6077 -32.1321 -1.7819 14.0326 -18.5044 1.0138 1.0057 1.0171 0.0010 0.0057 -0.0057
+ - cost(#17)=-0.017580 [o=70 t=17]
+ -- Parameters = 87.0306 -14.2621 -34.6583 -9.0179 9.8610 3.8227 0.9879 0.9809 0.9564 0.0087 0.0030 -0.0024
+ - param set #9 has cost=-0.017437 [o=2 t=3]
+ -- Parameters = 111.6816 -15.6843 -34.1914 -34.1112 21.7730 3.2434 0.9812 0.9743 0.9979 0.0080 0.0050 0.0001
+ - cost(#18)=-0.016272 [o=3 t=27]
+ -- Parameters = 92.1936 -30.6225 24.8890 5.7653 -33.6379 -32.8354 0.9906 0.9904 0.9919 0.0296 0.0046 -0.0038
+ - param set #10 has cost=-0.016749 [o=73 t=18]
+ -- Parameters = 80.3420 -6.1586 -27.5898 0.8852 31.2736 -21.4545 1.0392 1.0086 0.9963 0.0028 -0.0008 0.0019
+ - cost(#19)=-0.018831 [o=8 t=25]
+ -- Parameters = 92.0665 -2.7610 29.1632 -37.9380 15.4778 -34.5080 0.9731 0.9648 0.9439 -0.0023 -0.0035 0.0016
+ - param set #11 has cost=-0.018271 [o=72 t=4]
+ -- Parameters = 94.7323 -6.2371 -31.3001 -5.7165 7.6807 21.3001 0.9867 1.0148 0.9844 -0.0003 0.0122 -0.0047
+ - cost(#20)=-0.016643 [o=43 t=26]
+ -- Parameters = 66.9145 8.7317 -38.5499 19.9377 18.3452 -6.1108 1.0030 0.9976 0.9713 0.0006 0.0051 0.0221
+ - param set #12 has cost=-0.016191 [o=83 t=19]
+ -- Parameters = 82.6362 -5.6353 -31.4927 -9.9597 14.1660 -6.1758 1.0364 0.9938 1.0009 0.0067 0.0016 -0.0005
+ - cost(#21)=-0.015076 [o=33 t=16]
+ -- Parameters = 110.5064 17.5324 -34.6949 -44.8090 11.7870 22.7514 0.9787 1.0007 0.9652 -0.0008 0.0293 0.0042
+ - param set #13 has cost=-0.015853 [o=7 t=28]
+ - cost(#22)=-0.015064 [o=68 t=10]
+ -- Parameters = 94.9821 11.4850 -41.7087 -27.1684 18.0621 -14.7438 0.9953 1.0047 1.0033 0.0097 -0.0014 -0.0005
+ -- Parameters = 61.5351 31.3431 -33.9048 44.3326 -40.5053 -29.3835 1.0250 1.0014 0.9832 -0.0010 -0.0034 0.0147
+ - cost(#23)=-0.015215 [o=87 t=8]
+ -- Parameters = 80.8903 10.8767 31.8345 38.9750 -23.2805 -12.6024 0.9964 0.9868 0.9912 -0.0075 -0.0002 0.0015
+ - param set #14 has cost=-0.016259 [o=13 t=9]
+ -- Parameters = 64.0981 24.7000 -31.6466 20.0503 -12.2969 -33.3030 1.0101 0.9893 1.0080 0.0001 0.0003 0.0025
+ - cost(#24)=-0.014807 [o=55 t=13]
+ -- Parameters = 101.9003 23.5725 -40.5280 -5.6755 -15.6652 40.9313 1.0032 0.9991 1.0112 0.0085 -0.0004 -0.0001
+ - param set #15 has cost=-0.016768 [o=49 t=8]
+ -- Parameters = 83.8847 1.3558 -39.8692 -10.0807 14.1002 -7.9357 1.0024 0.9934 0.9828 -0.0011 -0.0014 -0.0041
+ - cost(#25)=-0.014391 [o=23 t=9]
+ -- Parameters = 88.9471 -11.7913 -34.3018 -12.1401 11.8714 4.7590 1.0001 0.9921 0.9903 0.0145 0.0001 -0.0000
+ - cost(#26)=-0.012283 [o=26 t=12]
+ - param set #16 has cost=-0.015523 [o=66 t=23]
+ -- Parameters = 112.7510 -14.0798 -34.9248 -28.0911 24.1450 4.0337 0.9952 1.0025 1.0059 0.0123 0.0083 -0.0004
+ -- Parameters = 80.3921 9.9749 -34.8137 3.8113 31.1348 -30.5749 1.0126 1.0033 1.0196 0.0012 -0.0001 -0.0009
+ - param set #17 has cost=-0.016673 [o=19 t=2]
+ - cost(#27)=-0.012050 [o=53 t=28]
+ -- Parameters = 107.9775 -14.6868 -35.7018 -31.4971 21.6043 -5.6362 1.0031 0.9959 1.0111 0.0002 0.0033 0.0091
+ -- Parameters = 119.0245 -8.2729 -38.8807 -34.4827 24.2436 -0.6653 1.0003 1.0042 1.0074 -0.0013 0.0015 0.0001
+ - param set #18 has cost=-0.015912 [o=76 t=7]
+ -- Parameters = 111.2374 21.3375 -29.0974 -36.7486 20.7390 12.5540 0.9807 0.9803 0.9858 0.0050 0.0043 0.0043
+ - param set #19 has cost=-0.014648 [o=10 t=10]
+ -- Parameters = 84.7452 -4.9497 -33.9636 -13.6488 11.8193 -2.0966 0.9990 0.9988 1.0001 -0.0000 -0.0006 0.0010
+ - cost(#28)=-0.011891 [o=24 t=23]
+ -- Parameters = 119.1164 -6.9467 -37.4876 -11.0812 5.1131 -12.3826 0.9973 1.0034 0.9823 -0.0065 0.0021 -0.0026
+ - param set #20 has cost=-0.016779 [o=67 t=22]
+ -- Parameters = 70.3578 21.0734 -34.5994 37.4589 6.8237 4.3216 0.9923 0.9938 0.9829 -0.0013 0.0088 -0.0022
+ - param set #21 has cost=-0.014144 [o=3 t=15]
+ -- Parameters = 68.5323 17.2552 -33.7320 35.3305 15.3195 -17.8718 0.9984 0.9932 0.9922 -0.0055 -0.0012 0.0027
+ - cost(#29)=-0.011366 [o=74 t=19]
+ -- Parameters = 66.4278 14.9361 -34.0231 33.9244 20.6755 -10.4931 0.9986 1.0021 0.9994 0.0001 0.0002 0.0008
+ - param set #22 has cost=-0.014284 [o=46 t=5]
+ -- Parameters = 106.1957 -10.7457 -34.4768 -25.4659 13.1058 -4.2697 0.9869 1.0073 1.0021 0.0071 0.0005 -0.0008
+ - cost(#30)=-0.012654 [o=15 t=22]
+ -- Parameters = 118.6806 -0.2779 -39.9209 -10.4236 10.9901 -14.6070 0.9859 0.9982 1.0056 0.0036 -0.0026 0.0003
+ - param set #23 has cost=-0.014837 [o=43 t=14]
+ -- Parameters = 119.5785 -33.0555 -39.5767 5.2598 32.2736 -21.0898 0.9911 0.9939 1.0024 0.0008 0.0054 0.0035
+ - cost(#31)=-0.226142 [o=-2 t=-2]
+ - param set #24 has cost=-0.016408 [o=78 t=27]
+ -- Parameters = 86.2853 -3.2417 0.0041 -0.6043 -1.3783 0.2327 0.9846 1.0247 0.9738 -0.0073 -0.0072 -0.0283
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.229705
+ - Initial fine Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ -- Parameters = 102.2802 30.8182 -39.9754 -7.5097 -13.9483 43.9346 0.9985 0.9663 0.9934 0.0020 0.0121 0.0043
+ - param set #25 has cost=-0.013574 [o=56 t=16]
+ -- Parameters = 59.4919 7.5457 -30.4650 40.3137 -7.1803 -30.8818 1.0036 1.0109 0.9786 0.0021 -0.0003 0.0011
+ - param set #26 has cost=-0.014020 [o=52 t=20]
+ -- Parameters = 60.6204 6.1029 -27.3132 39.2064 1.6237 -31.2036 0.9849 1.0067 0.9911 0.0095 0.0039 -0.0018
+ - param set #27 has cost=-0.013994 [o=31 t=13]
+ -- Parameters = 117.9574 -9.4925 -32.8757 -7.4021 6.5560 -8.5198 1.0020 1.0092 1.0178 0.0074 -0.0005 -0.0038
+ - Finalish cost = -0.230087 ; 443 funcs
+ - ini Finalish Parameters = 86.4243 -3.3934 0.0579 -0.6907 -2.4008 -0.2686 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
+ - param set #28 has cost=-0.013460 [o=24 t=25]
+ -- Parameters = 87.7214 -9.9050 -25.8087 -6.3015 31.4129 8.0160 0.9976 1.0201 1.0136 0.0004 0.0096 -0.0039
+ - param set #29 has cost=-0.012810 [o=40 t=11]
+ -- Parameters = 118.4645 -9.4135 -36.5992 -0.8830 -1.1261 -14.1717 0.9927 1.0050 0.9991 -0.0002 0.0001 -0.0022
+ - Final cost = -0.230087 ; 283 funcs
+ Final fine fit Parameters:
x-shift= 86.4239 y-shift= -3.3912 z-shift= 0.0590 ... enorm= 86.4904 mm
z-angle= -0.6917 x-angle= -2.3988 y-angle= -0.2681 ... total= 2.5094 deg
x-scale= 0.9907 y-scale= 1.0261 z-scale= 0.9755 ... vol3D= 0.9917 = base bigger than source
y/x-shear= -0.0090 z/x-shear= 0.0076 z/y-shear= -0.0512
+ - param set #30 has cost=-0.009779 [o=70 t=26]
+ -- Parameters = 63.8024 8.1917 -30.6542 10.0511 25.8948 8.7807 1.0013 1.0041 0.9883 0.0005 0.0009 -0.0025
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0578
+ --- dist(#3,#1) = 0.492
+ - Fine net CPU time = 0.0 s
+ --- dist(#4,#1) = 0.319
++ Computing output image
+ --- dist(#5,#1) = 0.482
+ --- dist(#6,#1) = 0.365
+ --- dist(#7,#1) = 0.333
+ --- dist(#8,#1) = 0.413
+ --- dist(#9,#1) = 0.402
+ --- dist(#10,#1) = 0.438
+ --- dist(#11,#1) = 0.443
++ image warp: parameters = 86.4239 -3.3912 0.0590 -0.6917 -2.3988 -0.2681 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
+ --- dist(#12,#1) = 0.509
+ --- dist(#13,#1) = 0.369
+ --- dist(#14,#1) = 0.457
+ --- dist(#15,#1) = 0.511
+ --- dist(#16,#1) = 0.406
+ --- dist(#17,#1) = 0.405
+ --- dist(#18,#1) = 0.402
+ --- dist(#19,#1) = 0.499
+ --- dist(#20,#1) = 0.446
+ --- dist(#21,#1) = 0.506
+ --- dist(#22,#1) = 0.435
+ --- dist(#23,#1) = 0.442
+ --- dist(#24,#1) = 0.433
+ --- dist(#25,#1) = 0.442
+ --- dist(#26,#1) = 0.422
+ --- dist(#27,#1) = 0.454
+ --- dist(#28,#1) = 0.371
+ --- dist(#29,#1) = 0.468
+ --- dist(#30,#1) = 0.394
+ - Total coarse refinement net CPU time = 0.0 s; 9571 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=52 usiz=160
++ Unloading unneeded data
++ Output dataset ./sub-08_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 25.2
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/__tt_sub-08_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/sub-08_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-08_T1w_ns+orig sub-08_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 32673 total points stored in 68 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.233161 * [o=0 t=0]
+ -- Parameters = 86.7063 -2.0554 0.8724 -0.5856 -1.9323 -1.2648 0.9893 1.0180 0.9736 -0.0076 0.0106 -0.0273
+ - cost(#2)=-0.231003 [o=-1 t=-1]
+ -- Parameters = 86.6848 -2.0691 0.8802 -0.5271 -1.3065 -1.3295 0.9893 1.0196 0.9779 -0.0049 0.0117 -0.0145
+ - cost(#3)=-0.019837 [o=22 t=1]
+ -- Parameters = 106.5914 20.1549 -36.2959 -44.8689 12.8005 -8.9907 0.9892 0.9951 0.9825 -0.0029 0.0007 0.0136
+ - cost(#4)=-0.017361 [o=37 t=17]
+ -- Parameters = 81.8583 -1.9729 -24.6700 -0.9716 24.9751 -18.3067 1.0111 1.0015 0.9891 -0.0017 -0.0012 -0.0010
+ - cost(#5)=-0.017714 [o=71 t=12]
+ -- Parameters = 106.1047 20.6599 -31.9738 -43.9775 13.0946 -5.6192 1.0004 0.9539 1.0404 0.0078 0.0169 0.0259
+ - cost(#6)=-0.016374 [o=65 t=21]
+ -- Parameters = 110.0321 17.7037 -28.3614 -32.4523 15.1376 9.4693 0.9565 0.8369 0.8625 -0.0210 0.0333 -0.0251
+ - cost(#7)=-0.017666 [o=17 t=6]
+ -- Parameters = 82.6351 -14.6963 -22.6522 5.8338 28.0412 -25.8251 0.9860 0.9667 0.9910 -0.0068 -0.0030 0.0013
+ - cost(#8)=-0.018229 [o=63 t=24]
+ -- Parameters = 80.5173 -8.6077 -32.1321 -1.7819 14.0326 -18.5044 1.0138 1.0057 1.0171 0.0010 0.0057 -0.0057
+ - cost(#9)=-0.016642 [o=72 t=4]
+ -- Parameters = 94.7323 -6.2371 -31.3001 -5.7165 7.6807 21.3001 0.9867 1.0148 0.9844 -0.0003 0.0122 -0.0047
+ - cost(#10)=-0.015085 [o=2 t=3]
+ -- Parameters = 111.6816 -15.6843 -34.1914 -34.1112 21.7730 3.2434 0.9812 0.9743 0.9979 0.0080 0.0050 0.0001
+ - cost(#11)=-0.015429 [o=67 t=22]
+ -- Parameters = 70.3578 21.0734 -34.5994 37.4589 6.8237 4.3216 0.9923 0.9938 0.9829 -0.0013 0.0088 -0.0022
+ - cost(#12)=-0.014227 [o=49 t=8]
+ -- Parameters = 83.8847 1.3558 -39.8692 -10.0807 14.1002 -7.9357 1.0024 0.9934 0.9828 -0.0011 -0.0014 -0.0041
+ - cost(#13)=-0.014841 [o=73 t=18]
+ -- Parameters = 80.3420 -6.1586 -27.5898 0.8852 31.2736 -21.4545 1.0392 1.0086 0.9963 0.0028 -0.0008 0.0019
+ - cost(#14)=-0.013490 [o=19 t=2]
+ -- Parameters = 107.9775 -14.6868 -35.7018 -31.4971 21.6043 -5.6362 1.0031 0.9959 1.0111 0.0002 0.0033 0.0091
+ - cost(#15)=-0.014651 [o=78 t=27]
+ -- Parameters = 102.2802 30.8182 -39.9754 -7.5097 -13.9483 43.9346 0.9985 0.9663 0.9934 0.0020 0.0121 0.0043
+ - cost(#16)=-0.014743 [o=13 t=9]
+ -- Parameters = 64.0981 24.7000 -31.6466 20.0503 -12.2969 -33.3030 1.0101 0.9893 1.0080 0.0001 0.0003 0.0025
+ - cost(#17)=-0.015521 [o=83 t=19]
+ -- Parameters = 82.6362 -5.6353 -31.4927 -9.9597 14.1660 -6.1758 1.0364 0.9938 1.0009 0.0067 0.0016 -0.0005
+ - cost(#18)=-0.014142 [o=76 t=7]
+ -- Parameters = 111.2374 21.3375 -29.0974 -36.7486 20.7390 12.5540 0.9807 0.9803 0.9858 0.0050 0.0043 0.0043
+ - cost(#19)=-0.014340 [o=7 t=28]
+ -- Parameters = 61.5351 31.3431 -33.9048 44.3326 -40.5053 -29.3835 1.0250 1.0014 0.9832 -0.0010 -0.0034 0.0147
+ - cost(#20)=-0.013096 [o=66 t=23]
+ -- Parameters = 80.3921 9.9749 -34.8137 3.8113 31.1348 -30.5749 1.0126 1.0033 1.0196 0.0012 -0.0001 -0.0009
+ - cost(#21)=-0.012618 [o=43 t=14]
+ -- Parameters = 119.5785 -33.0555 -39.5767 5.2598 32.2736 -21.0898 0.9911 0.9939 1.0024 0.0008 0.0054 0.0035
+ - cost(#22)=-0.012947 [o=10 t=10]
+ -- Parameters = 84.7452 -4.9497 -33.9636 -13.6488 11.8193 -2.0966 0.9990 0.9988 1.0001 -0.0000 -0.0006 0.0010
+ - cost(#23)=-0.011261 [o=46 t=5]
+ -- Parameters = 106.1957 -10.7457 -34.4768 -25.4659 13.1058 -4.2697 0.9869 1.0073 1.0021 0.0071 0.0005 -0.0008
+ - cost(#24)=-0.012283 [o=3 t=15]
+ -- Parameters = 68.5323 17.2552 -33.7320 35.3305 15.3195 -17.8718 0.9984 0.9932 0.9922 -0.0055 -0.0012 0.0027
+ - cost(#25)=-0.012514 [o=52 t=20]
+ -- Parameters = 60.6204 6.1029 -27.3132 39.2064 1.6237 -31.2036 0.9849 1.0067 0.9911 0.0095 0.0039 -0.0018
+ - cost(#26)=-0.012114 [o=31 t=13]
+ -- Parameters = 117.9574 -9.4925 -32.8757 -7.4021 6.5560 -8.5198 1.0020 1.0092 1.0178 0.0074 -0.0005 -0.0038
+ - cost(#27)=-0.011281 [o=56 t=16]
+ -- Parameters = 59.4919 7.5457 -30.4650 40.3137 -7.1803 -30.8818 1.0036 1.0109 0.9786 0.0021 -0.0003 0.0011
+ - cost(#28)=-0.011841 [o=24 t=25]
+ -- Parameters = 87.7214 -9.9050 -25.8087 -6.3015 31.4129 8.0160 0.9976 1.0201 1.0136 0.0004 0.0096 -0.0039
+ - cost(#29)=-0.011241 [o=40 t=11]
+ -- Parameters = 118.4645 -9.4135 -36.5992 -0.8830 -1.1261 -14.1717 0.9927 1.0050 0.9991 -0.0002 0.0001 -0.0022
+ - cost(#30)=-0.008977 [o=70 t=26]
+ -- Parameters = 63.8024 8.1917 -30.6542 10.0511 25.8948 8.7807 1.0013 1.0041 0.9883 0.0005 0.0009 -0.0025
+ - cost(#31)=-0.168560 [o=-2 t=-2]
+ -- Parameters = 86.2547 -1.6575 -0.0199 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.233361 * [o=0 t=0]
+ -- Parameters = 86.6848 -2.0657 0.8745 -0.5732 -1.9348 -1.2473 0.9910 1.0174 0.9739 -0.0082 0.0101 -0.0272
+ - cost(#2)=-0.232622 [o=-1 t=-1]
+ -- Parameters = 86.7255 -2.0266 0.8487 -0.6381 -1.5388 -1.3075 0.9907 1.0168 0.9736 -0.0100 0.0109 -0.0179
+ - cost(#3)=-0.019928 [o=22 t=1]
+ -- Parameters = 106.5189 20.1739 -36.2639 -44.8611 12.8475 -8.9440 0.9890 0.9958 0.9830 -0.0030 -0.0004 0.0143
+ - cost(#4)=-0.017565 [o=37 t=17]
+ -- Parameters = 81.7177 -2.0410 -24.6487 -0.9999 24.9875 -18.1815 1.0088 1.0023 0.9888 0.0001 -0.0010 -0.0009
+ - cost(#5)=-0.017930 [o=71 t=12]
+ -- Parameters = 106.0384 20.6675 -31.8491 -43.8927 13.1469 -5.6153 1.0000 0.9543 1.0397 0.0078 0.0170 0.0187
+ - cost(#6)=-0.016430 [o=65 t=21]
+ -- Parameters = 110.0331 17.7430 -28.3685 -32.4911 15.1825 9.5033 0.9566 0.8371 0.8621 -0.0219 0.0342 -0.0250
+ - cost(#7)=-0.017748 [o=17 t=6]
+ -- Parameters = 82.6445 -14.5856 -22.7192 5.7547 28.1249 -25.7555 0.9858 0.9682 0.9906 -0.0073 -0.0022 0.0011
+ - cost(#8)=-0.018393 [o=63 t=24]
+ -- Parameters = 80.4755 -8.6910 -32.0149 -1.7603 14.0106 -18.5259 1.0102 1.0062 1.0208 0.0009 0.0062 -0.0059
+ - cost(#9)=-0.017475 [o=72 t=4]
+ -- Parameters = 94.8254 -6.2634 -31.3755 -5.5902 7.6433 21.0777 0.9910 1.0139 0.9784 -0.0012 0.0120 -0.0144
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-08_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-08_T1w_al_junk+orig
+ - cost(#10)=-0.015522 [o=2 t=3]
+ -- Parameters = 111.6237 -15.9836 -33.9976 -34.0059 21.6707 3.4295 0.9774 0.9705 1.0011 0.0080 0.0055 0.0004
+ - cost(#11)=-0.015606 [o=67 t=22]
+ -- Parameters = 70.4833 21.1295 -34.6357 37.4262 6.8235 4.2216 0.9894 0.9960 0.9817 -0.0016 0.0097 -0.0035
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
+ - cost(#12)=-0.014261 [o=49 t=8]
+ -- Parameters = 83.8674 1.3643 -39.8477 -10.1024 14.1222 -7.9057 1.0018 0.9939 0.9830 -0.0018 -0.0010 -0.0046
#Script is running:
\rm -f ./__tt_sub-08_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
+ - cost(#13)=-0.014918 [o=73 t=18]
+ -- Parameters = 80.4225 -6.0916 -27.4774 0.9061 31.3592 -21.4803 1.0385 1.0097 0.9967 0.0027 -0.0007 0.0019
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-08_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
+ - cost(#14)=-0.016098 [o=19 t=2]
+ -- Parameters = 108.7916 -14.5577 -32.1457 -31.3129 21.4651 -5.9172 1.0043 0.9933 1.0076 0.0000 0.0038 0.0099
+ - cost(#15)=-0.014684 [o=78 t=27]
+ -- Parameters = 102.2904 30.7916 -39.9555 -7.5106 -13.9550 43.9311 0.9984 0.9661 0.9936 0.0020 0.0128 0.0039
+ - cost(#16)=-0.014793 [o=13 t=9]
+ -- Parameters = 64.1020 24.6882 -31.6672 20.0731 -12.3766 -33.3152 1.0086 0.9887 1.0092 0.0001 0.0003 0.0026
+ - cost(#17)=-0.015547 [o=83 t=19]
+ -- Parameters = 82.5791 -5.6434 -31.4093 -10.0069 14.1115 -6.1369 1.0361 0.9940 1.0028 0.0060 0.0017 -0.0005
+ - cost(#18)=-0.014650 [o=76 t=7]
+ -- Parameters = 111.0543 21.3662 -29.1096 -36.6641 20.9018 12.8033 0.9807 0.9865 0.9856 0.0056 0.0047 -0.0057
+ - cost(#19)=-0.014497 [o=7 t=28]
+ -- Parameters = 61.4954 31.4727 -33.9190 44.2397 -40.4944 -29.4068 1.0276 1.0003 0.9831 -0.0013 -0.0042 0.0154
+ - cost(#20)=-0.013194 [o=66 t=23]
+ -- Parameters = 80.4260 9.9559 -34.8388 3.8945 31.1140 -30.6435 1.0162 1.0055 1.0216 0.0012 -0.0008 -0.0005
+ - cost(#21)=-0.012903 [o=43 t=14]
+ -- Parameters = 119.5012 -33.2194 -39.4750 5.2035 32.0180 -21.1048 0.9901 0.9910 1.0032 0.0013 0.0137 0.0042
+ - cost(#22)=-0.013022 [o=10 t=10]
+ -- Parameters = 84.6679 -4.9553 -33.9474 -13.5697 11.8268 -2.1092 1.0004 0.9989 1.0003 0.0001 -0.0006 0.0008
+ - cost(#23)=-0.011278 [o=46 t=5]
+ -- Parameters = 106.2272 -10.7177 -34.4898 -25.4454 13.0796 -4.2667 0.9875 1.0075 1.0020 0.0076 0.0005 -0.0009
+ - cost(#24)=-0.012428 [o=3 t=15]
+ -- Parameters = 68.5446 17.2289 -33.7460 35.3344 15.2603 -17.9269 0.9983 0.9931 0.9938 -0.0051 -0.0010 -0.0041
+ - cost(#25)=-0.012570 [o=52 t=20]
+ -- Parameters = 60.6436 6.1088 -27.3498 39.2074 1.5923 -31.2612 0.9849 1.0063 0.9910 0.0096 0.0034 -0.0008
+ - cost(#26)=-0.012419 [o=31 t=13]
+ -- Parameters = 117.8880 -9.5370 -32.8037 -7.4891 6.4431 -8.3727 1.0027 1.0095 1.0223 0.0086 -0.0084 -0.0049
+ - cost(#27)=-0.015380 [o=56 t=16]
+ -- Parameters = 57.0060 1.5833 -29.0556 40.6691 -7.4393 -31.1530 1.0074 1.0040 0.9808 0.0005 0.0045 0.0033
+ - cost(#28)=-0.014995 [o=24 t=25]
+ -- Parameters = 87.2779 -13.7473 -25.1673 -6.7723 30.0955 8.3092 0.9886 1.0169 1.0161 0.0019 0.0104 -0.0041
+ - cost(#29)=-0.011297 [o=40 t=11]
+ -- Parameters = 118.4431 -9.4594 -36.6066 -0.8270 -1.1364 -14.1521 0.9945 1.0048 0.9984 -0.0005 0.0005 -0.0020
+ - cost(#30)=-0.009245 [o=70 t=26]
+ -- Parameters = 63.8099 8.5775 -30.7656 10.0236 26.0464 8.8006 1.0010 1.0021 0.9860 -0.0001 0.0011 -0.0022
+ - cost(#31)=-0.231227 [o=-2 t=-2]
+ -- Parameters = 86.6568 -2.1432 0.8901 -0.6518 -1.6355 -0.6395 0.9930 1.0195 0.9785 -0.0136 -0.0053 -0.0205
+ - case #1 [o=0 t=0] is now the best
+ - Initial cost = -0.233361
+ - Initial fine Parameters = 86.6848 -2.0657 0.8745 -0.5732 -1.9348 -1.2473 0.9910 1.0174 0.9739 -0.0082 0.0101 -0.0272
+ - Finalish cost = -0.233515 ; 371 funcs
+ - ini Finalish Parameters = 86.6965 -2.0664 0.8727 -0.6341 -1.9511 -1.1562 0.9911 1.0175 0.9738 -0.0105 0.0078 -0.0273
+ - Final cost = -0.233516 ; 342 funcs
+ Final fine fit Parameters:
x-shift= 86.6976 y-shift= -2.0675 z-shift= 0.8719 ... enorm= 86.7266 mm
z-angle= -0.6350 x-angle= -1.9506 y-angle= -1.1514 ... total= 2.3471 deg
x-scale= 0.9911 y-scale= 1.0176 z-scale= 0.9739 ... vol3D= 0.9822 = base bigger than source
y/x-shear= -0.0105 z/x-shear= 0.0077 z/y-shear= -0.0272
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.6976 -2.0675 0.8719 -0.6350 -1.9506 -1.1514 0.9911 1.0176 0.9739 -0.0105 0.0077 -0.0272
++ Unloading unneeded data
++ Output dataset ./sub-07_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-07_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 25.7
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/__tt_sub-07_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/sub-07_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-07_T1w_ns+orig sub-07_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
Performing center alignment with @Align_Centers
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-07_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-07_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-07_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-07_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-08_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
Performing center alignment with @Align_Centers
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-08_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-08_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-08_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-08_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-08_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-08_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-07_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-07_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-07_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-07_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-07_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-07_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-07_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Center distance of 0.000000 mm
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
Resampling ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
Center distance of 0.000000 mm
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
Resampling ...
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1336.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-08_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-08_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/125.00=4.2e+02
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1458.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-07_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-07_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/125.00=4.2e+02
RMS[0] = 0.283712 0.155217 ITER = 8/50
0.283712
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-08_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-08_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-08_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-08_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-08_T1w_ns+orig.BRIK
RMS[0] = 0.292904 0.152726 ITER = 7/50
0.292904
Warping has converged.
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-08_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-08_T1w_ns+orig __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc.HEAD -> sub-08_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc.BRIK -> sub-08_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-08_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-08_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-08.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-08.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
#Script is running (command trimmed):
3dinfo ./__tt_sub-09_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/__tt_sub-09_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
+ Automask has 40918 voxels
#Script is running (command trimmed):
+ 5959 voxels left in -maxdisp mask after erosion
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ Initializing alignment base
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 319.078796
+ Used gradual clip level = 308.024994 .. 332.049988
+ Number voxels above clip level = 39009
+ Clustering voxels ...
+ Largest cluster has 38460 voxels
+ Clustering voxels ...
+ Largest cluster has 37120 voxels
+ Filled 311 voxels in small holes; now have 37431 voxels
+ Filled 1 voxels in large holes; now have 37432 voxels
+ Clustering voxels ...
+ Largest cluster has 37432 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 126408 voxels
+ Mask now has 37432 voxels
++ 37432 voxels in the mask [out of 163840: 22.85%]
++ first 10 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 3 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 3 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 767.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/767.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-09_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-09_T1w_ns+orig -prefix ./sub-09_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-09_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-09_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-07_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-07_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-07_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-07_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-07_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-07_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-07_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-07_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-07_T1w_ns+orig __ats_tmp___upad15_pre.sub-07_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-07_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-07_T1w_ns+tlrc.HEAD -> sub-07_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-07_T1w_ns+tlrc.BRIK -> sub-07_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-07_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-07_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-07.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-07.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 42417 voxels
+ 6211 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ 1487610 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=5 ytop=1
++ Zero-pad: zbot=8 ztop=5
++ 37432 voxels [15.8%] in weight mask
++ Output dataset ./__tt_sub-09_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 37432
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 30.977 32.886 28.502 (index)
+ source center of mass = 88.642 114.506 129.787 (index)
+ source-target CM = 86.350 -2.184 -2.444 (xyz)
+ estimated center of mass shifts = 86.350 -2.184 -2.444
++ shift param auto-range: 25.7..147.0 -68.6..64.3 -69.2..64.3
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.349770 .. 96.349770 center = 86.349770
+ Range param#2 [y-shift] = -12.184143 .. 7.815857 center = -2.184143
+ Range param#3 [z-shift] = -12.443642 .. 7.556358 center = -2.443642
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.349770 .. 126.349770 center = 86.349770
+ Range param#2 [y-shift] = -42.184143 .. 37.815857 center = -2.184143
+ Range param#3 [z-shift] = -42.443642 .. 37.556358 center = -2.443642
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000089
++ Final parameter search ranges:
+ x-shift = 46.350 .. 126.350
+ y-shift = -42.184 .. 37.816
+ z-shift = -42.444 .. 37.556
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1487610 [out of 11534336] voxels
+ base mask has 50525 [out of 237440] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=36 usiz=142
+ - copying weight image
+ - using 37432 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 31183 total points stored in 61 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.014 pitch=-0.044 yaw=-0.059 dS=-0.092 dL=-0.041 dP=-0.083
++ Mean: roll=+0.022 pitch=+0.035 yaw=+0.065 dS=+0.016 dL=+0.029 dP=+0.013
++ Max : roll=+0.119 pitch=+0.184 yaw=+0.253 dS=+0.181 dL=+0.123 dP=+0.235
++ Max displacements (mm) for each sub-brick:
0.15(0.00) 0.09(0.16) 0.06(0.09) 0.05(0.08) 0.07(0.08) 0.07(0.03) 0.08(0.05) 0.06(0.05) 0.15(0.13) 0.09(0.12) 0.15(0.12) 0.09(0.13) 0.11(0.10) 0.09(0.06) 0.09(0.05) 0.11(0.04) 0.04(0.10) 0.08(0.08) 0.07(0.05) 0.13(0.10) 0.06(0.11) 0.06(0.09) 0.07(0.09) 0.03(0.05) 0.09(0.07) 0.06(0.13) 0.07(0.09) 0.05(0.08) 0.00(0.05) 0.06(0.06) 0.06(0.08) 0.05(0.06) 0.10(0.07) 0.06(0.06) 0.07(0.09) 0.06(0.10) 0.08(0.08) 0.08(0.07) 0.12(0.10) 0.08(0.11) 0.09(0.08) 0.11(0.07) 0.05(0.08) 0.08(0.05) 0.03(0.08) 0.07(0.06) 0.08(0.09) 0.06(0.05) 0.10(0.07) 0.08(0.09) 0.12(0.10) 0.10(0.08) 0.06(0.07) 0.14(0.14) 0.09(0.11) 0.11(0.10) 0.14(0.06) 0.09(0.12) 0.12(0.11) 0.10(0.09) 0.10(0.06) 0.15(0.07) 0.09(0.12) 0.11(0.10) 0.09(0.15) 0.14(0.16) 0.09(0.09) 0.10(0.03) 0.11(0.07) 0.08(0.07) 0.13(0.11) 0.09(0.11) 0.13(0.11) 0.09(0.08) 0.10(0.06) 0.13(0.10) 0.10(0.09) 0.14(0.11) 0.11(0.14) 0.11(0.10) 0.09(0.07) 0.10(0.08) 0.11(0.06) 0.10(0.10) 0.16(0.16) 0.10(0.16) 0.13(0.14) 0.13(0.11) 0.17(0.07) 0.18(0.07) 0.15(0.09) 0.15(0.07) 0.12(0.10) 0.17(0.10) 0.15(0.05) 0.16(0.07) 0.14(0.06) 0.16(0.10) 0.24(0.15) 0.14(0.17) 0.21(0.14) 0.16(0.12) 0.29(0.22) 0.20(0.16) 0.23(0.11) 0.21(0.07) 0.21(0.10) 0.23(0.05) 0.19(0.12) 0.22(0.08) 0.18(0.09) 0.23(0.10) 0.19(0.11) 0.28(0.14) 0.21(0.10) 0.23(0.06) 0.23(0.07) 0.29(0.15) 0.22(0.17) 0.29(0.10) 0.30(0.04) 0.27(0.07) 0.31(0.08) 0.28(0.06) 0.34(0.11) 0.30(0.07) 0.37(0.11) 0.28(0.15) 0.36(0.14) 0.29(0.14) 0.36(0.15) 0.32(0.30) 0.41(0.27) 0.49(0.29) 0.46(0.11) 0.49(0.18) 0.56(0.18) 0.55(0.14) 0.55(0.10) 0.54(0.03) 0.52(0.12) 0.51(0.14) 0.48(0.15) 0.41(0.17) 0.41(0.22) 0.42(0.12)
++ Max displacement in automask = 0.56 (mm) at sub-brick 136
++ Max delta displ in automask = 0.30 (mm) at sub-brick 131
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-08.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb00.sub-08.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-08.r01.tcat+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 0.027 -8.241 -29.822
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.0% 11.9% 51.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
+ - Test (64+191)*64 params [top5=*o+-.]:#ooo...+.-+-ooo+o.++-+-++o-o.++o++oo$*[#4199=-0.023561] *.+.--+
+ - best 88 costs found:
o= 0 v=-0.023561: 115.36 -29.33 -36.06 7.44 27.42 -25.63 [rand]
o= 1 v=-0.022396: 86.35 -2.18 -2.44 0.00 0.00 0.00 [grid]
o= 2 v=-0.021601: 69.55 16.28 -34.80 -10.10 -16.32 -39.31 [rand]
o= 3 v=-0.021424: 111.32 -31.24 -37.89 17.69 14.58 -34.92 [rand]
o= 4 v=-0.019160: 88.64 -9.36 -34.14 -10.97 18.37 -5.04 [rand]
o= 5 v=-0.019095: 118.56 -31.23 33.70 -40.41 -19.09 -40.75 [rand]
o= 6 v=-0.018885: 113.02 -28.85 -29.11 7.50 30.00 -30.00 [grid]
o= 7 v=-0.018433: 107.79 -22.17 -26.06 -27.55 25.32 -7.37 [rand]
o= 8 v=-0.018328: 79.30 -22.00 -38.19 -25.15 27.05 -27.90 [rand]
o= 9 v=-0.017356: 97.65 -36.32 -8.42 -4.11 32.66 25.74 [rand]
o=10 v=-0.016848: 59.68 24.48 24.22 -7.50 7.50 30.00 [grid]
o=11 v=-0.016431: 93.40 -22.00 -38.19 -25.15 27.05 -27.90 [rand]
o=12 v=-0.016364: 115.36 -29.33 -36.06 -7.44 27.42 -25.63 [rand]
o=13 v=-0.015800: 84.06 -9.36 -34.14 -10.97 18.37 -5.04 [rand]
o=14 v=-0.015337: 69.55 -20.65 -34.80 -10.10 16.32 -39.31 [rand]
o=15 v=-0.015333: 96.34 -36.23 -8.96 -9.05 41.60 20.37 [rand]
o=16 v=-0.014867: 121.96 -6.60 -39.31 -37.76 24.22 3.86 [rand]
o=17 v=-0.014825: 101.64 -23.07 -32.83 -14.56 29.40 -15.92 [rand]
o=18 v=-0.014252: 69.58 18.59 -31.72 12.96 3.05 9.17 [rand]
o=19 v=-0.013920: 109.62 -32.87 -8.07 20.48 28.84 27.87 [rand]
o=20 v=-0.013902: 69.55 16.28 -34.80 10.10 -16.32 -39.31 [rand]
o=21 v=-0.013895: 103.15 -20.65 -34.80 -10.10 16.32 -39.31 [rand]
o=22 v=-0.013825: 120.58 -26.50 -26.22 -40.77 21.40 13.61 [rand]
o=23 v=-0.013754: 69.30 20.51 -26.69 -12.15 -12.44 -39.75 [rand]
o=24 v=-0.013392: 114.17 -25.40 -28.98 -3.35 33.01 -26.09 [rand]
o=25 v=-0.013374: 101.57 -38.62 -12.95 -3.49 -11.25 35.41 [rand]
o=26 v=-0.013312: 114.17 -25.40 -28.98 3.35 33.01 -26.09 [rand]
o=27 v=-0.013181: 121.96 -6.60 -39.31 -37.76 24.22 -3.86 [rand]
o=28 v=-0.013114: 94.86 -31.19 -33.81 12.98 30.57 -5.92 [rand]
o=29 v=-0.012798: 100.06 -4.87 -36.74 -41.38 19.47 -31.64 [rand]
o=30 v=-0.012757: 118.84 -29.17 -34.34 9.69 21.75 -40.77 [rand]
o=31 v=-0.012553: 107.32 -30.57 -34.69 -3.65 28.33 -14.78 [rand]
o=32 v=-0.012511: 93.02 -28.85 -29.11 -7.50 7.50 -7.50 [grid]
o=33 v=-0.012436: 120.41 -36.46 -14.38 17.98 28.73 -5.91 [rand]
o=34 v=-0.012431: 71.35 -39.13 -20.69 34.16 13.44 -39.30 [rand]
o=35 v=-0.012424: 77.61 -39.46 -10.55 -29.17 22.04 -11.69 [rand]
o=36 v=-0.012363: 59.42 25.95 35.50 -15.05 -4.71 12.78 [rand]
o=37 v=-0.012291: 113.19 10.91 -35.03 -39.07 28.37 8.01 [rand]
o=38 v=-0.012126: 77.84 -31.19 -33.81 -12.98 30.57 -5.92 [rand]
o=39 v=-0.012119: 67.61 34.21 30.61 24.90 4.74 17.23 [rand]
o=40 v=-0.012087: 101.57 -38.62 -12.95 3.49 -11.25 35.41 [rand]
o=41 v=-0.011985: 84.02 -34.55 -29.15 4.84 28.19 -20.57 [rand]
o=42 v=-0.011946: 75.05 -36.32 -8.42 -4.11 32.66 -25.74 [rand]
o=43 v=-0.011940: 100.06 0.50 -36.74 -41.38 19.47 -31.64 [rand]
o=44 v=-0.011927: 94.86 -31.19 -33.81 12.98 30.57 5.92 [rand]
o=45 v=-0.011925: 77.69 21.97 34.13 -27.41 -42.10 24.35 [rand]
o=46 v=-0.011909: 100.35 -17.72 -38.13 -14.61 39.00 28.71 [rand]
o=47 v=-0.011902: 77.84 -31.19 -33.81 -12.98 30.57 5.92 [rand]
o=48 v=-0.011821: 59.68 4.48 -29.11 30.00 7.50 -7.50 [grid]
o=49 v=-0.011782: 69.58 18.59 -31.72 12.96 -3.05 9.17 [rand]
o=50 v=-0.011599: 65.38 -30.57 -34.69 -3.65 28.33 14.78 [rand]
o=51 v=-0.011565: 77.75 -23.81 -31.15 -14.86 11.51 -26.00 [rand]
o=52 v=-0.011402: 59.68 24.48 -29.11 30.00 -30.00 -30.00 [grid]
o=53 v=-0.011368: 58.33 -6.31 29.47 18.18 -25.37 33.48 [rand]
o=54 v=-0.011360: 74.86 -18.49 -36.20 19.17 13.96 2.56 [rand]
o=55 v=-0.011347: 107.32 -30.57 -34.69 3.65 28.33 -14.78 [rand]
o=56 v=-0.011289: 79.93 31.76 -39.02 19.28 -27.13 15.13 [rand]
o=57 v=-0.011188: 111.72 -29.44 -39.95 41.23 -14.90 -41.72 [rand]
o=58 v=-0.011106: 97.84 -18.49 -36.20 -19.17 13.96 -2.56 [rand]
o=59 v=-0.011096: 77.93 -37.22 -28.92 -3.45 23.10 -5.60 [rand]
o=60 v=-0.011051: 84.06 5.00 -34.14 -10.97 -18.37 5.04 [rand]
o=61 v=-0.010952: 92.76 -36.13 34.13 19.28 -27.13 -15.13 [rand]
o=62 v=-0.010903: 76.36 -36.23 -8.96 -9.05 41.60 -20.37 [rand]
o=63 v=-0.010865: 92.17 12.01 -32.08 -38.21 15.41 -13.51 [rand]
o=64 v=-0.010816: 101.29 -26.27 -32.37 -19.13 33.53 -22.48 [rand]
o=65 v=-0.010813: 97.65 -36.32 -8.42 4.11 32.66 25.74 [rand]
o=66 v=-0.010777: 84.02 -34.55 -29.15 -4.84 28.19 -20.57 [rand]
o=67 v=-0.010646: 71.20 34.11 -18.66 -13.02 24.31 -24.93 [rand]
o=68 v=-0.010542: 113.02 4.48 -29.11 -30.00 7.50 7.50 [grid]
o=69 v=-0.010414: 70.71 6.57 -36.28 23.16 17.17 3.59 [rand]
o=70 v=-0.010383: 97.84 -18.49 -36.20 -19.17 13.96 2.56 [rand]
o=71 v=-0.010338: 97.84 14.12 -36.20 19.17 -13.96 -2.56 [rand]
o=72 v=-0.010330: 119.38 -7.20 -39.26 11.98 -4.28 -19.20 [rand]
o=73 v=-0.010327: 101.37 -33.35 -20.66 -14.81 37.49 36.90 [rand]
o=74 v=-0.010294: 72.35 13.35 33.24 -14.61 -39.00 28.71 [rand]
o=75 v=-0.010276: 65.38 -30.57 -34.69 3.65 28.33 14.78 [rand]
o=76 v=-0.010272: 79.68 -8.85 -29.11 -7.50 7.50 -7.50 [grid]
o=77 v=-0.010270: 113.02 -28.85 -29.11 -7.50 30.00 -30.00 [grid]
o=78 v=-0.010269: 71.13 -38.62 -12.95 -3.49 11.25 -35.41 [rand]
o=79 v=-0.010268: 57.33 24.96 -36.06 7.44 -27.42 -25.63 [rand]
o=80 v=-0.010263: 79.93 31.76 -39.02 -19.28 -27.13 -15.13 [rand]
o=81 v=-0.010158: 112.42 -22.63 -30.03 2.86 40.29 -21.85 [rand]
o=82 v=-0.010080: 59.42 -30.32 -40.39 -15.05 4.71 12.78 [rand]
o=83 v=-0.010032: 112.42 -22.63 -30.03 -2.86 40.29 -21.85 [rand]
o=84 v=-0.010027: 79.68 -8.85 -29.11 -7.50 7.50 -30.00 [grid]
o=85 v=-0.010016: 69.55 16.28 29.91 -10.10 -16.32 -39.31 [rand]
o=86 v=-0.010007: 113.13 -16.81 28.90 -16.09 -31.75 -34.71 [rand]
o=87 v=-0.009978: 113.19 -15.28 -35.03 -39.07 28.37 8.01 [rand]
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.163 pitch=-0.676 yaw=-0.417 dS=+0.177 dL=-0.165 dP=+0.554
++ Mean: roll=-0.093 pitch=-0.447 yaw=-0.237 dS=+0.318 dL=-0.081 dP=+0.644
++ Max : roll=-0.044 pitch=-0.168 yaw=-0.151 dS=+0.437 dL=-0.032 dP=+0.708
++ Max displacements (mm) for each sub-brick:
1.65(0.00) 1.70(0.10) 1.67(0.06) 1.72(0.09) 1.72(0.07) 1.69(0.04) 1.71(0.02) 1.76(0.06) 1.72(0.09) 1.73(0.06) 1.76(0.03) 1.75(0.03) 1.75(0.05) 1.75(0.03) 1.71(0.05) 1.74(0.02) 1.75(0.05) 1.75(0.03) 1.75(0.06) 1.76(0.04) 1.76(0.07) 1.76(0.04) 1.77(0.03) 1.77(0.04) 1.75(0.04) 1.76(0.05) 1.78(0.05) 1.80(0.06) 1.72(0.11) 1.74(0.10) 1.78(0.07) 1.78(0.04) 1.76(0.05) 1.79(0.04) 1.77(0.04) 1.75(0.05) 1.74(0.05) 1.69(0.10) 1.68(0.04) 1.73(0.08) 1.72(0.05) 1.65(0.11) 1.76(0.18) 1.72(0.10) 1.70(0.03) 1.69(0.04) 1.68(0.07) 1.65(0.05) 1.68(0.04) 1.69(0.06) 1.69(0.05) 1.63(0.07) 1.70(0.10) 1.60(0.19) 1.62(0.11) 1.57(0.08) 1.63(0.14) 1.66(0.05) 1.67(0.02) 1.64(0.04) 1.59(0.08) 1.54(0.04) 1.60(0.09) 1.62(0.07) 1.62(0.04) 1.65(0.05) 1.59(0.11) 1.57(0.08) 1.55(0.05) 1.54(0.04) 1.58(0.06) 1.62(0.05) 1.54(0.13) 1.53(0.07) 1.51(0.08) 1.48(0.03) 1.54(0.12) 1.45(0.16) 1.48(0.06) 1.47(0.04) 1.46(0.03) 1.46(0.05) 1.42(0.09) 1.39(0.05) 1.33(0.06) 1.32(0.14) 1.29(0.09) 1.28(0.03) 1.24(0.08) 1.19(0.07) 1.17(0.10) 1.13(0.10) 1.10(0.04) 1.11(0.09) 1.13(0.05) 1.06(0.10) 1.02(0.04) 1.06(0.06) 1.08(0.05) 1.03(0.08) 1.02(0.06) 1.05(0.04) 1.05(0.06) 1.11(0.07) 1.04(0.13) 1.02(0.02) 1.04(0.06) 1.06(0.09) 1.08(0.03) 1.09(0.03) 1.08(0.05) 1.09(0.05) 1.09(0.07) 1.07(0.05) 1.02(0.07) 1.07(0.12) 1.07(0.05) 1.00(0.09) 1.03(0.10) 1.05(0.08) 1.01(0.05) 1.04(0.05) 0.97(0.13) 1.06(0.15) 1.05(0.05) 1.01(0.06) 0.95(0.10) 0.98(0.07) 1.03(0.05) 1.04(0.07) 1.04(0.04) 1.03(0.06) 1.01(0.04) 0.96(0.06) 1.01(0.08) 0.99(0.08) 1.07(0.08) 1.03(0.10) 1.01(0.05) 1.03(0.04) 1.01(0.06) 0.98(0.04) 1.02(0.08) 1.06(0.06) 1.05(0.01) 1.08(0.22)
++ Max displacement in automask = 1.80 (mm) at sub-brick 27
++ Max delta displ in automask = 0.22 (mm) at sub-brick 145
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-07.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
+ - A little optimization:*[#16325=-0.0239536] *[#16330=-0.0247026] *[#16336=-0.0248057] *[#16338=-0.025367] *[#16339=-0.0254096] *[#16341=-0.0265901] *[#16345=-0.0270257] *[#16347=-0.0270448] *[#16349=-0.0274629] *[#16356=-0.0275082] *[#16360=-0.0275403] *[#16362=-0.0276122] .*[#16370=-0.109248] *[#16379=-0.113685] *[#16382=-0.117702] *[#16385=-0.124022] *[#16386=-0.126948] *[#16393=-0.128074] *[#16396=-0.128591] *[#16397=-0.128909] *[#16398=-0.128976] *[#16401=-0.129162] *[#16402=-0.129207] *[#16403=-0.129256] *[#16405=-0.129262] *[#16408=-0.129307] *[#16410=-0.129324] *[#16411=-0.12934] .......................................................................................
+ - costs of the above after a little optimization:
o= 0 v=-0.027612: 115.72 -30.91 -36.65 11.95 23.73 -28.88 [rand] [f=45]
*o= 1 v=-0.129340: 86.40 -0.89 0.80 -0.31 -0.43 -0.50 [grid] [f=45]
o= 2 v=-0.027503: 68.99 18.64 -33.60 -8.62 -14.25 -33.22 [rand] [f=64]
o= 3 v=-0.027674: 112.58 -31.84 -39.37 14.98 22.44 -37.73 [rand] [f=59]
o= 4 v=-0.024495: 88.86 -13.51 -34.36 -10.89 18.30 -4.77 [rand] [f=36]
o= 5 v=-0.020914: 117.95 -31.98 33.77 -40.44 -19.22 -39.38 [rand] [f=46]
o= 6 v=-0.026692: 115.98 -29.65 -34.97 9.19 25.99 -27.92 [grid] [f=64]
o= 7 v=-0.033633: 106.88 -22.69 -28.32 -31.23 16.78 -7.08 [rand] [f=52]
o= 8 v=-0.036713: 84.26 -16.78 -40.16 -25.47 21.21 -38.17 [rand] [f=83]
o= 9 v=-0.024640: 102.18 -36.66 -11.18 -3.95 31.69 28.67 [rand] [f=54]
o=10 v=-0.021665: 59.68 25.02 23.62 -7.27 12.04 29.99 [grid] [f=43]
o=11 v=-0.031615: 91.10 -17.27 -41.82 -27.83 24.89 -31.85 [rand] [f=59]
o=12 v=-0.019992: 114.57 -29.24 -36.36 -6.67 27.98 -22.01 [rand] [f=42]
o=13 v=-0.024761: 87.78 -11.78 -34.16 -11.23 15.72 -6.49 [rand] [f=56]
o=14 v=-0.027274: 75.76 -21.46 -37.33 -12.75 15.18 -42.60 [rand] [f=50]
o=15 v=-0.027834: 98.55 -40.98 -10.54 -7.09 39.44 23.93 [rand] [f=65]
o=16 v=-0.022594: 123.38 -9.09 -35.73 -38.71 23.89 3.80 [rand] [f=45]
o=17 v=-0.020874: 101.31 -23.30 -35.19 -17.52 25.14 -17.34 [rand] [f=64]
o=18 v=-0.020638: 69.56 19.15 -32.85 17.19 3.18 7.57 [rand] [f=60]
o=19 v=-0.028524: 108.12 -38.52 -11.15 20.35 27.09 19.02 [rand] [f=63]
o=20 v=-0.023976: 68.59 16.81 -36.15 5.32 -19.14 -41.37 [rand] [f=42]
o=21 v=-0.020394: 102.71 -19.16 -38.73 -13.38 15.56 -31.56 [rand] [f=63]
o=22 v=-0.033412: 119.34 -19.69 -30.78 -44.68 22.43 7.89 [rand] [f=77]
o=23 v=-0.031912: 67.49 21.55 -34.93 -1.53 -20.65 -44.98 [rand] [f=62]
o=24 v=-0.021709: 114.93 -25.55 -32.61 2.09 29.76 -24.74 [rand] [f=49]
o=25 v=-0.017621: 102.01 -42.18 -13.16 -3.51 -11.73 35.15 [rand] [f=43]
o=26 v=-0.024775: 116.22 -29.72 -28.27 6.08 32.68 -25.56 [rand] [f=55]
o=27 v=-0.022633: 122.55 -10.94 -35.38 -42.82 27.34 6.50 [rand] [f=60]
o=28 v=-0.016382: 91.16 -31.39 -33.92 13.07 30.55 -5.88 [rand] [f=41]
o=29 v=-0.021693: 100.26 -3.26 -37.88 -40.67 19.39 -24.98 [rand] [f=48]
o=30 v=-0.028019: 115.96 -32.23 -36.69 15.19 25.18 -36.19 [rand] [f=47]
o=31 v=-0.021781: 111.84 -28.29 -36.09 -5.16 32.32 -16.00 [rand] [f=38]
o=32 v=-0.023904: 91.59 -22.73 -31.26 -8.73 4.99 -8.95 [grid] [f=56]
o=33 v=-0.015583: 122.19 -35.65 -12.09 17.36 28.14 -7.30 [rand] [f=45]
o=34 v=-0.027691: 70.32 -39.77 -20.09 35.84 2.73 -44.52 [rand] [f=70]
o=35 v=-0.021676: 77.75 -38.91 -3.17 -28.28 38.60 -8.11 [rand] [f=43]
o=36 v=-0.015398: 59.80 25.84 36.28 -14.47 -4.21 12.22 [rand] [f=43]
o=37 v=-0.018571: 111.45 7.83 -35.32 -39.87 25.81 8.98 [rand] [f=55]
o=38 v=-0.017765: 75.71 -31.70 -32.19 -8.10 31.11 -5.51 [rand] [f=63]
o=39 v=-0.020276: 65.72 35.03 31.62 26.37 14.05 18.40 [rand] [f=40]
o=40 v=-0.017208: 100.82 -42.18 -13.24 -0.96 -12.81 33.60 [rand] [f=58]
o=41 v=-0.020104: 76.97 -35.45 -31.98 2.63 32.02 -22.43 [rand] [f=55]
o=42 v=-0.016634: 71.72 -35.87 -6.83 -4.14 32.99 -25.94 [rand] [f=53]
o=43 v=-0.022202: 100.05 -0.51 -37.94 -41.09 19.23 -24.03 [rand] [f=69]
o=44 v=-0.015991: 94.86 -32.03 -34.69 7.63 29.67 3.28 [rand] [f=59]
o=45 v=-0.014612: 77.68 22.20 33.89 -23.01 -42.84 24.17 [rand] [f=36]
o=46 v=-0.012891: 100.60 -16.97 -38.25 -15.42 39.07 29.28 [rand] [f=57]
o=47 v=-0.016930: 81.48 -32.60 -33.98 -14.57 28.15 4.66 [rand] [f=48]
o=48 v=-0.017892: 61.12 7.99 -30.43 29.69 6.61 -7.22 [grid] [f=37]
o=49 v=-0.019772: 69.90 17.02 -34.20 14.97 3.00 7.51 [rand] [f=53]
o=50 v=-0.024966: 61.99 -30.12 -38.32 -3.87 29.86 12.53 [rand] [f=67]
o=51 v=-0.027698: 78.48 -18.75 -33.20 -15.20 15.69 -30.81 [rand] [f=43]
o=52 v=-0.019910: 60.79 25.98 -33.60 23.53 -29.62 -31.20 [grid] [f=55]
o=53 v=-0.014564: 58.50 -5.69 28.97 17.73 -18.17 34.69 [rand] [f=45]
o=54 v=-0.018199: 74.19 -18.49 -36.66 20.74 13.58 8.10 [rand] [f=59]
o=55 v=-0.025217: 117.51 -34.12 -34.94 1.00 28.98 -16.55 [rand] [f=51]
o=56 v=-0.029445: 82.69 24.58 -39.23 16.80 -18.72 17.86 [rand] [f=64]
o=57 v=-0.025320: 114.58 -26.93 -42.42 42.41 -8.13 -42.78 [rand] [f=58]
o=58 v=-0.029655: 99.02 -22.39 -29.89 -21.05 11.96 -8.12 [rand] [f=55]
o=59 v=-0.019903: 79.59 -38.38 -30.76 -0.89 32.77 -8.00 [rand] [f=68]
o=60 v=-0.034932: 90.75 11.11 -42.42 -12.80 -14.47 -1.31 [rand] [f=71]
o=61 v=-0.015988: 92.30 -35.91 37.55 17.65 -26.24 -15.48 [rand] [f=44]
o=62 v=-0.021411: 75.47 -36.21 -4.20 -15.85 39.35 -19.11 [rand] [f=52]
o=63 v=-0.024890: 95.58 11.57 -35.22 -31.70 16.00 -8.72 [rand] [f=56]
o=64 v=-0.022840: 102.79 -24.34 -32.59 -24.71 26.18 -12.18 [rand] [f=76]
o=65 v=-0.026416: 102.36 -36.01 -11.09 2.31 30.66 23.33 [rand] [f=43]
o=66 v=-0.020661: 80.21 -35.35 -31.42 0.21 32.86 -17.45 [rand] [f=64]
o=67 v=-0.014511: 71.14 37.79 -17.69 -13.02 23.96 -23.96 [rand] [f=46]
o=68 v=-0.021508: 110.22 6.01 -32.89 -33.52 8.83 0.87 [grid] [f=55]
o=69 v=-0.028469: 66.58 4.48 -37.75 22.84 6.82 8.44 [rand] [f=72]
o=70 v=-0.018983: 91.45 -18.24 -38.57 -18.02 12.46 2.51 [rand] [f=38]
o=71 v=-0.029058: 91.48 13.60 -39.39 17.89 -10.23 -1.45 [rand] [f=52]
o=72 v=-0.018012: 122.04 -7.30 -36.15 13.24 -8.07 -23.76 [rand] [f=72]
o=73 v=-0.031108: 99.55 -38.49 -13.14 -11.36 39.67 29.07 [rand] [f=59]
o=74 v=-0.013788: 72.33 17.24 33.82 -14.27 -38.87 28.96 [rand] [f=35]
o=75 v=-0.020479: 61.65 -29.49 -37.93 0.52 28.98 11.73 [rand] [f=83]
o=76 v=-0.028228: 83.90 -6.08 -38.64 -9.25 5.29 -7.06 [grid] [f=52]
o=77 v=-0.019696: 113.67 -29.94 -33.82 -0.80 29.49 -25.51 [grid] [f=38]
o=78 v=-0.016833: 70.54 -36.19 -10.53 -3.88 9.94 -34.18 [rand] [f=61]
o=79 v=-0.018436: 59.71 24.49 -38.90 8.48 -27.62 -28.03 [rand] [f=40]
o=80 v=-0.029277: 81.75 26.75 -41.99 -9.89 -25.58 -13.00 [rand] [f=53]
o=81 v=-0.020335: 116.51 -25.34 -32.08 -1.16 35.90 -16.17 [rand] [f=72]
o=82 v=-0.016846: 61.71 -29.02 -41.43 -14.66 12.00 13.02 [rand] [f=61]
o=83 v=-0.024442: 116.43 -29.76 -26.61 5.00 34.63 -22.29 [rand] [f=68]
o=84 v=-0.022908: 77.83 -11.37 -37.13 -6.05 5.36 -14.94 [grid] [f=67]
o=85 v=-0.013851: 69.55 16.89 33.37 -9.95 -16.13 -40.27 [rand] [f=44]
o=86 v=-0.017656: 113.05 -17.16 33.43 -17.32 -31.77 -37.18 [rand] [f=55]
o=87 v=-0.035332: 114.98 -19.61 -28.94 -43.72 20.01 0.05 [rand] [f=60]
+ - saving # 1 for use with twobest
+ - saving # 8 for use with twobest
+ - saving #87 for use with twobest
+ - saving #60 for use with twobest
+ - saving # 7 for use with twobest
+ - saving #22 for use with twobest
+ - saving #23 for use with twobest
+ - saving #11 for use with twobest
+ - saving #73 for use with twobest
+ - saving #58 for use with twobest
+ - saving #56 for use with twobest
+ - saving #80 for use with twobest
+ - saving #71 for use with twobest
+ - saving #19 for use with twobest
+ - saving #69 for use with twobest
+ - saving #76 for use with twobest
+ - saving #30 for use with twobest
+ - saving #15 for use with twobest
+ - saving #51 for use with twobest
+ - saving #34 for use with twobest
+ - skip # 3 for twobest: too close to set #30
+ - saving # 0 for use with twobest
+ - saving # 2 for use with twobest
+ - saving #14 for use with twobest
+ - skip # 6 for twobest: too close to set # 0
+ - saving #65 for use with twobest
+ - saving #57 for use with twobest
+ - saving #55 for use with twobest
+ - saving #50 for use with twobest
+ - saving #63 for use with twobest
+ - saving #26 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=36 usiz=142
+ - retaining old weight image
+ - using 37432 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30431 total points stored in 60 'TOHD(17.3748)' bloks (0 duplicates)
*[#21223=-0.194947] *[#21249=-0.196785] *[#21252=-0.197421] *[#21253=-0.198285] *[#21254=-0.199191] *[#21256=-0.200021] *[#21257=-0.200971] *[#21258=-0.201931] *[#21259=-0.202325] *[#21262=-0.202651] *[#21263=-0.20282] *[#21266=-0.203201] *[#21267=-0.204566] *[#21268=-0.205105] *[#21277=-0.205198] *[#21281=-0.205891] *[#21282=-0.205894] *[#21283=-0.206076] *[#21286=-0.206186] *[#21290=-0.206235] *[#21293=-0.206576] *[#21298=-0.206725] *[#21303=-0.207025] *[#21304=-0.207172] *[#21305=-0.207426] *[#21310=-0.207573] *[#21311=-0.207616] *[#21316=-0.207708] *[#21317=-0.207818] *[#21318=-0.208025] *[#21319=-0.208094]
+ - param set #1 has cost=-0.208094 [o=1 t=0]
+ -- Parameters = 86.0974 -1.4271 0.7463 0.4440 -1.0596 -0.9170 0.9916 1.0247 0.9764 0.0137 0.0041 -0.0124
+ - param set #2 has cost=-0.027583 [o=8 t=1]
+ -- Parameters = 84.2865 -16.7371 -40.1155 -25.4177 21.1804 -38.2227 1.0012 0.9982 1.0031 0.0124 -0.0007 -0.0011
+ - param set #3 has cost=-0.025993 [o=87 t=2]
+ -- Parameters = 114.6941 -20.1788 -28.7208 -43.0894 19.6971 -0.5094 0.9997 0.9995 0.9939 -0.0005 -0.0003 0.0018
+ - param set #4 has cost=-0.029904 [o=60 t=3]
+ -- Parameters = 86.5309 10.7203 -42.4428 -12.6185 -14.1260 -2.0429 1.0078 0.9950 0.9980 0.0015 0.0003 0.0030
+ - param set #5 has cost=-0.024730 [o=7 t=4]
+ -- Parameters = 106.7865 -22.6553 -28.5218 -31.3768 16.7018 -6.8846 1.0075 1.0023 0.9899 0.0000 -0.0003 0.0026
+ - param set #6 has cost=-0.022784 [o=22 t=5]
+ -- Parameters = 118.7420 -19.7074 -30.4598 -44.6166 22.8511 8.0528 1.0013 1.0020 1.0060 0.0024 0.0167 0.0035
+ - param set #7 has cost=-0.031003 [o=23 t=6]
+ -- Parameters = 66.9436 20.8546 -35.4103 -2.2891 -24.0116 -43.6610 0.9867 0.9886 0.9630 0.0004 0.0049 -0.0095
+ - param set #8 has cost=-0.023679 [o=11 t=7]
+ -- Parameters = 91.0545 -17.2840 -41.7698 -27.7338 24.7472 -31.7441 1.0028 0.9983 1.0064 0.0009 -0.0014 0.0004
+ - param set #9 has cost=-0.022274 [o=73 t=8]
+ -- Parameters = 99.3132 -38.3052 -12.9901 -10.8460 40.0972 28.9157 0.9961 0.9949 0.9965 0.0011 0.0008 -0.0009
+ - param set #10 has cost=-0.019803 [o=58 t=9]
+ -- Parameters = 98.2286 -22.1561 -30.0723 -20.2934 12.1981 -8.4463 1.0412 0.9962 1.0029 0.0007 0.0002 0.0011
+ - param set #11 has cost=-0.021323 [o=56 t=10]
+ -- Parameters = 82.3435 24.6173 -39.0773 17.0356 -18.3470 18.1181 0.9923 1.0051 1.0012 -0.0003 -0.0011 -0.0009
+ - param set #12 has cost=-0.024291 [o=80 t=11]
+ -- Parameters = 82.9880 24.1506 -42.4283 -8.9972 -25.1699 -14.2877 1.0017 1.0071 0.9805 0.0005 0.0022 -0.0041
+ - param set #13 has cost=-0.023520 [o=71 t=12]
+ -- Parameters = 91.7086 13.7855 -38.9879 13.2854 -8.6294 -1.3706 1.0073 1.0012 1.0025 0.0010 -0.0011 -0.0016
+ - param set #14 has cost=-0.021310 [o=19 t=13]
+ -- Parameters = 107.2910 -39.7042 -10.5091 20.5632 25.7603 17.2052 0.9967 0.9990 0.9927 0.0001 -0.0007 0.0008
+ - param set #15 has cost=-0.021981 [o=69 t=14]
+ -- Parameters = 66.4457 4.3631 -37.7182 22.9008 6.5885 8.5107 1.0006 0.9977 1.0048 -0.0008 -0.0006 -0.0010
+ - param set #16 has cost=-0.023824 [o=76 t=15]
+ -- Parameters = 83.3261 -6.0724 -38.9034 -8.5154 4.8785 -6.7462 1.0081 1.0375 1.0088 -0.0003 -0.0006 0.0004
+ - param set #17 has cost=-0.020384 [o=30 t=16]
+ -- Parameters = 116.0543 -32.3977 -36.6748 15.0832 25.1260 -36.2094 0.9977 1.0041 1.0042 -0.0015 0.0117 -0.0008
+ - param set #18 has cost=-0.021054 [o=15 t=17]
+ -- Parameters = 98.6498 -41.2052 -11.2930 -6.7847 39.1224 24.5710 0.9935 1.0156 1.0021 -0.0019 -0.0017 -0.0004
+ - param set #19 has cost=-0.024323 [o=51 t=18]
+ -- Parameters = 77.8843 -18.7174 -33.2491 -14.4825 15.9178 -31.2811 0.9955 1.0070 1.0002 0.0128 -0.0002 0.0002
+ - param set #20 has cost=-0.022527 [o=34 t=19]
+ -- Parameters = 70.0142 -39.3668 -19.4822 35.0448 2.3799 -44.9947 0.9959 0.9987 1.0078 0.0002 -0.0010 0.0108
+ - param set #21 has cost=-0.018662 [o=0 t=20]
+ -- Parameters = 115.7372 -31.0853 -36.5407 11.8368 23.5351 -28.7772 0.9996 0.9999 0.9970 -0.0008 0.0002 0.0005
+ - param set #22 has cost=-0.026780 [o=2 t=21]
+ -- Parameters = 67.4843 18.5901 -33.6098 -7.7480 -16.0887 -37.5992 0.9594 0.9861 0.9741 0.0058 0.0229 0.0015
+ - param set #23 has cost=-0.019831 [o=14 t=22]
+ -- Parameters = 75.7949 -21.4387 -37.4471 -12.5993 15.0908 -42.5064 0.9974 0.9983 1.0000 -0.0006 0.0010 -0.0001
+ - param set #24 has cost=-0.018888 [o=65 t=23]
+ -- Parameters = 102.4525 -36.4282 -10.9836 2.5523 30.7193 23.2301 0.9997 0.9908 1.0034 -0.0016 0.0132 0.0023
+ - param set #25 has cost=-0.018087 [o=57 t=24]
+ -- Parameters = 114.4980 -26.7962 -42.4363 42.3424 -8.1039 -42.7825 0.9988 1.0003 1.0009 -0.0003 0.0002 0.0129
++ Output dataset ./rm.epi.all1+orig.BRIK
+ - param set #26 has cost=-0.018057 [o=55 t=25]
+ -- Parameters = 117.4109 -34.1078 -34.8488 0.9823 28.9112 -16.4005 1.0027 0.9998 1.0044 -0.0004 0.0004 -0.0007
cat_matvec -ONELINE sub-07_T1w_ns+tlrc::WARP_DATA -I sub-07_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
+ - param set #27 has cost=-0.020836 [o=50 t=26]
+ -- Parameters = 62.0150 -30.0517 -38.3462 -3.8282 29.8585 12.6340 1.0073 0.9950 0.9926 -0.0006 -0.0015 0.0003
+ - param set #28 has cost=-0.019960 [o=63 t=27]
+ -- Parameters = 94.9997 11.6160 -35.8609 -31.8556 15.6896 -8.9586 0.9919 1.0116 0.9855 0.0006 -0.0027 -0.0000
+ - param set #29 has cost=-0.018014 [o=26 t=28]
+ -- Parameters = 116.2325 -29.6665 -28.2147 6.3837 32.9971 -25.8429 0.9983 1.0029 0.9986 0.0001 -0.0012 0.0122
+ - param set #30 has cost=-0.207262 [o=-1 t=-1]
+ -- Parameters = 86.2545 -1.1303 0.6993 -0.0340 -0.4104 -0.4107 0.9859 1.0123 0.9761 -0.0048 -0.0063 0.0018
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0829
+ --- dist(#3,#1) = 0.475
+ --- dist(#4,#1) = 0.54
+ --- dist(#5,#1) = 0.511
+ --- dist(#6,#1) = 0.429
+ --- dist(#7,#1) = 0.484
+ --- dist(#8,#1) = 0.366
+ --- dist(#9,#1) = 0.425
+ --- dist(#10,#1) = 0.54
+ --- dist(#11,#1) = 0.496
+ --- dist(#12,#1) = 0.531
+ --- dist(#13,#1) = 0.497
+ --- dist(#14,#1) = 0.501
+ --- dist(#15,#1) = 0.49
+ --- dist(#16,#1) = 0.461
+ --- dist(#17,#1) = 0.481
+ --- dist(#18,#1) = 0.498
+ --- dist(#19,#1) = 0.478
3dAllineate -base sub-07_T1w_ns+tlrc -input pb00.sub-07.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
+ --- dist(#20,#1) = 0.497
+ --- dist(#21,#1) = 0.489
+ --- dist(#22,#1) = 0.468
+ --- dist(#23,#1) = 0.458
+ --- dist(#24,#1) = 0.477
+ --- dist(#25,#1) = 0.385
+ --- dist(#26,#1) = 0.438
+ --- dist(#27,#1) = 0.466
+ --- dist(#28,#1) = 0.54
+ --- dist(#29,#1) = 0.445
+ --- dist(#30,#1) = 0.378
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-07.r01.tcat+orig.HEAD
++ Base dataset: ./sub-07_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
+ !source mask fill: ubot=36 usiz=142
+ - retaining old weight image
+ - using 37432 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30725 total points stored in 62 'TOHD(17.2644)' bloks (0 duplicates)
*[#24096=-0.228962] *[#24124=-0.22929] *[#24129=-0.229334] *[#24130=-0.229457] *[#24133=-0.229502] *[#24135=-0.229609] *[#24140=-0.229743] *[#24141=-0.229781] *[#24144=-0.230102] *[#24151=-0.230287] *[#24158=-0.230347] *[#24159=-0.230433] *[#24162=-0.230463] *[#24163=-0.230506] *[#24170=-0.230573] *[#24171=-0.2306] *[#24174=-0.230647] *[#24180=-0.230655] *[#24181=-0.230678] *[#24185=-0.230715] *[#24186=-0.230737] *[#24187=-0.23078] *[#24188=-0.230802] *[#24189=-0.230831] *[#24190=-0.230832] *[#24191=-0.230838] *[#24193=-0.230845] *[#24195=-0.230893] *[#24196=-0.230907] *[#24197=-0.230936] *[#24198=-0.230942] *[#24199=-0.230942] *[#24201=-0.230982] *[#24202=-0.230991] *[#24203=-0.231036] *[#24204=-0.231077] *[#24205=-0.231095]
+ - param set #1 has cost=-0.231095 [o=1 t=0]
+ -- Parameters = 86.0700 -1.2335 0.8450 0.4957 -1.1754 -0.5761 0.9922 1.0182 0.9796 0.0133 0.0003 -0.0143
+ - param set #2 has cost=-0.230749 [o=-1 t=-1]
+ -- Parameters = 86.2236 -1.1868 0.7687 -0.0206 -0.8842 -0.7609 0.9915 1.0157 0.9748 -0.0068 0.0018 -0.0067
+ - param set #3 has cost=-0.028419 [o=23 t=6]
+ -- Parameters = 66.9933 20.5409 -35.4528 -2.4788 -23.8461 -44.1597 0.9690 0.9857 0.9472 -0.0000 0.0006 -0.0103
+ - param set #4 has cost=-0.024841 [o=60 t=3]
+ -- Parameters = 86.4669 10.7627 -42.4431 -12.6620 -14.1498 -1.8926 1.0090 0.9945 0.9977 0.0012 -0.0014 0.0037
+ - param set #5 has cost=-0.020444 [o=8 t=1]
+ -- Parameters = 84.3134 -16.7680 -40.1309 -25.3674 21.1341 -38.4043 1.0017 0.9985 1.0080 0.0109 0.0023 -0.0009
+ - param set #6 has cost=-0.023049 [o=2 t=21]
+ -- Parameters = 67.3156 18.4444 -33.7079 -7.6025 -16.3422 -37.7865 0.9577 0.9837 0.9729 0.0064 0.0236 0.0014
+ - param set #7 has cost=-0.019282 [o=87 t=2]
+ -- Parameters = 114.7373 -20.3098 -28.7563 -43.0807 19.8587 -0.4469 1.0002 0.9978 0.9904 -0.0001 -0.0005 0.0017
+ - param set #8 has cost=-0.020016 [o=7 t=4]
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
+ -- Parameters = 106.0361 -23.0184 -29.2041 -31.4828 15.9732 -6.8857 1.0076 1.0044 0.9789 0.0099 -0.0028 0.0058
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 1.466 -6.559 -27.812
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.5% 9.5% 48.1%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
+ - param set #9 has cost=-0.020316 [o=51 t=18]
+ -- Parameters = 77.6478 -18.7165 -33.4061 -14.3080 15.6489 -31.1904 0.9911 1.0056 1.0003 0.0211 -0.0002 0.0014
+ - param set #10 has cost=-0.020443 [o=80 t=11]
+ -- Parameters = 83.1562 24.2774 -42.3526 -9.0699 -24.9608 -14.2509 1.0003 1.0065 0.9772 -0.0000 0.0027 -0.0058
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
+ - param set #11 has cost=-0.016891 [o=76 t=15]
+ -- Parameters = 83.1771 -5.9624 -38.8901 -8.3907 4.9544 -6.7313 1.0085 1.0372 1.0066 -0.0006 -0.0010 0.0007
+ - param set #12 has cost=-0.018266 [o=11 t=7]
+ -- Parameters = 91.0011 -17.3563 -41.4749 -27.7847 24.7022 -31.7281 1.0039 1.0004 1.0119 0.0015 -0.0017 0.0005
+ - param set #13 has cost=-0.021927 [o=71 t=12]
+ -- Parameters = 92.5349 13.5045 -39.9597 11.7660 -7.8838 -1.3090 1.0146 1.0089 0.9983 -0.0016 -0.0057 -0.0033
+ - param set #14 has cost=-0.016670 [o=22 t=5]
+ -- Parameters = 118.3535 -20.0463 -30.9244 -44.9849 23.3157 8.3330 1.0034 1.0027 1.0075 0.0036 0.0273 0.0047
+ - param set #15 has cost=-0.019429 [o=34 t=19]
+ -- Parameters = 70.0154 -39.3816 -19.4905 34.9740 2.4098 -44.9954 0.9955 0.9991 1.0085 0.0002 -0.0010 0.0117
+ - param set #16 has cost=-0.016082 [o=73 t=8]
+ -- Parameters = 99.1694 -38.4283 -12.8484 -10.8532 40.0486 28.7928 0.9970 0.9927 0.9915 0.0012 0.0092 -0.0011
+ - param set #17 has cost=-0.016576 [o=69 t=14]
+ -- Parameters = 66.3527 4.0542 -37.5306 22.7745 6.2604 8.5442 1.0039 0.9929 1.0106 0.0017 -0.0024 -0.0026
+ - param set #18 has cost=-0.017206 [o=56 t=10]
+ -- Parameters = 82.1946 24.6323 -38.7027 17.0170 -18.0370 18.1772 0.9971 1.0058 1.0063 0.0009 -0.0034 -0.0019
+ - param set #19 has cost=-0.018305 [o=19 t=13]
+ -- Parameters = 107.1633 -39.8516 -10.5419 20.5047 25.6369 17.0579 0.9986 0.9944 0.9840 0.0004 0.0007 -0.0004
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
+ - param set #20 has cost=-0.015831 [o=15 t=17]
+ -- Parameters = 99.0127 -41.0706 -11.5525 -6.6728 39.1688 24.3797 0.9890 1.0179 0.9931 -0.0025 -0.0013 -0.0015
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 6.4
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
+ - param set #21 has cost=-0.015800 [o=50 t=26]
+ -- Parameters = 61.9574 -29.9676 -38.3838 -3.7368 29.9211 12.5449 1.0077 0.9975 0.9905 -0.0011 -0.0030 -0.0011
+ - param set #22 has cost=-0.014911 [o=30 t=16]
+ -- Parameters = 116.0779 -32.4184 -36.6600 14.8585 25.0837 -36.0714 0.9976 1.0041 1.0061 -0.0014 0.0201 -0.0009
+ - param set #23 has cost=-0.017974 [o=63 t=27]
+ -- Parameters = 94.7607 11.6467 -38.9750 -31.1138 14.8155 -8.8414 0.9949 1.0127 0.9777 0.0011 -0.0008 0.0014
+ - param set #24 has cost=-0.017025 [o=14 t=22]
+ -- Parameters = 75.2796 -21.0521 -37.4261 -12.3184 15.8740 -41.7684 0.9724 0.9908 1.0050 -0.0013 -0.0056 0.0168
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
+ - param set #25 has cost=-0.014484 [o=58 t=9]
+ -- Parameters = 98.1076 -22.1853 -30.0608 -20.3083 12.1856 -8.3296 1.0443 0.9954 1.0016 0.0001 -0.0012 0.0013
+ - param set #26 has cost=-0.013213 [o=65 t=23]
+ -- Parameters = 102.3703 -36.5348 -11.0112 2.2023 30.3916 23.4165 1.0026 0.9839 1.0088 -0.0020 0.0125 0.0030
+ - param set #27 has cost=-0.016172 [o=0 t=20]
+ -- Parameters = 115.0968 -31.5095 -39.7590 11.7169 22.6645 -28.1755 0.9980 0.9978 0.9932 -0.0001 0.0005 0.0015
+ - param set #28 has cost=-0.016392 [o=57 t=24]
+ -- Parameters = 114.8846 -26.6019 -42.4361 41.5813 -9.7031 -41.0852 0.9590 0.9617 0.9814 0.0032 0.0004 0.0169
+ - param set #29 has cost=-0.012843 [o=55 t=25]
+ -- Parameters = 117.2955 -34.0762 -34.8397 1.0902 28.9624 -16.4351 1.0062 1.0012 1.0097 -0.0008 0.0012 0.0067
+ - param set #30 has cost=-0.012906 [o=26 t=28]
+ -- Parameters = 116.1713 -29.6973 -28.1432 6.3785 32.8639 -25.9784 0.9968 1.0030 0.9991 -0.0001 -0.0017 0.0113
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0904
+ --- dist(#3,#1) = 0.484
+ --- dist(#4,#1) = 0.541
+ --- dist(#5,#1) = 0.432
+ --- dist(#6,#1) = 0.51
+ --- dist(#7,#1) = 0.512
+ --- dist(#8,#1) = 0.54
+ --- dist(#9,#1) = 0.428
+ --- dist(#10,#1) = 0.376
+ --- dist(#11,#1) = 0.494
+ --- dist(#12,#1) = 0.484
+ --- dist(#13,#1) = 0.483
+ --- dist(#14,#1) = 0.529
+ --- dist(#15,#1) = 0.498
+ --- dist(#16,#1) = 0.494
+ --- dist(#17,#1) = 0.478
+ --- dist(#18,#1) = 0.497
+ --- dist(#19,#1) = 0.505
+ --- dist(#20,#1) = 0.48
+ --- dist(#21,#1) = 0.541
+ --- dist(#22,#1) = 0.508
+ --- dist(#23,#1) = 0.465
+ --- dist(#24,#1) = 0.498
+ --- dist(#25,#1) = 0.49
+ --- dist(#26,#1) = 0.469
+ --- dist(#27,#1) = 0.386
+ --- dist(#28,#1) = 0.441
+ --- dist(#29,#1) = 0.378
+ --- dist(#30,#1) = 0.446
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.280 -18.654 -32.063
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.2% 26.9% 55.5%
+ !source mask fill: ubot=36 usiz=142
+ - retaining old weight image
+ - using 37432 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 30797 total points stored in 63 'TOHD(17.1973)' bloks (0 duplicates)
*[#27275=-0.243688] *[#27296=-0.244261] *[#27297=-0.244332] *[#27298=-0.244436] *[#27299=-0.244842] *[#27300=-0.244969] *[#27301=-0.245172] *[#27302=-0.245532] *[#27305=-0.245579] *[#27306=-0.245836] *[#27313=-0.246105] *[#27316=-0.246269] *[#27321=-0.246324] *[#27322=-0.246483] *[#27323=-0.246649] *[#27324=-0.24667] *[#27326=-0.246701] *[#27327=-0.246717] *[#27329=-0.24678] *[#27332=-0.246849] *[#27333=-0.246931] *[#27334=-0.246996] *[#27337=-0.24707] *[#27340=-0.247076] *[#27342=-0.247304] *[#27345=-0.247362] *[#27346=-0.247414] *[#27347=-0.247436] *[#27348=-0.247549] *[#27357=-0.247577] *[#27361=-0.247673] *[#27362=-0.247921] *[#27363=-0.247936] *[#27365=-0.248049] *[#27366=-0.248064] *[#27367=-0.248077] *[#27370=-0.248113] *[#27371=-0.248176] *[#27380=-0.248226] *[#27381=-0.248256] *[#27384=-0.248261] *[#27387=-0.248263] *[#27388=-0.248285] *[#27389=-0.248309] *[#27394=-0.248384]
+ - param set #1 has cost=-0.248384 [o=1 t=0]
+ -- Parameters = 86.1944 -1.0930 0.8868 0.1834 -1.6496 -0.7057 0.9921 1.0138 0.9761 -0.0028 0.0003 -0.0263
+ - param set #2 has cost=-0.248292 [o=-1 t=-1]
+ -- Parameters = 86.3279 -1.0994 0.8727 -0.1092 -1.5791 -0.2743 0.9912 1.0144 0.9770 -0.0125 -0.0087 -0.0244
+ - param set #3 has cost=-0.023256 [o=23 t=6]
+ -- Parameters = 67.0415 20.6003 -35.4082 -2.5354 -23.6785 -44.2066 0.9683 0.9857 0.9480 -0.0004 0.0002 -0.0097
+ - param set #4 has cost=-0.024001 [o=60 t=3]
+ -- Parameters = 85.8306 14.5315 -42.4387 -12.2868 -14.0112 -2.0217 1.0096 1.0038 1.0005 0.0033 -0.0014 0.0041
+ - param set #5 has cost=-0.019012 [o=2 t=21]
+ -- Parameters = 67.3290 18.4963 -33.7193 -7.6211 -16.3778 -37.8008 0.9602 0.9845 0.9727 0.0075 0.0229 0.0012
+ - param set #6 has cost=-0.018337 [o=71 t=12]
+ -- Parameters = 92.2449 13.4816 -40.0132 11.7825 -7.8925 -1.2794 1.0211 1.0100 0.9919 -0.0019 -0.0059 -0.0049
+ - param set #7 has cost=-0.019816 [o=8 t=1]
+ -- Parameters = 85.8774 -16.2607 -39.8684 -25.0739 20.9043 -38.5525 1.0084 1.0086 1.0296 0.0107 0.0030 -0.0020
+ - param set #8 has cost=-0.016829 [o=80 t=11]
+ -- Parameters = 83.0425 24.2655 -42.4408 -9.0660 -25.1603 -14.0028 1.0007 1.0000 0.9751 0.0004 0.0022 -0.0064
+ - param set #9 has cost=-0.016849 [o=51 t=18]
+ -- Parameters = 77.6930 -18.7582 -33.3354 -14.2854 15.5151 -31.2591 0.9938 1.0049 1.0008 0.0218 -0.0004 0.0015
+ - param set #10 has cost=-0.017023 [o=7 t=4]
+ -- Parameters = 106.3127 -23.2953 -29.3893 -31.6195 16.0869 -7.0148 1.0290 1.0027 0.9790 0.0099 -0.0038 0.0061
+ - param set #11 has cost=-0.016084 [o=34 t=19]
+ -- Parameters = 70.0319 -39.4338 -19.4792 34.9632 2.3668 -44.9996 0.9972 0.9984 1.0088 -0.0001 -0.0009 0.0118
+ - param set #12 has cost=-0.015457 [o=87 t=2]
+ -- Parameters = 114.9813 -20.3796 -28.7550 -43.0773 19.9714 -0.4166 1.0045 0.9960 0.9916 -0.0008 -0.0013 0.0011
+ - param set #13 has cost=-0.014745 [o=19 t=13]
+ -- Parameters = 107.2671 -40.0041 -10.0364 21.1657 25.4499 16.7391 0.9961 0.9912 0.9785 0.0011 0.0007 0.0003
+ - param set #14 has cost=-0.015368 [o=11 t=7]
+ -- Parameters = 90.8589 -17.3500 -41.1585 -28.1247 24.0824 -31.3804 1.0106 1.0045 1.0359 0.0068 -0.0054 0.0012
+ - param set #15 has cost=-0.016696 [o=63 t=27]
+ -- Parameters = 94.1629 11.6896 -39.6182 -30.8646 14.7365 -8.4448 0.9924 1.0084 0.9638 0.0029 -0.0066 0.0022
+ - param set #16 has cost=-0.016452 [o=56 t=10]
+ -- Parameters = 79.8421 23.2739 -38.6888 17.1000 -17.6298 18.7518 0.9977 1.0237 1.0118 0.0049 -0.0072 -0.0039
+ - param set #17 has cost=-0.014630 [o=14 t=22]
+ -- Parameters = 75.0905 -21.4305 -37.4649 -12.3673 15.2064 -41.6188 0.9687 0.9731 1.0018 -0.0017 -0.0047 0.0170
+ - param set #18 has cost=-0.014288 [o=76 t=15]
+ -- Parameters = 82.8456 -5.6638 -39.0002 -8.3674 4.7023 -5.7031 1.0142 1.0387 0.9956 -0.0027 0.0002 0.0037
+ - param set #19 has cost=-0.013132 [o=22 t=5]
+ -- Parameters = 118.3713 -20.0176 -30.9282 -44.9530 23.2519 8.2996 1.0055 1.0014 1.0074 0.0035 0.0266 0.0048
+ - param set #20 has cost=-0.015901 [o=69 t=14]
+ -- Parameters = 65.7127 1.7019 -38.2841 22.5871 6.0819 9.3209 0.9999 0.9870 1.0086 -0.0016 -0.0053 -0.0020
+ - param set #21 has cost=-0.013806 [o=57 t=24]
+ -- Parameters = 115.0151 -26.6841 -42.4377 41.9844 -9.8112 -40.9910 0.9598 0.9422 0.9736 0.0025 0.0004 0.0178
+ - param set #22 has cost=-0.014080 [o=0 t=20]
+ -- Parameters = 115.1288 -31.4611 -39.7860 11.6523 22.7690 -28.0410 0.9981 1.0095 0.9909 0.0000 0.0006 0.0018
+ - param set #23 has cost=-0.014095 [o=73 t=8]
+ -- Parameters = 98.9391 -38.2663 -12.7341 -10.5526 39.9784 28.6960 1.0010 0.9828 0.9898 -0.0008 0.0086 -0.0004
+ - param set #24 has cost=-0.012095 [o=15 t=17]
+ -- Parameters = 98.9339 -41.2234 -11.5339 -6.7721 39.1075 24.5054 0.9894 1.0150 0.9948 -0.0029 -0.0022 -0.0034
+ - param set #25 has cost=-0.012551 [o=50 t=26]
+ -- Parameters = 61.9433 -29.9463 -38.4075 -3.7593 29.9047 12.4886 1.0047 0.9979 0.9901 -0.0012 -0.0033 -0.0015
+ - param set #26 has cost=-0.012899 [o=30 t=16]
+ -- Parameters = 116.0172 -32.4761 -36.7416 14.8334 25.1096 -36.0573 0.9962 1.0037 1.0064 -0.0014 0.0201 -0.0009
+ - param set #27 has cost=-0.012616 [o=58 t=9]
+ -- Parameters = 97.3908 -21.9443 -31.3257 -20.8354 13.3939 -6.5179 1.0367 0.9989 0.9646 -0.0015 -0.0009 0.0003
+ - param set #28 has cost=-0.010784 [o=65 t=23]
+ -- Parameters = 102.3612 -36.7618 -10.7898 2.3767 30.0769 23.1473 1.0046 0.9604 1.0111 -0.0020 0.0115 0.0031
+ - param set #29 has cost=-0.009521 [o=26 t=28]
+ -- Parameters = 116.3879 -29.7252 -28.2347 6.3938 32.7518 -26.1078 0.9973 1.0022 1.0001 0.0001 -0.0019 0.0112
+ - param set #30 has cost=-0.010186 [o=55 t=25]
+ -- Parameters = 117.2725 -34.0185 -34.7876 1.0632 28.8306 -16.3143 1.0069 0.9990 1.0092 -0.0004 0.0009 0.0071
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0437
+ --- dist(#3,#1) = 0.542
+ --- dist(#4,#1) = 0.483
+ --- dist(#5,#1) = 0.509
+ --- dist(#6,#1) = 0.433
+ --- dist(#7,#1) = 0.511
+ --- dist(#8,#1) = 0.378
+ --- dist(#9,#1) = 0.428
+ --- dist(#10,#1) = 0.542
+ --- dist(#11,#1) = 0.506
+ --- dist(#12,#1) = 0.495
+ --- dist(#13,#1) = 0.492
+ --- dist(#14,#1) = 0.49
+ --- dist(#15,#1) = 0.481
+ --- dist(#16,#1) = 0.526
+ --- dist(#17,#1) = 0.486
+ --- dist(#18,#1) = 0.479
+ --- dist(#19,#1) = 0.499
+ --- dist(#20,#1) = 0.465
+ --- dist(#21,#1) = 0.508
+ --- dist(#22,#1) = 0.542
+ --- dist(#23,#1) = 0.502
+ --- dist(#24,#1) = 0.47
+ --- dist(#25,#1) = 0.403
+ --- dist(#26,#1) = 0.491
+ --- dist(#27,#1) = 0.502
+ --- dist(#28,#1) = 0.446
+ --- dist(#29,#1) = 0.446
+ --- dist(#30,#1) = 0.382
+ - Total coarse refinement net CPU time = 0.0 s; 9475 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=36 usiz=142
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 30846 total points stored in 64 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.247935 * [o=1 t=0]
+ -- Parameters = 86.1944 -1.0930 0.8868 0.1834 -1.6496 -0.7057 0.9921 1.0138 0.9761 -0.0028 0.0003 -0.0263
+ - cost(#2)=-0.247229 [o=-1 t=-1]
+ -- Parameters = 86.3279 -1.0994 0.8727 -0.1092 -1.5791 -0.2743 0.9912 1.0144 0.9770 -0.0125 -0.0087 -0.0244
+ - cost(#3)=-0.021662 [o=60 t=3]
+ -- Parameters = 85.8306 14.5315 -42.4387 -12.2868 -14.0112 -2.0217 1.0096 1.0038 1.0005 0.0033 -0.0014 0.0041
+ - cost(#4)=-0.021326 [o=23 t=6]
+ -- Parameters = 67.0415 20.6003 -35.4082 -2.5354 -23.6785 -44.2066 0.9683 0.9857 0.9480 -0.0004 0.0002 -0.0097
+ - cost(#5)=-0.016192 [o=8 t=1]
+ -- Parameters = 85.8774 -16.2607 -39.8684 -25.0739 20.9043 -38.5525 1.0084 1.0086 1.0296 0.0107 0.0030 -0.0020
+ - cost(#6)=-0.016624 [o=2 t=21]
+ -- Parameters = 67.3290 18.4963 -33.7193 -7.6211 -16.3778 -37.8008 0.9602 0.9845 0.9727 0.0075 0.0229 0.0012
+ - cost(#7)=-0.016889 [o=71 t=12]
+ -- Parameters = 92.2449 13.4816 -40.0132 11.7825 -7.8925 -1.2794 1.0211 1.0100 0.9919 -0.0019 -0.0059 -0.0049
+ - cost(#8)=-0.015336 [o=7 t=4]
+ -- Parameters = 106.3127 -23.2953 -29.3893 -31.6195 16.0869 -7.0148 1.0290 1.0027 0.9790 0.0099 -0.0038 0.0061
+ - cost(#9)=-0.014875 [o=51 t=18]
+ -- Parameters = 77.6930 -18.7582 -33.3354 -14.2854 15.5151 -31.2591 0.9938 1.0049 1.0008 0.0218 -0.0004 0.0015
+ - cost(#10)=-0.014950 [o=80 t=11]
+ -- Parameters = 83.0425 24.2655 -42.4408 -9.0660 -25.1603 -14.0028 1.0007 1.0000 0.9751 0.0004 0.0022 -0.0064
+ - cost(#11)=-0.016436 [o=63 t=27]
+ -- Parameters = 94.1629 11.6896 -39.6182 -30.8646 14.7365 -8.4448 0.9924 1.0084 0.9638 0.0029 -0.0066 0.0022
+ - cost(#12)=-0.015037 [o=56 t=10]
+ -- Parameters = 79.8421 23.2739 -38.6888 17.1000 -17.6298 18.7518 0.9977 1.0237 1.0118 0.0049 -0.0072 -0.0039
+ - cost(#13)=-0.014331 [o=34 t=19]
+ -- Parameters = 70.0319 -39.4338 -19.4792 34.9632 2.3668 -44.9996 0.9972 0.9984 1.0088 -0.0001 -0.0009 0.0118
+ - cost(#14)=-0.014740 [o=69 t=14]
+ -- Parameters = 65.7127 1.7019 -38.2841 22.5871 6.0819 9.3209 0.9999 0.9870 1.0086 -0.0016 -0.0053 -0.0020
+ - cost(#15)=-0.013150 [o=87 t=2]
+ -- Parameters = 114.9813 -20.3796 -28.7550 -43.0773 19.9714 -0.4166 1.0045 0.9960 0.9916 -0.0008 -0.0013 0.0011
+ - cost(#16)=-0.013561 [o=11 t=7]
+ -- Parameters = 90.8589 -17.3500 -41.1585 -28.1247 24.0824 -31.3804 1.0106 1.0045 1.0359 0.0068 -0.0054 0.0012
+ - cost(#17)=-0.013200 [o=19 t=13]
+ -- Parameters = 107.2671 -40.0041 -10.0364 21.1657 25.4499 16.7391 0.9961 0.9912 0.9785 0.0011 0.0007 0.0003
+ - cost(#18)=-0.013773 [o=14 t=22]
+ -- Parameters = 75.0905 -21.4305 -37.4649 -12.3673 15.2064 -41.6188 0.9687 0.9731 1.0018 -0.0017 -0.0047 0.0170
+ - cost(#19)=-0.012271 [o=76 t=15]
+ -- Parameters = 82.8456 -5.6638 -39.0002 -8.3674 4.7023 -5.7031 1.0142 1.0387 0.9956 -0.0027 0.0002 0.0037
+ - cost(#20)=-0.011488 [o=73 t=8]
+ -- Parameters = 98.9391 -38.2663 -12.7341 -10.5526 39.9784 28.6960 1.0010 0.9828 0.9898 -0.0008 0.0086 -0.0004
+ - cost(#21)=-0.012133 [o=0 t=20]
+ -- Parameters = 115.1288 -31.4611 -39.7860 11.6523 22.7690 -28.0410 0.9981 1.0095 0.9909 0.0000 0.0006 0.0018
+ - cost(#22)=-0.010666 [o=57 t=24]
+ -- Parameters = 115.0151 -26.6841 -42.4377 41.9844 -9.8112 -40.9910 0.9598 0.9422 0.9736 0.0025 0.0004 0.0178
+ - cost(#23)=-0.011176 [o=22 t=5]
+ -- Parameters = 118.3713 -20.0176 -30.9282 -44.9530 23.2519 8.2996 1.0055 1.0014 1.0074 0.0035 0.0266 0.0048
+ - cost(#24)=-0.011393 [o=30 t=16]
+ -- Parameters = 116.0172 -32.4761 -36.7416 14.8334 25.1096 -36.0573 0.9962 1.0037 1.0064 -0.0014 0.0201 -0.0009
+ - cost(#25)=-0.012197 [o=58 t=9]
+ -- Parameters = 97.3908 -21.9443 -31.3257 -20.8354 13.3939 -6.5179 1.0367 0.9989 0.9646 -0.0015 -0.0009 0.0003
+ - cost(#26)=-0.010773 [o=50 t=26]
+ -- Parameters = 61.9433 -29.9463 -38.4075 -3.7593 29.9047 12.4886 1.0047 0.9979 0.9901 -0.0012 -0.0033 -0.0015
+ - cost(#27)=-0.009462 [o=15 t=17]
+ -- Parameters = 98.9339 -41.2234 -11.5339 -6.7721 39.1075 24.5054 0.9894 1.0150 0.9948 -0.0029 -0.0022 -0.0034
+ - cost(#28)=-0.008462 [o=65 t=23]
+ -- Parameters = 102.3612 -36.7618 -10.7898 2.3767 30.0769 23.1473 1.0046 0.9604 1.0111 -0.0020 0.0115 0.0031
+ - cost(#29)=-0.008424 [o=55 t=25]
+ -- Parameters = 117.2725 -34.0185 -34.7876 1.0632 28.8306 -16.3143 1.0069 0.9990 1.0092 -0.0004 0.0009 0.0071
+ - cost(#30)=-0.007322 [o=26 t=28]
+ -- Parameters = 116.3879 -29.7252 -28.2347 6.3938 32.7518 -26.1078 0.9973 1.0022 1.0001 0.0001 -0.0019 0.0112
+ - cost(#31)=-0.082426 [o=-2 t=-2]
+ -- Parameters = 86.3498 -2.1841 -2.4436 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.248210 * [o=1 t=0]
+ -- Parameters = 86.1974 -1.0666 0.9082 0.1420 -1.6590 -0.6734 0.9929 1.0125 0.9782 -0.0031 -0.0002 -0.0266
*[#30976=-0.248403] *[#30981=-0.248417] *[#30982=-0.24842] *[#30983=-0.24844] *[#30988=-0.248447] *[#30989=-0.248461] *[#30990=-0.248468] *[#30993=-0.248469] *[#30995=-0.248474] *[#31000=-0.24848] *[#31003=-0.248482] *[#31008=-0.248485] *[#31013=-0.248489] *[#31016=-0.248492]
+ - cost(#2)=-0.248492 * [o=-1 t=-1]
+ -- Parameters = 86.2590 -1.1481 0.9271 0.0689 -1.5312 -0.3582 0.9937 1.0147 0.9793 -0.0051 -0.0082 -0.0245
+ - cost(#3)=-0.022848 [o=60 t=3]
+ -- Parameters = 85.7755 14.4944 -42.2576 -12.4318 -13.8582 -1.9745 1.0102 1.0059 1.0383 0.0058 -0.0018 0.0037
+ - cost(#4)=-0.021418 [o=23 t=6]
+ -- Parameters = 67.0038 20.5890 -35.3618 -2.5245 -23.6737 -44.1648 0.9677 0.9860 0.9493 -0.0016 0.0003 -0.0098
+ - cost(#5)=-0.016332 [o=8 t=1]
+ -- Parameters = 85.7996 -16.2974 -39.8999 -25.0753 20.7890 -38.4821 1.0099 1.0079 1.0307 0.0105 0.0032 -0.0024
+ - cost(#6)=-0.016640 [o=2 t=21]
+ -- Parameters = 67.3163 18.5070 -33.7298 -7.6156 -16.3725 -37.8032 0.9604 0.9842 0.9723 0.0076 0.0234 0.0014
+ - cost(#7)=-0.016999 [o=71 t=12]
+ -- Parameters = 92.2619 13.5171 -40.1388 11.7585 -7.9200 -1.1761 1.0216 1.0095 0.9944 -0.0020 -0.0053 -0.0053
+ - cost(#8)=-0.015386 [o=7 t=4]
+ -- Parameters = 106.3367 -23.2841 -29.4356 -31.5903 16.0758 -7.0149 1.0290 1.0010 0.9794 0.0092 -0.0039 0.0060
+ - cost(#9)=-0.014978 [o=51 t=18]
+ -- Parameters = 77.6605 -18.7340 -33.2648 -14.2947 15.4942 -31.1176 0.9941 1.0043 1.0007 0.0234 -0.0008 0.0018
+ - cost(#10)=-0.016946 [o=80 t=11]
+ -- Parameters = 82.0976 24.0490 -42.4071 -8.9448 -25.0150 -12.5391 1.0184 0.9993 0.9586 0.0005 0.0028 -0.0138
+ - cost(#11)=-0.016701 [o=63 t=27]
+ -- Parameters = 94.1766 11.6912 -39.5829 -30.9725 14.6677 -8.4562 0.9919 1.0078 0.9658 -0.0046 -0.0066 0.0027
+ - cost(#12)=-0.015609 [o=56 t=10]
+ -- Parameters = 79.8786 23.1978 -38.9638 17.2742 -17.8286 18.9307 0.9989 1.0253 1.0094 0.0059 -0.0140 -0.0045
+ - cost(#13)=-0.014350 [o=34 t=19]
+ -- Parameters = 70.0057 -39.4279 -19.4773 34.9692 2.4225 -44.9998 0.9973 0.9982 1.0087 -0.0001 -0.0010 0.0119
+ - cost(#14)=-0.014831 [o=69 t=14]
+ -- Parameters = 65.7334 1.6443 -38.3205 22.5967 6.0091 9.3009 0.9997 0.9868 1.0078 -0.0018 -0.0051 -0.0020
+ - cost(#15)=-0.013200 [o=87 t=2]
+ -- Parameters = 115.0261 -20.3361 -28.6950 -43.0492 19.9747 -0.4866 1.0045 0.9955 0.9913 -0.0009 -0.0016 0.0012
+ - cost(#16)=-0.013617 [o=11 t=7]
+ -- Parameters = 90.8387 -17.3445 -41.1682 -28.1451 24.0757 -31.3948 1.0111 1.0031 1.0358 0.0070 -0.0053 0.0012
+ - cost(#17)=-0.013229 [o=19 t=13]
+ -- Parameters = 107.2639 -40.0044 -10.0172 21.1362 25.4385 16.7437 0.9961 0.9913 0.9786 0.0011 0.0000 0.0003
+ - cost(#18)=-0.014075 [o=14 t=22]
+ -- Parameters = 75.1602 -21.5980 -37.4271 -12.4683 15.1563 -41.5184 0.9686 0.9727 1.0021 -0.0084 -0.0042 0.0176
+ - cost(#19)=-0.012365 [o=76 t=15]
+ -- Parameters = 82.8347 -5.6278 -38.9680 -8.4485 4.6890 -5.6870 1.0135 1.0381 0.9953 -0.0030 -0.0004 0.0028
+ - cost(#20)=-0.011529 [o=73 t=8]
+ -- Parameters = 98.9278 -38.3346 -12.6573 -10.5431 39.8927 28.6116 1.0027 0.9835 0.9914 -0.0013 0.0087 -0.0006
+ - cost(#21)=-0.012366 [o=0 t=20]
+ -- Parameters = 115.2326 -31.4078 -39.7518 11.5730 22.8005 -28.0368 0.9946 1.0082 0.9857 -0.0008 0.0018 -0.0003
+ - cost(#22)=-0.010819 [o=57 t=24]
+ -- Parameters = 115.0554 -26.7733 -42.4436 42.0254 -9.8113 -40.9755 0.9603 0.9436 0.9734 0.0107 0.0002 0.0182
+ - cost(#23)=-0.011345 [o=22 t=5]
+ -- Parameters = 118.3325 -19.7729 -30.9625 -44.9987 23.4952 8.2990 1.0054 1.0004 1.0073 0.0033 0.0341 0.0051
+ - cost(#24)=-0.011572 [o=30 t=16]
+ -- Parameters = 116.0278 -32.4342 -36.6849 14.8129 25.0690 -36.0994 0.9961 1.0037 1.0069 -0.0014 0.0202 -0.0084
+ - cost(#25)=-0.012969 [o=58 t=9]
+ -- Parameters = 97.8204 -21.9399 -31.5635 -20.8335 13.5917 -6.8115 1.0403 1.0020 0.9604 -0.0018 -0.0012 0.0014
+ - cost(#26)=-0.010815 [o=50 t=26]
+ -- Parameters = 61.9216 -29.9287 -38.4431 -3.7981 29.9163 12.5114 1.0040 0.9986 0.9895 -0.0022 -0.0034 -0.0014
+ - cost(#27)=-0.009499 [o=15 t=17]
+ -- Parameters = 98.9397 -41.2185 -11.5621 -6.8046 39.0677 24.5470 0.9896 1.0132 0.9948 -0.0027 -0.0016 -0.0035
+ - cost(#28)=-0.008678 [o=65 t=23]
+ -- Parameters = 102.4250 -36.8903 -10.7446 2.4435 29.9970 23.1146 1.0038 0.9593 1.0118 -0.0021 0.0115 0.0028
+ - cost(#29)=-0.011826 [o=55 t=25]
+ -- Parameters = 116.2609 -39.1000 -30.7880 1.7620 28.3207 -17.4486 1.0164 1.0109 0.9970 0.0017 -0.0002 0.0025
+ - cost(#30)=-0.007352 [o=26 t=28]
+ -- Parameters = 116.3966 -29.6726 -28.2263 6.3777 32.7735 -26.1220 0.9982 1.0027 1.0002 -0.0002 -0.0019 0.0111
+ - cost(#31)=-0.245047 [o=-2 t=-2]
+ -- Parameters = 86.2119 -1.3112 0.7659 0.1340 -1.0097 -0.6979 0.9958 1.0183 0.9879 -0.0029 0.0022 -0.0114
+ - case #2 [o=-1 t=-1] is now the best
+ - Initial cost = -0.248492
+ - Initial fine Parameters = 86.2590 -1.1481 0.9271 0.0689 -1.5312 -0.3582 0.9937 1.0147 0.9793 -0.0051 -0.0082 -0.0245
*[#34061=-0.248495] *[#34062=-0.248495] *[#34066=-0.248497] *[#34067=-0.2485] *[#34070=-0.248505] *[#34071=-0.248505] *[#34073=-0.248506] *[#34081=-0.248507] *[#34086=-0.24851] *[#34087=-0.248517] *[#34088=-0.248528] *[#34091=-0.248531] *[#34092=-0.248535] *[#34093=-0.248541] *[#34094=-0.248547] *[#34095=-0.248553] *[#34096=-0.248561] *[#34099=-0.248579] *[#34100=-0.248595] *[#34101=-0.248617] *[#34104=-0.248634] *[#34109=-0.248637] *[#34110=-0.248655] *[#34111=-0.248656] *[#34113=-0.248662] *[#34115=-0.248687] *[#34116=-0.248694] *[#34117=-0.248697] *[#34118=-0.248701] *[#34122=-0.248701] *[#34123=-0.248714] *[#34124=-0.248715] *[#34128=-0.24872] *[#34129=-0.248724] *[#34130=-0.248725] *[#34131=-0.248727] *[#34140=-0.248733] *[#34141=-0.248734] *[#34142=-0.248734] *[#34143=-0.248736] *[#34144=-0.248737] *[#34145=-0.24874] *[#34150=-0.248745] *[#34151=-0.248748] *[#34154=-0.248753] *[#34155=-0.248753] *[#34156=-0.248754] *[#34159=-0.248758] *[#34160=-0.24876] *[#34165=-0.24876] *[#34166=-0.248761] *[#34169=-0.248767] *[#34170=-0.248767] *[#34172=-0.248771] *[#34175=-0.248773] *[#34176=-0.248776] *[#34183=-0.248777] *[#34198=-0.248778] *[#34202=-0.248779] *[#34208=-0.248779] *[#34209=-0.24878] *[#34213=-0.24878] *[#34216=-0.24878] *[#34218=-0.24878] *[#34219=-0.24878] *[#34222=-0.24878] *[#34223=-0.24878] *[#34224=-0.248781] *[#34227=-0.248781] *[#34228=-0.248782] *[#34238=-0.248782] *[#34239=-0.248782] *[#34240=-0.248782] *[#34241=-0.248782] *[#34242=-0.248783] *[#34243=-0.248783] *[#34244=-0.248783] *[#34245=-0.248783] *[#34248=-0.248784] *[#34249=-0.248784] *[#34252=-0.248785] *[#34256=-0.248785] *[#34259=-0.248785] *[#34267=-0.248785] *[#34268=-0.248785] *[#34269=-0.248786] *[#34270=-0.248786] *[#34272=-0.248786] *[#34277=-0.248786] *[#34278=-0.248787] *[#34279=-0.248787] *[#34280=-0.248788] *[#34281=-0.248788] *[#34283=-0.248788] *[#34284=-0.248788] *[#34287=-0.248789] *[#34288=-0.248789] *[#34289=-0.248789] *[#34290=-0.248789] *[#34291=-0.248789] *[#34295=-0.24879] *[#34300=-0.24879] *[#34301=-0.24879] *[#34302=-0.248791] *[#34305=-0.248791] *[#34306=-0.248791] *[#34308=-0.248791] *[#34309=-0.248792] *[#34310=-0.248792] *[#34313=-0.248792] *[#34315=-0.248792] *[#34320=-0.248792] *[#34321=-0.248793] *[#34322=-0.248793] *[#34325=-0.248794] *[#34327=-0.248794] *[#34330=-0.248794] *[#34331=-0.248795] *[#34332=-0.248795] *[#34334=-0.248795] *[#34337=-0.248796] *[#34338=-0.248796] *[#34339=-0.248796] *[#34340=-0.248796] *[#34343=-0.248796] *[#34345=-0.248796] *[#34346=-0.248797] *[#34347=-0.248797] *[#34348=-0.248797] *[#34351=-0.248797] *[#34353=-0.248798] *[#34354=-0.248798] *[#34355=-0.248798] *[#34356=-0.248798] *[#34363=-0.248799] *[#34364=-0.248799] *[#34365=-0.248799] *[#34366=-0.2488] *[#34367=-0.2488] *[#34368=-0.2488] *[#34372=-0.248801] *[#34373=-0.248801] *[#34374=-0.248801] *[#34378=-0.248802] *[#34379=-0.248802] *[#34380=-0.248803] *[#34381=-0.248803] *[#34384=-0.248804] *[#34385=-0.248804] *[#34386=-0.248805] *[#34387=-0.248806] *[#34392=-0.248806] *[#34394=-0.248806] *[#34395=-0.248806] *[#34397=-0.248806] *[#34400=-0.248807] *[#34401=-0.248807] *[#34402=-0.248807] *[#34403=-0.248808] *[#34404=-0.248808] *[#34407=-0.248808] *[#34409=-0.248808] *[#34410=-0.248808] *[#34411=-0.248809] *[#34412=-0.248809] *[#34413=-0.248809] *[#34416=-0.248809] *[#34418=-0.248809] *[#34420=-0.24881] *[#34422=-0.24881] *[#34423=-0.24881] *[#34426=-0.24881] *[#34428=-0.248811] *[#34429=-0.248811] *[#34430=-0.248811] *[#34432=-0.248812] *[#34435=-0.248812] *[#34437=-0.248812] *[#34438=-0.248813] *[#34439=-0.248814] *[#34440=-0.248815] *[#34441=-0.248817] *[#34446=-0.248818] *[#34447=-0.24882] *[#34454=-0.24882] *[#34458=-0.248821] *[#34459=-0.248821] *[#34466=-0.248821] *[#34467=-0.248821] *[#34468=-0.248821] *[#34469=-0.248822] *[#34470=-0.248822] *[#34473=-0.248823] *[#34484=-0.248823] *[#34485=-0.248823] *[#34486=-0.248823] *[#34487=-0.248823] *[#34488=-0.248823] *[#34499=-0.248824] *[#34502=-0.248824] *[#34503=-0.248824] *[#34504=-0.248824] *[#34511=-0.248824] *[#34513=-0.248825] *[#34520=-0.248825] *[#34521=-0.248825] *[#34522=-0.248825] *[#34525=-0.248825] *[#34529=-0.248826] *[#34530=-0.248826] *[#34531=-0.248826] *[#34536=-0.248826] *[#34537=-0.248826] *[#34538=-0.248827] *[#34563=-0.248827] *[#34569=-0.248827] *[#34572=-0.248827]
+ - Finalish cost = -0.248827 ; 593 funcs
+ - ini Finalish Parameters = 86.2359 -1.0776 0.9676 0.1222 -1.8006 -0.3840 0.9938 1.0136 0.9799 -0.0031 -0.0068 -0.0310
*[#34796=-0.248827] *[#34817=-0.248827]
+ - Final cost = -0.248827 ; 266 funcs
+ Final fine fit Parameters:
x-shift= 86.2386 y-shift= -1.0777 z-shift= 0.9677 ... enorm= 86.2508 mm
z-angle= 0.1201 x-angle= -1.8004 y-angle= -0.3834 ... total= 1.8452 deg
x-scale= 0.9938 y-scale= 1.0135 z-scale= 0.9800 ... vol3D= 0.9871 = base bigger than source
y/x-shear= -0.0031 z/x-shear= -0.0068 z/y-shear= -0.0311
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.2386 -1.0777 0.9677 0.1201 -1.8004 -0.3834 0.9938 1.0135 0.9800 -0.0031 -0.0068 -0.0311
++ Unloading unneeded data
++ Output dataset ./sub-09_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-09_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 19.9
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/__tt_sub-09_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/sub-09_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-09_T1w_ns+orig sub-09_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 7.0
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-07_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 6.0
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-09_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-09_T1w_al_junk+orig
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-09_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-09_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.724 -14.468 -26.047
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.3% 20.9% 45.1%
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-08.r02.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-08.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40888 voxels
+ 5952 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
Performing center alignment with @Align_Centers
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-09_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-09_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-09_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-09_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-09_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-09_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-09_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.0
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
Center distance of 0.000000 mm
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-07.r02.tcat+orig
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
++ Reading input dataset ./pb00.sub-07.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
Resampling ...
+ Automask has 41830 voxels
+ 5970 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1245.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-09_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-09_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/105.00=3.5e+02
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=+0.065 pitch=-0.196 yaw=-0.295 dS=-0.284 dL=+0.045 dP=-0.220
++ Mean: roll=+0.160 pitch=+0.114 yaw=+0.164 dS=-0.032 dL=+0.132 dP=-0.070
++ Max : roll=+0.233 pitch=+0.250 yaw=+0.335 dS=+0.142 dL=+0.306 dP=+0.067
++ Max displacements (mm) for each sub-brick:
0.70(0.00) 0.61(0.16) 0.62(0.09) 0.60(0.07) 0.58(0.06) 0.63(0.08) 0.61(0.07) 0.62(0.08) 0.60(0.07) 0.55(0.07) 0.62(0.08) 0.61(0.11) 0.61(0.07) 0.59(0.05) 0.50(0.10) 0.54(0.06) 0.52(0.08) 0.57(0.09) 0.58(0.09) 0.56(0.09) 0.58(0.06) 0.56(0.07) 0.59(0.10) 0.57(0.09) 0.57(0.07) 0.59(0.05) 0.56(0.05) 0.59(0.08) 0.60(0.08) 0.58(0.07) 0.58(0.07) 0.58(0.06) 0.58(0.02) 0.59(0.05) 0.57(0.34) 0.38(0.39) 0.47(0.48) 0.90(0.47) 0.59(0.35) 0.49(0.19) 0.50(0.16) 0.46(0.12) 0.48(0.16) 0.46(0.17) 0.48(0.13) 0.47(0.10) 0.48(0.10) 0.49(0.09) 0.51(0.10) 0.53(0.06) 0.51(0.09) 0.53(0.09) 0.50(0.10) 0.50(0.09) 0.50(0.08) 0.49(0.07) 0.52(0.06) 0.46(0.10) 0.46(0.21) 0.52(0.33) 0.50(0.20) 0.54(0.12) 0.54(0.09) 0.60(0.12) 0.55(0.14) 0.59(0.12) 0.58(0.10) 0.62(0.13) 0.53(0.11) 0.49(0.14) 0.49(0.04) 0.46(0.07) 0.55(0.11) 0.48(0.14) 0.57(0.15) 0.46(0.14) 0.54(0.11) 0.44(0.13) 0.51(0.11) 0.47(0.10) 0.55(0.15) 0.59(0.11) 0.56(0.13) 0.53(0.13) 0.60(0.14) 0.58(0.10) 0.55(0.04) 0.53(0.07) 0.59(0.11) 0.49(0.15) 0.57(0.14) 0.52(0.17) 0.77(0.46) 0.58(0.27) 0.57(0.20) 0.57(0.10) 0.58(0.13) 0.56(0.14) 0.57(0.14) 0.55(0.09) 0.60(0.10) 0.55(0.09) 0.55(0.05) 0.55(0.04) 0.57(0.07) 0.61(0.06) 0.53(0.11) 0.67(0.16) 0.55(0.16) 0.59(0.11) 0.53(0.10) 0.54(0.06) 0.57(0.07) 0.51(0.12) 0.56(0.12) 0.57(0.17) 0.64(0.20) 0.58(0.21) 0.56(0.09) 0.60(0.07) 0.55(0.07) 0.61(0.08) 0.56(0.12) 0.60(0.14) 0.59(0.10) 0.61(0.07) 0.59(0.06) 0.56(0.08) 0.67(0.14) 0.61(0.12) 0.64(0.14) 0.64(0.04) 0.64(0.19) 0.57(0.25) 0.54(0.10) 0.54(0.07) 0.53(0.05) 0.57(0.11) 0.53(0.11) 0.58(0.12) 0.53(0.09) 0.55(0.06) 0.52(0.11) 0.56(0.13) 0.55(0.10) 0.53(0.06)
++ Max displacement in automask = 0.90 (mm) at sub-brick 37
++ Max delta displ in automask = 0.48 (mm) at sub-brick 36
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-08.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb00.sub-08.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-08.r02.tcat+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -0.046 -8.175 -29.810
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.1% 11.8% 51.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.142 pitch=-0.397 yaw=-0.235 dS=-0.061 dL=-0.085 dP=-0.055
++ Mean: roll=-0.061 pitch=-0.095 yaw=-0.112 dS=+0.039 dL=-0.031 dP=+0.007
++ Max : roll=+0.046 pitch=+0.133 yaw=+0.015 dS=+0.150 dL=+0.062 dP=+0.095
++ Max displacements (mm) for each sub-brick:
0.84(0.00) 0.81(0.07) 0.75(0.07) 0.76(0.03) 0.81(0.06) 0.73(0.11) 0.74(0.08) 0.70(0.07) 0.71(0.04) 0.59(0.13) 0.56(0.06) 0.57(0.03) 0.55(0.08) 0.57(0.04) 0.57(0.02) 0.51(0.07) 0.52(0.09) 0.54(0.06) 0.51(0.06) 0.51(0.06) 0.48(0.05) 0.47(0.02) 0.48(0.06) 0.50(0.06) 0.56(0.07) 0.63(0.09) 0.57(0.11) 0.59(0.03) 0.58(0.04) 0.57(0.02) 0.56(0.06) 0.55(0.03) 0.51(0.07) 0.54(0.05) 0.51(0.04) 0.44(0.07) 0.52(0.09) 0.53(0.07) 0.48(0.06) 0.48(0.03) 0.45(0.04) 0.49(0.04) 0.44(0.09) 0.47(0.06) 0.45(0.06) 0.44(0.04) 0.47(0.05) 0.52(0.10) 0.44(0.11) 0.36(0.11) 0.37(0.05) 0.43(0.09) 0.40(0.07) 0.36(0.06) 0.27(0.12) 0.34(0.12) 0.29(0.09) 0.33(0.08) 0.29(0.07) 0.35(0.11) 0.37(0.03) 0.34(0.06) 0.28(0.09) 0.30(0.06) 0.32(0.08) 0.30(0.09) 0.30(0.04) 0.28(0.04) 0.30(0.04) 0.32(0.04) 0.32(0.05) 0.32(0.05) 0.30(0.03) 0.29(0.06) 0.31(0.06) 0.28(0.06) 0.31(0.04) 0.31(0.05) 0.32(0.05) 0.31(0.07) 0.31(0.10) 0.29(0.09) 0.28(0.04) 0.28(0.02) 0.24(0.06) 0.23(0.10) 0.23(0.09) 0.26(0.08) 0.25(0.05) 0.24(0.05) 0.23(0.03) 0.24(0.06) 0.27(0.06) 0.27(0.02) 0.23(0.06) 0.26(0.09) 0.23(0.08) 0.24(0.06) 0.22(0.06) 0.21(0.07) 0.21(0.13) 0.19(0.04) 0.17(0.04) 0.18(0.04) 0.15(0.08) 0.19(0.10) 0.10(0.11) 0.09(0.03) 0.08(0.04) 0.15(0.12) 0.09(0.07) 0.11(0.04) 0.07(0.05) 0.07(0.07) 0.08(0.02) 0.07(0.06) 0.05(0.08) 0.08(0.09) 0.05(0.04) 0.04(0.03) 0.06(0.06) 0.04(0.05) 0.07(0.05) 0.00(0.07) 0.05(0.05) 0.08(0.04) 0.11(0.05) 0.12(0.04) 0.10(0.03) 0.14(0.08) 0.16(0.04) 0.17(0.04) 0.17(0.04) 0.17(0.04) 0.18(0.03) 0.13(0.07) 0.18(0.07) 0.17(0.05) 0.17(0.05) 0.19(0.03) 0.19(0.03) 0.22(0.04) 0.22(0.03) 0.25(0.03) 0.23(0.03) 0.19(0.08)
++ Max displacement in automask = 0.84 (mm) at sub-brick 0
++ Max delta displ in automask = 0.13 (mm) at sub-brick 9
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-07.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
RMS[0] = 0.315618 0.170838 ITER = 7/50
0.315618
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-09_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-09_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-09_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-09_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-09_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-09_T1w_ns+orig.BRIK
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-07_T1w_ns+tlrc::WARP_DATA -I sub-07_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-09_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-09_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-09_T1w_ns+orig __ats_tmp___upad15_pre.sub-09_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-09_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
3dAllineate -base sub-07_T1w_ns+tlrc -input pb00.sub-07.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-09_T1w_ns+tlrc.HEAD -> sub-09_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-09_T1w_ns+tlrc.BRIK -> sub-09_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-09_T1w_ns+tlrc::WARP_DATA -I
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-07.r02.tcat+orig.HEAD
++ Base dataset: ./sub-07_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
if ( ! -f sub-09_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb00.sub-09.r01.tcat+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb00.sub-09.r01.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 1.425 -6.091 -27.697
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.5% 8.8% 47.9%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
+ Automask has 40262 voxels
+ 6092 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.7
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.280 -18.654 -32.063
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.2% 26.9% 55.5%
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.2
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-07_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.4
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 0.724 -14.468 -26.047
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.3% 20.9% 45.1%
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-08.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.052 pitch=-0.174 yaw=-0.094 dS=-0.444 dL=-0.249 dP=-0.079
++ Mean: roll=-0.000 pitch=-0.087 yaw=-0.021 dS=-0.123 dL=-0.096 dP=+0.034
++ Max : roll=+0.049 pitch=+0.073 yaw=+0.050 dS=+0.037 dL=+0.066 dP=+0.202
++ Max displacements (mm) for each sub-brick:
0.77(0.00) 0.69(0.11) 0.67(0.07) 0.64(0.09) 0.65(0.08) 0.64(0.08) 0.64(0.04) 0.63(0.05) 0.61(0.04) 0.60(0.05) 0.58(0.05) 0.57(0.06) 0.55(0.09) 0.62(0.18) 0.55(0.09) 0.55(0.05) 0.55(0.05) 0.59(0.15) 0.55(0.16) 0.52(0.15) 0.54(0.11) 0.54(0.05) 0.59(0.05) 0.61(0.09) 0.55(0.11) 0.52(0.07) 0.52(0.13) 0.47(0.09) 0.48(0.05) 0.50(0.09) 0.43(0.13) 0.45(0.07) 0.43(0.08) 0.44(0.02) 0.41(0.06) 0.42(0.08) 0.40(0.11) 0.40(0.03) 0.40(0.11) 0.34(0.10) 0.38(0.10) 0.40(0.04) 0.39(0.05) 0.37(0.04) 0.39(0.08) 0.35(0.10) 0.37(0.04) 0.42(0.12) 0.37(0.08) 0.34(0.04) 0.34(0.05) 0.32(0.10) 0.33(0.08) 0.35(0.05) 0.32(0.06) 0.37(0.11) 0.33(0.07) 0.36(0.12) 0.36(0.13) 0.34(0.05) 0.33(0.06) 0.33(0.10) 0.27(0.12) 0.34(0.11) 0.33(0.06) 0.25(0.19) 0.31(0.16) 0.26(0.07) 0.28(0.04) 0.33(0.09) 0.28(0.08) 0.26(0.06) 0.30(0.09) 0.28(0.10) 0.28(0.06) 0.27(0.11) 0.22(0.10) 0.27(0.10) 0.26(0.08) 0.29(0.07) 0.31(0.10) 0.28(0.11) 0.28(0.07) 0.28(0.03) 0.27(0.08) 0.30(0.11) 0.20(0.15) 0.22(0.07) 0.26(0.07) 0.25(0.10) 0.26(0.11) 0.21(0.06) 0.19(0.09) 0.24(0.12) 0.23(0.09) 0.24(0.09) 0.25(0.07) 0.23(0.03) 0.20(0.07) 0.24(0.06) 0.25(0.05) 0.20(0.09) 0.19(0.06) 0.17(0.03) 0.18(0.09) 0.24(0.11) 0.21(0.11) 0.21(0.09) 0.19(0.11) 0.15(0.07) 0.11(0.05) 0.11(0.08) 0.16(0.14) 0.20(0.29) 0.15(0.21) 0.13(0.05) 0.06(0.14) 0.06(0.07) 0.07(0.07) 0.08(0.05) 0.12(0.13) 0.06(0.06) 0.11(0.13) 0.19(0.17) 0.10(0.15) 0.13(0.16) 0.05(0.11) 0.00(0.05) 0.07(0.07) 0.07(0.09) 0.12(0.10) 0.07(0.09) 0.07(0.05) 0.11(0.12) 0.12(0.10) 0.12(0.04) 0.11(0.08) 0.10(0.05) 0.08(0.09) 0.09(0.08) 0.09(0.04) 0.08(0.09) 0.12(0.11) 0.10(0.09) 0.10(0.05) 0.14(0.12)
++ Max displacement in automask = 0.77 (mm) at sub-brick 0
++ Max delta displ in automask = 0.29 (mm) at sub-brick 113
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb00.sub-09.r01.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.6
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-09_T1w_ns+tlrc::WARP_DATA -I sub-09_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
3dAllineate -base sub-09_T1w_ns+tlrc -input pb00.sub-09.r01.tcat+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-09.r01.tcat+orig.HEAD
++ Base dataset: ./sub-09_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 8.337 -11.233 -16.690
+ shift search range is +/- = 57.780 69.336 57.780
+ 14.4% 16.2% 28.9%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb01.sub-08.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-08.r02.volreg
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-07.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.9
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-09_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Output dataset ./pb01.sub-08.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I
++ Output dataset ./pb01.sub-07.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-07.r02.volreg
3dAllineate -base sub-08_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 6.691 -18.484 -17.715
+ shift search range is +/- = 57.780 69.336 57.780
+ 11.6% 26.7% 30.7%
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 0.073 -8.252 -29.781
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.1% 11.9% 51.5%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-08_T1w_ns+tlrc anat_final.sub-08
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-08+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-08+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -0.040 1.128 0.173
+ shift search range is +/- = 66.447 79.929 68.373
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.102416
sp = 0.294745
mi = 2.69858
crM = 0.0196901
nmi = 0.820873
je = 2.69858
hel = -0.12172
crA = 0.141537
crU = 0.160044
lss = 0.897584
lpc = 0.647903
lpa = 0.352097
lpc+ = 0.746398
lpa+ = 0.450592
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.7
++ ###########################################################
3dAllineate -source sub-08_T1w+orig -master anat_final.sub-08+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-08_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.3
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-08.r01.blur pb01.sub-08.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-08.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-08.r02.blur pb01.sub-08.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
++ Output dataset ./pb01.sub-07.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-07_T1w_ns+tlrc::WARP_DATA -I sub-07_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-07_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.4
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-07_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 1.416 -6.146 -27.647
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.5% 8.9% 47.8%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-07_T1w_ns+tlrc anat_final.sub-07
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-07+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-07+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
..................................................................................................................................................
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
-- Wrote edited dataset: ./pb02.sub-08.r02.blur+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb00.sub-09.r02.tcat+orig
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-08.r01.blur+tlrc
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-08.r01.blur+tlrc
++ Forming automask
++ Reading input dataset ./pb00.sub-09.r02.tcat+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = 0.023 2.018 1.295
+ shift search range is +/- = 70.299 78.966 69.336
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ Automask has 40155 voxels
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.103334
+ 6126 voxels left in -maxdisp mask after erosion
sp = 0.304151
mi = 2.66643
++ Initializing alignment base
crM = 0.0165679
nmi = 0.814178
je = 2.66643
hel = -0.131312
crA = 0.129488
crU = 0.143607
lss = 0.896666
lpc = 0.761044
lpa = 0.238956
lpc+ = 0.843601
lpa+ = 0.321512
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.7
++ ###########################################################
3dAllineate -source sub-07_T1w+orig -master anat_final.sub-07+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-07_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ Fixed clip level = 343.540375
+ Used gradual clip level = 322.806030 .. 362.186707
+ Number voxels above clip level = 69428
+ Clustering voxels ...
+ Largest cluster has 68830 voxels
+ Clustering voxels ...
+ Largest cluster has 68733 voxels
+ Filled 139 voxels in small holes; now have 68872 voxels
+ Filled 2 voxels in large holes; now have 68874 voxels
+ Clustering voxels ...
+ Largest cluster has 68873 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202760 voxels
+ Mask now has 68873 voxels
++ 68873 voxels in the mask [out of 271633: 25.36%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-08.r02.blur+tlrc
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-08.r02.blur+tlrc
++ Forming automask
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
+ Fixed clip level = 342.217621
+ Used gradual clip level = 321.662537 .. 360.685547
+ Number voxels above clip level = 69449
+ Clustering voxels ...
+ Largest cluster has 68853 voxels
+ Clustering voxels ...
+ Largest cluster has 68749 voxels
+ Filled 134 voxels in small holes; now have 68883 voxels
+ Filled 2 voxels in large holes; now have 68885 voxels
+ Clustering voxels ...
+ Largest cluster has 68885 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202748 voxels
+ Mask now has 68885 voxels
++ 68885 voxels in the mask [out of 271633: 25.36%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.1
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-07.r01.blur pb01.sub-07.r01.volreg+tlrc
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 68948 survived, 202685 were zero
++ writing result full_mask.sub-08...
++ Output dataset ./full_mask.sub-08+tlrc.BRIK
3dresample -master full_mask.sub-08+tlrc -input sub-08_T1w_ns+tlrc -prefix rm.resam.anat
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-08
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76711 survived, 194922 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-08...
++ Output dataset ./mask_anat.sub-08+tlrc.BRIK
3dmask_tool -input full_mask.sub-08+tlrc mask_anat.sub-08+tlrc -inter -prefix mask_epi_anat.sub-08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 10363 clipped, 67648 survived, 193622 were zero
++ writing result mask_epi_anat.sub-08...
++ Output dataset ./mask_epi_anat.sub-08+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-08+tlrc mask_anat.sub-08+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-08+tlrc.BRIK B=./mask_anat.sub-08+tlrc.BRIK
..................................................................................................................................................
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
68948 76711 78011 67648 1300 9063 1.8855 11.8145 1.0652 0.9478 1.1189
3ddot -dodice full_mask.sub-08+tlrc mask_anat.sub-08+tlrc
tee out.mask_ae_dice.txt
-- Wrote edited dataset: ./pb02.sub-07.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-07.r02.blur pb01.sub-07.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
0.928854
3dresample -master full_mask.sub-08+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-08+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.960445
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-08.r01.blur+tlrc
..................................................................................................................................................
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
-- Wrote edited dataset: ./pb02.sub-07.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-07.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-07.r01.blur+tlrc
++ Forming automask
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-08.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-08.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Fixed clip level = 355.907684
+ Used gradual clip level = 340.100769 .. 371.265900
+ Number voxels above clip level = 72892
+ Clustering voxels ...
+ Largest cluster has 71734 voxels
+ Clustering voxels ...
+ Largest cluster has 71460 voxels
+ Filled 81 voxels in small holes; now have 71541 voxels
+ Filled 1 voxels in large holes; now have 71542 voxels
+ Clustering voxels ...
+ Largest cluster has 71542 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200091 voxels
+ Mask now has 71542 voxels
++ 71542 voxels in the mask [out of 271633: 26.34%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 3 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-07.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-07.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 355.232056
+ Used gradual clip level = 339.918793 .. 370.664093
+ Number voxels above clip level = 72836
+ Clustering voxels ...
+ Largest cluster has 71619 voxels
+ Clustering voxels ...
+ Largest cluster has 71446 voxels
+ Filled 98 voxels in small holes; now have 71544 voxels
+ Filled 1 voxels in large holes; now have 71545 voxels
+ Clustering voxels ...
+ Largest cluster has 71542 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200091 voxels
+ Mask now has 71542 voxels
++ 71542 voxels in the mask [out of 271633: 26.34%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 7 y-planes are zero [from P]
++ last 3 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 7 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-07
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 71768 survived, 199865 were zero
++ writing result full_mask.sub-07...
++ Output dataset ./full_mask.sub-07+tlrc.BRIK
3dresample -master full_mask.sub-07+tlrc -input sub-07_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-07
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 75947 survived, 195686 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-07...
++ Output dataset ./mask_anat.sub-07+tlrc.BRIK
3dmask_tool -input full_mask.sub-07+tlrc mask_anat.sub-07+tlrc -inter -prefix mask_epi_anat.sub-07
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 8411 clipped, 69652 survived, 193570 were zero
++ writing result mask_epi_anat.sub-07...
++ Output dataset ./mask_epi_anat.sub-07+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-07+tlrc mask_anat.sub-07+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-07+tlrc.BRIK B=./mask_anat.sub-07+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
71768 75947 78063 69652 2116 6295 2.9484 8.2887 1.0446 0.9605 1.0482
3ddot -dodice full_mask.sub-07+tlrc mask_anat.sub-07+tlrc
tee out.mask_ae_dice.txt
0.943059
3dresample -master full_mask.sub-07+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-07+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.96351
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-07.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-07.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-07.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.131 pitch=-0.323 yaw=-0.177 dS=-0.394 dL=-0.087 dP=-0.613
++ Mean: roll=-0.069 pitch=-0.249 yaw=-0.124 dS=-0.287 dL=-0.023 dP=-0.514
++ Max : roll=+0.016 pitch=-0.104 yaw=-0.076 dS=-0.217 dL=+0.028 dP=-0.450
++ Max displacements (mm) for each sub-brick:
0.84(0.00) 0.76(0.10) 0.77(0.10) 0.84(0.09) 0.89(0.07) 0.82(0.18) 0.88(0.07) 0.86(0.09) 0.88(0.09) 0.95(0.08) 0.90(0.06) 0.87(0.07) 0.95(0.09) 0.92(0.04) 0.87(0.06) 0.95(0.10) 0.94(0.04) 0.93(0.07) 0.96(0.08) 0.86(0.11) 0.89(0.08) 0.88(0.04) 0.87(0.07) 0.93(0.08) 0.90(0.13) 0.93(0.06) 0.89(0.07) 0.85(0.06) 0.89(0.06) 0.90(0.04) 0.84(0.07) 0.97(0.14) 0.90(0.08) 0.86(0.04) 0.90(0.08) 0.88(0.05) 0.88(0.03) 0.97(0.10) 0.90(0.08) 0.91(0.05) 0.94(0.04) 0.90(0.04) 0.91(0.03) 0.95(0.04) 0.87(0.10) 0.95(0.10) 0.92(0.04) 0.82(0.10) 0.92(0.10) 0.87(0.06) 0.89(0.08) 0.92(0.04) 0.86(0.07) 0.90(0.06) 0.96(0.06) 0.89(0.09) 0.94(0.06) 0.93(0.03) 0.89(0.06) 0.98(0.10) 0.87(0.12) 0.94(0.07) 0.98(0.06) 0.92(0.09) 0.98(0.07) 0.92(0.06) 0.89(0.04) 0.94(0.06) 0.95(0.05) 0.91(0.04) 0.98(0.08) 0.92(0.05) 0.95(0.04) 1.01(0.08) 0.92(0.10) 1.00(0.09) 0.99(0.04) 0.92(0.08) 1.04(0.12) 0.93(0.14) 0.97(0.07) 1.01(0.04) 0.92(0.10) 1.00(0.10) 1.01(0.02) 0.94(0.09) 1.04(0.10) 0.95(0.09) 0.98(0.07) 1.03(0.10) 0.95(0.08) 0.97(0.04) 1.01(0.04) 0.93(0.09) 1.03(0.12) 1.01(0.04) 0.92(0.09) 1.01(0.10) 0.91(0.10) 0.96(0.06) 1.01(0.09) 0.97(0.05) 0.95(0.03) 1.04(0.11) 0.98(0.06) 0.97(0.05) 0.99(0.02) 0.94(0.06) 0.99(0.09) 0.93(0.07) 0.97(0.04) 1.01(0.05) 0.91(0.10) 0.97(0.06) 0.94(0.03) 0.95(0.05) 0.96(0.03) 0.89(0.07) 0.96(0.07) 1.00(0.06) 0.93(0.09) 1.00(0.08) 0.89(0.13) 0.92(0.04) 0.94(0.07) 0.87(0.08) 0.95(0.09) 1.00(0.08) 0.88(0.12) 0.98(0.10) 0.93(0.06) 0.91(0.06) 1.02(0.12) 0.96(0.07) 0.88(0.10) 0.95(0.09) 0.86(0.09) 0.96(0.10) 0.93(0.06) 0.86(0.08) 0.96(0.10) 0.91(0.09) 0.91(0.05) 0.90(0.03) 0.79(0.12) 0.90(0.10)
++ Max displacement in automask = 1.04 (mm) at sub-brick 78
++ Max delta displ in automask = 0.18 (mm) at sub-brick 5
++ Output dataset ./pb03.sub-08.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-08.r02.blur+tlrc
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb00.sub-09.r02.tcat+orig -expr 1 -prefix rm.epi.all1
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-08.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-08.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-09_T1w_ns+tlrc::WARP_DATA -I sub-09_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-09_T1w_ns+tlrc -input pb00.sub-09.r02.tcat+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb00.sub-09.r02.tcat+orig.HEAD
++ Base dataset: ./sub-09_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 8.246 -10.864 -16.553
+ shift search range is +/- = 57.780 69.336 57.780
+ 14.3% 15.7% 28.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
++ Output dataset ./pb03.sub-07.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-07.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-07.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-07.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb03.sub-08.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-08
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 4
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-08_censor.1D -show_trs_uncensored encoded
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD -censor motion_sub-08_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-08 -errts errts.sub-08 -bucket stats.sub-08
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.0
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-09_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 6.691 -18.484 -17.715
+ shift search range is +/- = 57.780 69.336 57.780
+ 11.6% 26.7% 30.7%
++ STAT automask has 247109 voxels (out of 271633 = 91.0%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (before censor) ; 288 (after)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Output dataset ./pb03.sub-07.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-07
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-08_REML -Rvar stats.sub-08_REMLvar \
-Rfitts fitts.sub-08_REML -Rerrts errts.sub-08_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (288x20): 3.6161 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (288x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (288x18): 3.58511 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (288x12): 2.74051 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (288x6): 1.00864 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 1.85305e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.71 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=4.121
++ Job #2: processing voxels 67908 to 101861; elapsed time=4.142
++ Job #3: processing voxels 101862 to 135815; elapsed time=4.157
++ Job #4: processing voxels 135816 to 169769; elapsed time=4.171
++ Job #5: processing voxels 169770 to 203723; elapsed time=4.185
++ Job #6: processing voxels 203724 to 237677; elapsed time=4.200
++ Job #7: processing voxels 237678 to 271632; elapsed time=4.215
++ Job #0: processing voxels 0 to 33953; elapsed time=4.225
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 0
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-07_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-07.r01.scale+tlrc.HEAD pb03.sub-07.r02.scale+tlrc.HEAD -censor motion_sub-07_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-07 -errts errts.sub-07 -bucket stats.sub-07
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-07.r01.scale+tlrc.HEAD pb03.sub-07.r02.scale+tlrc.HEAD
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 5.8
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ STAT automask has 250914 voxels (out of 271633 = 92.4%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (no censoring)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-09.r01.volreg
++ voxel loop:0123456789.0123456789.0123456789.0123456789.01234567++ Job #1 finished; elapsed time=8.359
++ Job #2 finished; elapsed time=8.360
++ Job #4 finished; elapsed time=8.363
8++ Job #3 finished; elapsed time=8.395
++ Job #7 finished; elapsed time=8.396
++ Job #5 finished; elapsed time=8.460
9.
++ Job #0 waiting for children to finish; elapsed time=8.477
++ Job #6 finished; elapsed time=8.500
++ Job #0 now finishing up; elapsed time=8.509
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Smallest FDR q [0 Full_Fstat] = 3.75538e-12
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-07.r01.scale+tlrc.HEAD pb03.sub-07.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-07_REML -Rvar stats.sub-07_REMLvar \
-Rfitts fitts.sub-07_REML -Rerrts errts.sub-07_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (292x20): 4.3586 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (292x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (292x18): 4.35375 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (292x12): 3.90257 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (292x6): 1 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 2.91996e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.67 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=5.177
++ Job #2: processing voxels 67908 to 101861; elapsed time=5.189
++ Job #3: processing voxels 101862 to 135815; elapsed time=5.201
++ Job #4: processing voxels 135816 to 169769; elapsed time=5.213
++ Job #5: processing voxels 169770 to 203723; elapsed time=5.225
++ Job #6: processing voxels 203724 to 237677; elapsed time=5.237
++ Job #7: processing voxels 237678 to 271632; elapsed time=5.249
++ Job #0: processing voxels 0 to 33953; elapsed time=5.259
++ Smallest FDR q [2 congruent#0_Tstat] = 4.21698e-08
++ Smallest FDR q [3 congruent_Fstat] = 4.21697e-08
++ Smallest FDR q [5 incongruent#0_Tstat] = 1.15572e-11
++ Smallest FDR q [6 incongruent_Fstat] = 1.15564e-11
++ Output dataset ./pb01.sub-09.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb01.sub-09.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ voxel loop:0123456789.0123456789.0123456789.0123456789.012345678++ Job #1 finished; elapsed time=9.357
++ Job #3 finished; elapsed time=9.385
++ Job #6 finished; elapsed time=9.402
++ Job #2 finished; elapsed time=9.429
++ Job #7 finished; elapsed time=9.431
9.
++ Job #0 waiting for children to finish; elapsed time=9.445
++ Job #4 finished; elapsed time=9.490
++ Job #5 finished; elapsed time=9.514
++ Job #0 now finishing up; elapsed time=9.520
++ Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.00171179
++ Smallest FDR q [0 Full_Fstat] = 7.6772e-12
++ Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.00171185
++ Smallest FDR q [2 congruent#0_Tstat] = 5.35386e-08
++ Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.00171179
++ Smallest FDR q [3 congruent_Fstat] = 5.35399e-08
++ Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.00171185
++ Wrote bucket dataset into ./stats.sub-08+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Output dataset ./pb01.sub-09.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-09_T1w_ns+tlrc::WARP_DATA -I sub-09_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-09_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-09_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be large
*+ WARNING: - at least one is more than 20% of search range
+ -cmass x y z shifts = 8.244 -11.305 -16.795
+ shift search range is +/- = 57.780 69.336 57.780
+ 14.3% 16.3% 29.1%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.3
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-09_T1w_ns+tlrc anat_final.sub-09
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.378e-11
++ Wrote 3D+time dataset into ./fitts.sub-08+tlrc.BRIK
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Smallest FDR q [6 incongruent_Fstat] = 2.3779e-11
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub-09+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub-09+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = 0.019 1.592 -1.778
+ shift search range is +/- = 72.225 83.781 68.373
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.11883
sp = 0.307306
mi = 2.5645
crM = 0.0211913
nmi = 0.821578
je = 2.5645
hel = -0.116865
crA = 0.146486
crU = 0.162824
lss = 0.88117
++ Wrote 3D+time dataset into ./errts.sub-08+tlrc.BRIK
lpc = 0.426466
lpa = 0.573534
lpc+ = 0.530439
lpa+ = 0.677506
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.3
++ ###########################################################
3dAllineate -source sub-09_T1w+orig -master anat_final.sub-09+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ Program finished; elapsed time=20.015
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-09_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
++ Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.0633663
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
++ Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.0633636
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 4 : 1.4%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 24 : 8.2%
final DF : 268 : 91.8%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-08_REML -Rvar stats.sub-08_REMLvar -Rfitts fitts.sub-08_REML -Rerrts errts.sub-08_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
++ Authored by: RWCox
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.0
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-09.r01.blur pb01.sub-09.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-09.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb02.sub-09.r02.blur pb01.sub-09.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
++ Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.0633663
++ FDR automask has 247109 voxels (out of 271633 = 91.0%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (288x20): 3.6161 ++ VERY GOOD ++
++ Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.0633636
++ Wrote bucket dataset into ./stats.sub-07+tlrc.BRIK
+ created 9 FDR curves in bucket header
..................................................................................................................................................
-- Wrote edited dataset: ./pb02.sub-09.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb02.sub-09.r01.blur+tlrc
++ Wrote 3D+time dataset into ./fitts.sub-07+tlrc.BRIK
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-09.r01.blur+tlrc
++ Forming automask
+ masked off 47222 voxels for being all zero; 224411 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.39
+ X matrix: 78.924% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=288 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.40
+ average case bandwidth = 15.86
+ Fixed clip level = 318.385895
+ Used gradual clip level = 305.184967 .. 332.750641
+ Number voxels above clip level = 72567
+ Clustering voxels ...
+ Largest cluster has 72037 voxels
+ Clustering voxels ...
+ Largest cluster has 71232 voxels
+ Filled 129 voxels in small holes; now have 71361 voxels
+ Clustering voxels ...
+ Largest cluster has 71361 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200272 voxels
+ Mask now has 71361 voxels
++ 71361 voxels in the mask [out of 271633: 26.27%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb02.sub-09.r02.blur+tlrc
++ Wrote 3D+time dataset into ./errts.sub-07+tlrc.BRIK
++ Program finished; elapsed time=19.657
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb02.sub-09.r02.blur+tlrc
++ Forming automask
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
+ Fixed clip level = 318.771973
+ Used gradual clip level = 306.355438 .. 332.605286
+ Number voxels above clip level = 72428
+ Clustering voxels ...
+ Largest cluster has 71887 voxels
+ Clustering voxels ...
+ Largest cluster has 71056 voxels
+ Filled 137 voxels in small holes; now have 71193 voxels
+ Filled 2 voxels in large holes; now have 71195 voxels
+ Clustering voxels ...
+ Largest cluster has 71190 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 200443 voxels
+ Mask now has 71190 voxels
++ 71190 voxels in the mask [out of 271633: 26.21%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 6 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
++ CPU time = 0.000000 sec
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 0 : 0.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub-09
total DF used : 20 : 6.8%
final DF : 272 : 93.2%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-07.r01.scale+tlrc.HEAD pb03.sub-07.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-07_REML -Rvar stats.sub-07_REMLvar -Rfitts fitts.sub-07_REML -Rerrts errts.sub-07_REML -verb
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 71499 survived, 200134 were zero
++ writing result full_mask.sub-09...
++ Output dataset ./full_mask.sub-09+tlrc.BRIK
3dresample -master full_mask.sub-09+tlrc -input sub-09_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub-09
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76995 survived, 194638 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub-09...
++ Output dataset ./mask_anat.sub-09+tlrc.BRIK
3dmask_tool -input full_mask.sub-09+tlrc mask_anat.sub-09+tlrc -inter -prefix mask_epi_anat.sub-09
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 11174 clipped, 68660 survived, 191799 were zero
++ writing result mask_epi_anat.sub-09...
++ Output dataset ./mask_epi_anat.sub-09+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub-09+tlrc mask_anat.sub-09+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub-09+tlrc.BRIK B=./mask_anat.sub-09+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
71499 76995 79834 68660 2839 8335 3.9707 10.8254 1.0385 0.9833 1.0413
3ddot -dodice full_mask.sub-09+tlrc mask_anat.sub-09+tlrc
tee out.mask_ae_dice.txt
0.924751
3dresample -master full_mask.sub-09+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
++ FDR automask has 250914 voxels (out of 271633 = 92.4%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (292x20): 4.3586 ++ VERY GOOD ++
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub-09+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.957488
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb02.sub-09.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
+ masked off 45658 voxels for being all zero; 225975 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.53
+ X matrix: 78.938% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=292 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.55
+ average case bandwidth = 16.10
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb02.sub-09.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-09.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb03.sub-09.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb02.sub-09.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb02.sub-09.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb03.sub-09.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb03.sub-09.r02.scale+tlrc.BRIK
end
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -demean -write motion_demean.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile motion_demean.1D -set_nruns 2 -split_into_pad_runs mot_demean
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.01
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=19.81
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -show_censor_count -censor_prev_TR -censor_motion 0.3 motion_sub-09
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
total number of censored TRs (simple form) = 0
set ktrs = `1d_tool.py -infile motion_${subj}_censor.1D
-show_trs_uncensored encoded`
1d_tool.py -infile motion_sub-09_censor.1D -show_trs_uncensored encoded
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dDeconvolve -input pb03.sub-09.r01.scale+tlrc.HEAD pb03.sub-09.r02.scale+tlrc.HEAD -censor motion_sub-09_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym SYM: +incongruent -congruent -glt_label 1 incongruent-congruent -gltsym SYM: +congruent -incongruent -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-09 -errts errts.sub-09 -bucket stats.sub-09
++ 3dDeconvolve extending num_stimts from 2 to 14 due to -ortvec
++ 3dDeconvolve: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ loading dataset pb03.sub-09.r01.scale+tlrc.HEAD pb03.sub-09.r02.scale+tlrc.HEAD
++ Auto-catenated input datasets treated as multiple imaging runs
++ Auto-catenated datasets start at: 0 146
++ STAT automask has 253235 voxels (out of 271633 = 93.2%)
++ Skipping check for initial transients
++ Input polort=2; Longest run=292.0 s; Recommended minimum polort=2 ++ OK ++
++ -stim_times using TR=2 s for stimulus timing conversion
++ -stim_times using TR=2 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
------------------------------------------------------------
GLT matrix from 'SYM: +incongruent -congruent':
0 0 0 0 0 0 -1 1 0 0 0 0 0 0 0 0 0 0 0 0
------------------------------------------------------------
GLT matrix from 'SYM: +congruent -incongruent':
0 0 0 0 0 0 1 -1 0 0 0 0 0 0 0 0 0 0 0 0
++ Number of time points: 292 (no censoring)
+ Number of parameters: 20 [18 baseline ; 2 signal]
++ total shared memory needed = 648,659,604 bytes (about 649 million)
++ mmap() memory allocated: 648,659,604 bytes (about 649 million)
++ Memory required for output bricks = 648,659,604 bytes (about 649 million)
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.01
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=19.44
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ Wrote matrix image to file X.jpg
++ Wrote matrix values to file X.xmat.1D
++ ========= Things you can do with the matrix file =========
++ (a) Linear regression with ARMA(1,1) modeling of serial correlation:
3dREMLfit -matrix X.xmat.1D \
-input "pb03.sub-09.r01.scale+tlrc.HEAD pb03.sub-09.r02.scale+tlrc.HEAD" \
-fout -tout -Rbuck stats.sub-09_REML -Rvar stats.sub-09_REMLvar \
-Rfitts fitts.sub-09_REML -Rerrts errts.sub-09_REML -verb
++ N.B.: 3dREMLfit command above written to file stats.REML_cmd
++ (b) Visualization/analysis of the matrix via ExamineXmat.R
++ (c) Synthesis of sub-model datasets using 3dSynthesize
++ ==========================================================
++ Wrote matrix values to file X.nocensor.xmat.1D
++ ----- Signal+Baseline matrix condition [X] (292x20): 5.34683 ++ VERY GOOD ++
++ ----- Signal-only matrix condition [X] (292x2): 1 ++ VERY GOOD ++
++ ----- Baseline-only matrix condition [X] (292x18): 5.33616 ++ VERY GOOD ++
++ ----- stim_base-only matrix condition [X] (292x12): 3.61502 ++ VERY GOOD ++
++ ----- polort-only matrix condition [X] (292x6): 1 ++ VERY GOOD ++
++ +++++ Matrix inverse average error = 3.94502e-15 ++ VERY GOOD ++
++ Matrix setup time = 0.67 s
++ Voxels in dataset: 271633
++ Voxels per job: 33954
++ Job #1: processing voxels 33954 to 67907; elapsed time=3.728
++ Job #2: processing voxels 67908 to 101861; elapsed time=3.740
++ Job #3: processing voxels 101862 to 135815; elapsed time=3.752
++ Job #4: processing voxels 135816 to 169769; elapsed time=3.765
++ Job #5: processing voxels 169770 to 203723; elapsed time=3.777
++ Job #6: processing voxels 203724 to 237677; elapsed time=3.789
++ Job #7: processing voxels 237678 to 271632; elapsed time=3.801
++ Job #0: processing voxels 0 to 33953; elapsed time=3.811
++ voxel loop:0123456789.0123456789.0123456789.0123456789.01234567++ Job #6 finished; elapsed time=7.889
8++ Job #2 finished; elapsed time=7.938
++ Job #1 finished; elapsed time=7.965
++ Job #7 finished; elapsed time=7.983
++ Job #4 finished; elapsed time=7.985
++ Job #5 finished; elapsed time=7.996
9.
++ Job #0 waiting for children to finish; elapsed time=8.001
++ Job #3 finished; elapsed time=8.006
++ Job #0 now finishing up; elapsed time=8.012
++ Smallest FDR q [0 Full_Fstat] = 1.80696e-12
++ Smallest FDR q [2 congruent#0_Tstat] = 5.0961e-11
++ Smallest FDR q [3 congruent_Fstat] = 5.096e-11
++ Smallest FDR q [5 incongruent#0_Tstat] = 4.13492e-12
++ Smallest FDR q [6 incongruent_Fstat] = 4.13525e-12
*+ WARNING: Smallest FDR q [8 incongruent-congruent_GLT#0_Tstat] = 0.147261 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_GLT_Fstat] = 0.146895 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [11 congruent-incongruent_GLT#0_Tstat] = 0.147261 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_GLT_Fstat] = 0.146895 ==> few true single voxel detections
++ Wrote bucket dataset into ./stats.sub-09+tlrc.BRIK
+ created 9 FDR curves in bucket header
++ Wrote 3D+time dataset into ./fitts.sub-09+tlrc.BRIK
++ Wrote 3D+time dataset into ./errts.sub-09+tlrc.BRIK
++ Program finished; elapsed time=19.794
if ( 0 != 0 ) then
1d_tool.py -show_cormat_warnings -infile X.xmat.1D
tee out.cormat_warn.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- no warnings for correlation matrix (cut = 0.400) --
1d_tool.py -show_df_info -infile X.xmat.1D
tee out.df_info.txt
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
initial DF : 292 : 100.0%
DF used for regs of interest : 2 : 0.7%
DF used for censoring : 0 : 0.0%
DF used for polort : 6 : 2.1%
DF used for motion : 12 : 4.1%
total DF used : 20 : 6.8%
final DF : 272 : 93.2%
tcsh -x stats.REML_cmd
3dREMLfit -matrix X.xmat.1D -input pb03.sub-09.r01.scale+tlrc.HEAD pb03.sub-09.r02.scale+tlrc.HEAD -fout -tout -Rbuck stats.sub-09_REML -Rvar stats.sub-09_REMLvar -Rfitts fitts.sub-09_REML -Rerrts errts.sub-09_REML -verb
++ 3dREMLfit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWCox
++ Number of OpenMP threads = 15
++ No mask ==> computing for all 271633 voxels
++ FDR automask has 253235 voxels (out of 271633 = 93.2%)
++ shortest run = 146 longest run = 146
++ ----- matrix condition (292x20): 5.34683 ++ VERY GOOD ++
+ masked off 44398 voxels for being all zero; 227235 left in mask
++ starting REML setup calculations; total CPU=0.00 Elapsed=2.29
+ X matrix: 79.110% of elements are nonzero
+ starting 15 OpenMP threads for REML setup
+ REML setup finished: matrix rows=292 cols=20; 109*1 cases; total CPU=0.00 Elapsed=2.30
+ average case bandwidth = 16.10
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=45.95
++ Output dataset ./stats.sub-08_REMLvar+tlrc.BRIK
++ Output dataset ./fitts.sub-08_REML+tlrc.BRIK
++ Output dataset ./errts.sub-08_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-08_REML+tlrc
++ Smallest FDR q [0 Full_Fstat] = 5.40643e-12
++ Smallest FDR q [2 congruent#0_Tstat] = 1.58242e-09
++ Smallest FDR q [3 congruent_Fstat] = 1.58242e-09
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=46.17
++ Output dataset ./stats.sub-07_REMLvar+tlrc.BRIK
++ Smallest FDR q [5 incongruent#0_Tstat] = 2.02374e-10
++ Output dataset ./fitts.sub-07_REML+tlrc.BRIK
++ Smallest FDR q [6 incongruent_Fstat] = 2.02374e-10
++ Output dataset ./errts.sub-07_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-07_REML+tlrc
++ Smallest FDR q [0 Full_Fstat] = 8.17708e-12
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.200666 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.200666 ==> few true single voxel detections
++ Smallest FDR q [2 congruent#0_Tstat] = 6.00692e-09
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.200666 ==> few true single voxel detections
++ Smallest FDR q [3 congruent_Fstat] = 6.00692e-09
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.200666 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-08_REML+tlrc
++ Output dataset ./stats.sub-08_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=56.37
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-08 pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ REML voxel loop: [15 threads]0123456789.0123456789.0123456789.0123456789.01
+ ARMA voxel parameters estimated: total CPU=0.00 Elapsed=15.90
++ Ljung-Box max lag parameter h = 37 (35 chi-squared DOF)
++ elapsed time = 0.6 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-08+tlrc[0..180,183..236,239..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Smallest FDR q [5 incongruent#0_Tstat] = 9.6685e-11
++ Smallest FDR q [6 incongruent_Fstat] = 9.6685e-11
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-08_REML+tlrc[0..180,183..236,239..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-08
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-08+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-08_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.0655013
++ Smallest FDR q [9 incongruent-congruent_Fstat] = 0.0655013
++ Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.0655013
++ Smallest FDR q [12 congruent-incongruent_Fstat] = 0.0655013
+ Added 9 FDR curves to dataset stats.sub-07_REML+tlrc
++ Output dataset ./stats.sub-07_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=57.70
++ Output dataset ./rm.errts.unit+tlrc.BRIK
if ( 0 != 0 ) then
3dmaskave -quiet -mask full_mask.sub-08+tlrc rm.errts.unit+tlrc
3dTcat -prefix all_runs.sub-07 pb03.sub-07.r01.scale+tlrc.HEAD pb03.sub-07.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 68948 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.0804356
3dmaskave -quiet -mask full_mask.sub-08+tlrc errts.sub-08_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 68948 voxels survive the mask
++ elapsed time = 0.6 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-07+tlrc[0..291]
3dTcorr1D -prefix corr_brain errts.sub-08_REML+tlrc mean.errts.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-08_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-08_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-07_REML+tlrc[0..291]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: THD_Tcorr1D: 47222 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-07
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-08.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-07+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-07_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-08+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68948
++ detrending start: 11 baseline funcs, 146 time points
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-07+tlrc rm.errts.unit+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 71768 voxels survive the mask
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.0512396
3dmaskave -quiet -mask full_mask.sub-07+tlrc errts.sub-07_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 71768 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-07_REML+tlrc mean.errts.1D
+ detrending done (0.00 CPU s thus far)
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-07_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-07_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
++ start ACF calculations out to radius = 17.73 mm
*+ WARNING: THD_Tcorr1D: 45658 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-07.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-07+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71768
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..180,183..236,239..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-08+tlrc[146..180,183..236,239..291]
+ detrending done (0.00 CPU s thus far)
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68948
++ detrending start: 11 baseline funcs, 142 time points
++ start ACF calculations out to radius = 17.20 mm
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.90 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-07+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71768
++ detrending start: 11 baseline funcs, 146 time points
++ GLSQ loop:0123456789.0123456789.0123456789.0123456789.0123456789.
+ GLSQ regression done: total CPU=0.00 Elapsed=40.37
++ Output dataset ./stats.sub-09_REMLvar+tlrc.BRIK
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ Output dataset ./fitts.sub-09_REML+tlrc.BRIK
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.781039 3.35335 12.4825 8.57985
average epits ACF blurs: 0.781039 3.35335 12.4825 8.57985
echo 0.781039 3.35335 12.4825 8.57985 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
+ detrending done (0.00 CPU s thus far)
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-08+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Output dataset ./errts.sub-09_REML+tlrc.BRIK
++ creating FDR curves in dataset stats.sub-09_REML+tlrc
++ Number of voxels in mask = 68948
++ Smallest FDR q [0 Full_Fstat] = 2.0264e-12
++ start ACF calculations out to radius = 17.20 mm
++ detrending start: 11 baseline funcs, 146 time points
++ Smallest FDR q [2 congruent#0_Tstat] = 6.91554e-11
++ Smallest FDR q [3 congruent_Fstat] = 6.91554e-11
+ detrending done (0.00 CPU s thus far)
++ Smallest FDR q [5 incongruent#0_Tstat] = 8.08567e-12
++ Smallest FDR q [6 incongruent_Fstat] = 8.08567e-12
++ start ACF calculations out to radius = 17.68 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.870719 3.31332 13.6087 8.25561
average epits ACF blurs: 0.870719 3.31332 13.6087 8.25561
echo 0.870719 3.31332 13.6087 8.25561 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-07+tlrc[0..145]
*+ WARNING: Smallest FDR q [8 incongruent-congruent#0_Tstat] = 0.375836 ==> few true single voxel detections
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71768
*+ WARNING: Smallest FDR q [9 incongruent-congruent_Fstat] = 0.375837 ==> few true single voxel detections
++ detrending start: 11 baseline funcs, 146 time points
*+ WARNING: Smallest FDR q [11 congruent-incongruent#0_Tstat] = 0.375836 ==> few true single voxel detections
*+ WARNING: Smallest FDR q [12 congruent-incongruent_Fstat] = 0.375837 ==> few true single voxel detections
+ Added 9 FDR curves to dataset stats.sub-09_REML+tlrc
++ Output dataset ./stats.sub-09_REML+tlrc.BRIK
+ unloading input dataset and REML matrices
++ 3dREMLfit is all done! total CPU=0.00 Elapsed=50.52
if ( 0 != 0 ) then
3dTcat -prefix all_runs.sub-09 pb03.sub-09.r01.scale+tlrc.HEAD pb03.sub-09.r02.scale+tlrc.HEAD
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ detrending done (0.00 CPU s thus far)
++ elapsed time = 0.6 s
3dTstat -mean -prefix rm.signal.all all_runs.sub-09+tlrc[0..291]
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ start ACF calculations out to radius = 17.13 mm
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..180,183..236,239..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-08+tlrc[146..180,183..236,239..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68948
++ Output dataset ./rm.signal.all+tlrc.BRIK
3dTstat -stdev -prefix rm.noise.all errts.sub-09_REML+tlrc[0..291]
++ detrending start: 11 baseline funcs, 142 time points
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.noise.all+tlrc.BRIK
3dcalc -a rm.signal.all+tlrc -b rm.noise.all+tlrc -expr a/b -prefix TSNR.sub-09
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./TSNR.sub-09+tlrc.BRIK
3dTnorm -norm2 -prefix rm.errts.unit errts.sub-09_REML+tlrc
++ 3dTnorm: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.89 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ Output dataset ./rm.errts.unit+tlrc.BRIK
3dmaskave -quiet -mask full_mask.sub-09+tlrc rm.errts.unit+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-07+tlrc[146..291]
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
+++ 71499 voxels survive the mask
++ Number of voxels in mask = 71768
3dTstat -sos -prefix - mean.errts.unit.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo -- GCOR = `cat out.gcor.1D`
cat out.gcor.1D
-- GCOR = 0.037331
3dmaskave -quiet -mask full_mask.sub-09+tlrc errts.sub-09_REML+tlrc
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ detrending start: 11 baseline funcs, 146 time points
+++ 71499 voxels survive the mask
3dTcorr1D -prefix corr_brain errts.sub-09_REML+tlrc mean.errts.1D
++ 3dTcorr1D: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ reading dataset file errts.sub-09_REML+tlrc
+ reading 1D file mean.errts.1D
+ loading dataset ./errts.sub-09_REML+tlrc.BRIK into memory
++ Start correlations: 271633 voxels X 1 time series(292); 1 threads
*+ WARNING: THD_Tcorr1D: 44398 voxels skipped because were constant in time
++ Wrote dataset: ./corr_brain+tlrc.BRIK
1dcat X.nocensor.xmat.1D[6]
+ detrending done (0.00 CPU s thus far)
1dcat X.nocensor.xmat.1D[7]
1d_tool.py -infile X.nocensor.xmat.1D -write_xstim X.stim.xmat.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dTstat -sum -prefix sum_ideal.1D X.stim.xmat.1D
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Output dataset ./sum_ideal.1D
touch blur_est.sub-09.1D
mkdir files_ACF
touch blur.epits.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r01.1D all_runs.sub-09+tlrc[0..145]
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ start ACF calculations out to radius = 17.16 mm
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
++ Number of voxels in mask = 71499
echo average errts ACF blurs: 0.792866 3.35606 12.7234 8.55597
average errts ACF blurs: 0.792866 3.35606 12.7234 8.55597
echo 0.792866 3.35606 12.7234 8.55597 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
++ detrending start: 11 baseline funcs, 146 time points
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-08_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68948
++ detrending start: 11 baseline funcs, 146 time points
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.19 mm
++ start ACF calculations out to radius = 17.66 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.880099 3.31029 13.8739 8.22803
average errts ACF blurs: 0.880099 3.31029 13.8739 8.22803
echo 0.880099 3.31029 13.8739 8.22803 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-07_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71768
++ detrending start: 11 baseline funcs, 146 time points
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.epits.r02.1D all_runs.sub-09+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71499
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ detrending start: 11 baseline funcs, 146 time points
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..180,183..236,239..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-08+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-08_REML+tlrc[146..180,183..236,239..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 68948
++ detrending start: 11 baseline funcs, 142 time points
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.11 mm
+ detrending done (0.00 CPU s thus far)
+ detrending done (0.00 CPU s thus far)
++ start ACF calculations out to radius = 17.14 mm
++ start ACF calculations out to radius = 17.85 mm
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-07+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-07_REML+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.epits.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ Number of voxels in mask = 71768
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.epits.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{0..$(2)}'
++ detrending start: 11 baseline funcs, 146 time points
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits FWHM blurs: 0 0 0 0
average epits FWHM blurs: 0 0 0 0
echo 0 0 0 0 # epits FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.epits.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.epits.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average epits ACF blurs: 0.897711 3.32134 15.8151 8.22927
average epits ACF blurs: 0.897711 3.32134 15.8151 8.22927
echo 0.897711 3.32134 15.8151 8.22927 # epits ACF blur estimates
touch blur.errts.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r01.1D errts.sub-09+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71499
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ detrending start: 11 baseline funcs, 146 time points
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.793667 3.35254 12.6561 8.54195
average err_reml ACF blurs: 0.793667 3.35254 12.6561 8.54195
echo 0.793667 3.35254 12.6561 8.54195 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-08 -dsets pb00.sub-08.r01.tcat+orig.HEAD pb00.sub-08.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-08+tlrc.HEAD -ss_review_dset out.ss_review.sub-08.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-08.txt
subject ID : sub-08
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-08+tlrc.HEAD
final stats dset : stats.sub-08_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
+ detrending done (0.00 CPU s thus far)
num TRs above mot limit : 2
average motion (per TR) : 0.0767002
average censored motion : 0.0744091
++ start ACF calculations out to radius = 17.13 mm
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 0.868807
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 0.868807
++ start ACF calculations out to radius = 17.15 mm
average outlier frac (TR) : 0.00122644
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 142
num TRs per run (censored): 0 4
fraction censored per run : 0 0.0273973
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 288
degrees of freedom used : 20
degrees of freedom left : 268
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 4
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
censor fraction : 0.013699
num regs of interest : 2
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.880565 3.30767 13.8377 8.22071
average err_reml ACF blurs: 0.880565 3.30767 13.8377 8.22071
echo 0.880565 3.30767 13.8377 8.22071 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-07 -dsets pb00.sub-07.r01.tcat+orig.HEAD pb00.sub-07.r02.tcat+orig.HEAD
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-07+tlrc.HEAD -ss_review_dset out.ss_review.sub-07.txt -write_uvars_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.errts.r02.1D errts.sub-09+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71499
num TRs per stim (orig) : 120 110
num TRs censored per stim : 0 4
fraction TRs censored : 0.000 0.036
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
++ detrending start: 11 baseline funcs, 146 time points
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-07.txt
subject ID : sub-07
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-07+tlrc.HEAD
final stats dset : stats.sub-07_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 0
average motion (per TR) : 0.0422039
average censored motion : 0.0422039
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
ave mot per sresp (orig) : 0.070899 0.079812
max motion displacement : 1.18663
ave mot per sresp (cens) : 0.070899 0.073748
TSNR average : 139.227
global correlation (GCOR) : 0.0804356
anat/EPI mask Dice coef : 0.928854
anat/templ mask Dice coef : 0.960445
maximum F-stat (masked) : 55.4792
blur estimates (ACF) : 0.793667 3.35254 12.6561
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.18663
average outlier frac (TR) : 0.000896575
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 37 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
num runs found : 2
num TRs per run : 146 146
tcsh @ss_review_html
tee out.review_html
+ detrending done (0.00 CPU s thus far)
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ start ACF calculations out to radius = 17.10 mm
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
censor fraction : 0.000000
num regs of interest : 2
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 829.000000 -func_range 829.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-08/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-08/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-08/media/__tmp_chauf_udHP0cMFvlB
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 829.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_udHP0cMFvlB/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_udHP0cMFvlB/tmp_olay.nii
++ User-entered function range value value (829.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
num TRs per stim (orig) : 120 110
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..54 y=0..59 z=0..36
++ 3dAutobox: output dataset = QC_sub-08/media/__tmp_chauf_udHP0cMFvlB/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-08/media/__tmp_chauf_udHP0cMFvlB/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_YIss-dm9AsfUETtrDyCZLw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_YIss-dm9AsfUETtrDyCZLw+orig.BRIK
++ 99900 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 1.5 8.945793 -26.24097
++ Will have the ref box central gapord: 6 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :977
[1] 2582713
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
ave mot per sresp (orig) : 0.042234 0.041772
ave mot per sresp (cens) : 0.042234 0.041772
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.errts.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
TSNR average : 144.539
global correlation (GCOR) : 0.0512396
anat/EPI mask Dice coef : 0.943059
anat/templ mask Dice coef : 0.96351
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.errts.r02.1D.png
maximum F-stat (masked) : 54.439
blur estimates (ACF) : 0.880565 3.30767 13.8377
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
end
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{0..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts FWHM blurs: 0 0 0 0
average errts FWHM blurs: 0 0 0 0
echo 0 0 0 0 # errts FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.errts.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.errts.1D{1..$(2)}'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average errts ACF blurs: 0.906301 3.31954 16.4363 8.20629
average errts ACF blurs: 0.906301 3.31954 16.4363 8.20629
echo 0.906301 3.31954 16.4363 8.20629 # errts ACF blur estimates
touch blur.err_reml.1D
foreach run ( 01 02 )
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 01
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 0..145 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D errts.sub-09_REML+tlrc[0..145]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 37 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
++ Number of voxels in mask = 71499
++ detrending start: 11 baseline funcs, 146 time points
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 858.000000 -func_range 858.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-07/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-07/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-07/media/__tmp_chauf_VpA52C1KIKO
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 858.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_VpA52C1KIKO/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_VpA52C1KIKO/tmp_olay.nii
++ User-entered function range value value (858.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=8..54 y=2..59 z=0..37
++ 3dAutobox: output dataset = QC_sub-07/media/__tmp_chauf_VpA52C1KIKO/ulay_box_0.nii
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_udHP0cMFvlB++ Writing palette image to QC_sub-08/media/qc_00_vorig_EPI.pbar.jpg
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_00_vorig_EPI.sag.jpg'
+ detrending done (0.00 CPU s thus far)
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:6:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-08/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 829.000000 -com SET_FUNC_RANGE 829.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 1.5 8.945793 -26.24097 -com SAVE_JPEG sagittalimage QC_sub-08/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-08/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-08/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-08/media/__tmp_chauf_udHP0cMFvlB
+* Removing temporary image directory 'QC_sub-08/media/__tmp_chauf_udHP0cMFvlB'.
[1] Done Xvfb :977 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-08/media/qc_00_vorig_EPI*
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-07/media/__tmp_chauf_VpA52C1KIKO/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_5XrrkwBRZIQr9voTaOrt3Q argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_5XrrkwBRZIQr9voTaOrt3Q+orig.BRIK
++ 103588 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 4.5 10.2831 -17.18073
++ Will have the ref box central gapord: 6 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :498
[1] 2584501
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ start ACF calculations out to radius = 17.13 mm
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.3
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 125.049599
+ Used gradual clip level = 113.553802 .. 136.496994
+ Number voxels above clip level = 226551
+ Clustering voxels ...
+ Largest cluster has 226334 voxels
+ Clustering voxels ...
+ Largest cluster has 221333 voxels
+ Filled 8457 voxels in small holes; now have 229790 voxels
+ Filled 5358 voxels in large holes; now have 235148 voxels
+ Clustering voxels ...
+ Largest cluster has 235139 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 667426 voxels
+ Mask now has 235203 voxels
++ 235203 voxels in the mask [out of 902629: 26.06%]
++ first 9 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 10 y-planes are zero [from P]
++ last 8 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 11 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 38.126259 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_NiiqeAPQrDP
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.340431, 853.099365]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_NiiqeAPQrDP/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_NiiqeAPQrDP/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 99.547501
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_NiiqeAPQrDP/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_NiiqeAPQrDP/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_QX8CjFzRu8AUYnbM9UIj-A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_QX8CjFzRu8AUYnbM9UIj-A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :213
[1] 2585204
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_VpA52C1KIKO++ Writing palette image to QC_sub-07/media/qc_00_vorig_EPI.pbar.jpg
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:6:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-07/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 858.000000 -com SET_FUNC_RANGE 858.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 4.5 10.2831 -17.18073 -com SAVE_JPEG sagittalimage QC_sub-07/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-07/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-07/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-07/media/__tmp_chauf_VpA52C1KIKO
+* Removing temporary image directory 'QC_sub-07/media/__tmp_chauf_VpA52C1KIKO'.
[1] Done Xvfb :498 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-07/media/qc_00_vorig_EPI*
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r01.1D.png
end
set trs = `1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded
-show_trs_run $run`
1d_tool.py -infile X.xmat.1D -show_trs_uncensored encoded -show_trs_run 02
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
if ( 146..291 == ) continue
3dFWHMx -detrend -mask full_mask.sub-09+tlrc -ACF files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D errts.sub-09_REML+tlrc[146..291]
++ 3dFWHMx: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: The Bob
++ Number of voxels in mask = 71499
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ detrending start: 11 baseline funcs, 146 time points
++ Output dataset ./QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 125.069000
+ Used gradual clip level = 111.492500 .. 136.488007
+ Number voxels above clip level = 224405
+ Clustering voxels ...
+ Largest cluster has 224287 voxels
+ Clustering voxels ...
+ Largest cluster has 220438 voxels
+ Filled 9447 voxels in small holes; now have 229885 voxels
+ Filled 6071 voxels in large holes; now have 235956 voxels
+ Clustering voxels ...
+ Largest cluster has 235952 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 666422 voxels
+ Mask now has 236207 voxels
++ 236207 voxels in the mask [out of 902629: 26.17%]
++ first 9 x-planes are zero [from L]
++ last 8 x-planes are zero [from R]
++ first 10 y-planes are zero [from P]
++ last 7 y-planes are zero [from A]
++ first 2 z-planes are zero [from I]
++ last 11 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 37.695633 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_nWauLhOaCgW
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.302927, 886.176819]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_nWauLhOaCgW/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_nWauLhOaCgW/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 97.238632
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_nWauLhOaCgW/ulay_box_0.nii
+ detrending done (0.00 CPU s thus far)
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_nWauLhOaCgW/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_-cdJRGF5kb439SoVrFERSQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_-cdJRGF5kb439SoVrFERSQ+tlrc.BRIK
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_NiiqeAPQrDP++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
AFNI QUITTs!
------------------- end of optionizing -------------------
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.340431 853.099365 -com SET_FUNC_RANGE 99.547501 -com SET_THRESHNEW 38.126259 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_NiiqeAPQrDP
-- trying to start Xvfb :540
[1] 2586174
+* Removing temporary image directory '../__tmp_chauf_NiiqeAPQrDP'.
[1] Done Xvfb :213 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-08/media/__workdir_EAC_WrvHJCBfgNq*'
++ DONE! Image output:
QC_sub-08/media/qc_01_ve2a_epi2anat
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ start ACF calculations out to radius = 17.07 mm
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__workdir_EAC_W2nrR9q6CsF/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__workdir_EAC_W2nrR9q6CsF/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-08/media/__workdir_EAC_W2nrR9q6CsF/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_hhY4oNB9Acw
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 481.832880]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_hhY4oNB9Acw/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_hhY4oNB9Acw/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_hhY4oNB9Acw/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_hhY4oNB9Acw/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_ZSkMCkprRqfw66flgrNAWQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_ZSkMCkprRqfw66flgrNAWQ+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :687
[1] 2586939
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_nWauLhOaCgW++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.302927 886.176819 -com SET_FUNC_RANGE 97.238632 -com SET_THRESHNEW 37.695633 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_nWauLhOaCgW
+* Removing temporary image directory '../__tmp_chauf_nWauLhOaCgW'.
[1] Done Xvfb :540 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-07/media/__workdir_EAC_g4DqTV6ZB59*'
++ DONE! Image output:
QC_sub-07/media/qc_01_ve2a_epi2anat
+ ACF done (0.00 CPU s thus far)
++ ACF 1D file [radius ACF mixed_model gaussian_NEWmodel] written to files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D
++ 1dplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RWC et al.
+ and 1dplot-ed to file files_ACF/out.3dFWHMx.ACF.err_reml.r02.1D.png
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
end
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{0..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{0..$(2)}'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml FWHM blurs: 0 0 0 0
average err_reml FWHM blurs: 0 0 0 0
echo 0 0 0 0 # err_reml FWHM blur estimates
set blurs = ( `3dTstat -mean -prefix - blur.err_reml.1D'{1..$(2)}'\'` )
3dTstat -mean -prefix - blur.err_reml.1D{1..$(2)}'
++ Output dataset ./QC_sub-07/media/__workdir_EAC_JWINL19NdEE/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
echo average err_reml ACF blurs: 0.907516 3.31665 16.4517 8.19683
average err_reml ACF blurs: 0.907516 3.31665 16.4517 8.19683
echo 0.907516 3.31665 16.4517 8.19683 # err_reml ACF blur estimates
gen_epi_review.py -script @epi_review.sub-09 -dsets pb00.sub-09.r01.tcat+orig.HEAD pb00.sub-09.r02.tcat+orig.HEAD
++ Output dataset ./QC_sub-07/media/__workdir_EAC_JWINL19NdEE/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
gen_ss_review_scripts.py -exit0 -mot_limit 0.3 -mask_dset full_mask.sub-09+tlrc.HEAD -ss_review_dset out.ss_review.sub-09.txt -write_uvars_json out.ss_review_uvars.json
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-07/media/__workdir_EAC_JWINL19NdEE/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ writing ss review basic: @ss_review_basic
** no mask_dset dset, cannot drive view_stats, skipping...
++ writing ss review driver: @ss_review_driver
++ writing ss review drive commands: @ss_review_driver_commands
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.errts.unit+tlrc.BRIK rm.errts.unit+tlrc.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.noise.all+tlrc.BRIK rm.noise.all+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD rm.signal.all+tlrc.BRIK rm.signal.all+tlrc.HEAD
if ( -e @ss_review_basic ) then
./@ss_review_basic
tee out.ss_review.sub-09.txt
subject ID : sub-09
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-09+tlrc.HEAD
final stats dset : stats.sub-09_REML+tlrc.HEAD
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_YKe19jcmBi7
num TRs above mot limit : 0
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
average motion (per TR) : 0.057909
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 470.007051]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_YKe19jcmBi7/tmp_ulay.nii
average censored motion : 0.057909
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_YKe19jcmBi7/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_YKe19jcmBi7/ulay_box_0.nii
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 0.850377
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_YKe19jcmBi7/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_l13B6Wwl9No3MYxXIvJtnA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 0.850377
++ Output dataset /tmp/3dcalc_XYZ_l13B6Wwl9No3MYxXIvJtnA+tlrc.BRIK
average outlier frac (TR) : 0.000870959
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :671
[1] 2588414
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_hhY4oNB9Acw++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 481.832880 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_hhY4oNB9Acw
+* Removing temporary image directory '../__tmp_chauf_hhY4oNB9Acw'.
[1] Done Xvfb :687 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-08/media/__workdir_EAC_W2nrR9q6CsF*'
++ DONE! Image output:
QC_sub-08/media/qc_02_va2t_anat2temp
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-08_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 16.352385 -thr_olay 4.907798 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-08/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-08/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-08_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-08/media/__tmp_chauf_uemz0o9w0xw
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
censor fraction : 0.000000
num regs of interest : 2
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_uemz0o9w0xw/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_uemz0o9w0xw/tmp_olay.nii
++ User-entered function range value value (16.352385)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-08/media/__tmp_chauf_uemz0o9w0xw/ulay_box_0.nii
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-08/media/__tmp_chauf_uemz0o9w0xw/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_X7rBgPiiVM3aZujaLlCDrQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_X7rBgPiiVM3aZujaLlCDrQ+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :92
[1] 2590467
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
ave mot per sresp (orig) : 0.058195 0.056895
ave mot per sresp (cens) : 0.058195 0.056895
TSNR average : 133.674
global correlation (GCOR) : 0.037331
anat/EPI mask Dice coef : 0.924751
anat/templ mask Dice coef : 0.957488
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_YKe19jcmBi7++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 470.007051 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_YKe19jcmBi7
+* Removing temporary image directory '../__tmp_chauf_YKe19jcmBi7'.
[1] Done Xvfb :671 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-07/media/__workdir_EAC_JWINL19NdEE*'
++ DONE! Image output:
QC_sub-07/media/qc_02_va2t_anat2temp
maximum F-stat (masked) : 68.6935
blur estimates (ACF) : 0.907516 3.31665 16.4517
blur estimates (FWHM) : 0 0 0
apqc_make_tcsh.py -review_style pythonic -subj_dir . -uvar_json out.ss_review_uvars.json
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Found 38 files for QCing.
++ Done making (executable) script to generate HTML QC:
./@ss_review_html
tcsh @ss_review_html
tee out.review_html
*+ Found main dset (template): /opt/afni-latest/MNI_avg152T1+tlrc
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-07_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 17.298092 -thr_olay 6.020756 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-07/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-07/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-07_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
bc: Command not found.
bc: Command not found.
bc: Command not found.
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0/tmp_olay.nii
++ User-entered function range value value (17.298092)
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay vr_base_min_outlier+orig.HEAD -olay vr_base_min_outlier+orig.HEAD -ulay_range_nz 0 773.000000 -func_range 773.000000 -box_focus_slices AMASK_FOCUS_ULAY -cbar gray_scale -pbar_posonly -pbar_saveim QC_sub-09/media/qc_00_vorig_EPI.pbar.jpg -pbar_comm_range 98%ile in vol -pbar_for dset -prefix QC_sub-09/media/qc_00_vorig_EPI -save_ftype JPEG -blowup 4 -opacity 9 -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Found input file: vr_base_min_outlier+orig.HEAD
++ Using blowup factor: 4
++ Using opacity: 9
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Making temporary work directory to copy vis files: QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ
++ Converted 0 to labels
++ Final subbrick indices: -1 -1 -1
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0, 773.000000]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 3dAutobox: output dataset = QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0/ulay_box_0.nii
++ Output dataset ./QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ/tmp_olay.nii
++ User-entered function range value value (773.000000)
++ Dimensions (xyzt): 64 64 40 1
++ (initial) Slice spacing ordered (x,y,z) is: 9 9 5
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_BpYv0tsdt2p6xAJn7WfYaw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=7..56 y=3..61 z=0..36
++ 3dAutobox: output dataset = QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ/ulay_box_0.nii
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_BpYv0tsdt2p6xAJn7WfYaw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :300
[1] 2592839
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_uemz0o9w0xw++ Writing palette image to QC_sub-08/media/qc_03_vstat_Full_Fstat.pbar.jpg
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_G8QZpD7KCicGLGG5Onp69w argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_G8QZpD7KCicGLGG5Onp69w+orig.BRIK
++ 109150 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 7.283096 -0.054207 -13.95181
++ Will have the ref box central gapord: 7 8 5
------------------- end of optionizing -------------------
-- trying to start Xvfb :790
[1] 2592969
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-08/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 16.352385 -com SET_THRESHNEW 4.907798 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-08/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-08/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-08/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-08/media/__tmp_chauf_uemz0o9w0xw
+* Removing temporary image directory 'QC_sub-08/media/__tmp_chauf_uemz0o9w0xw'.
[1] Done Xvfb :92 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-08/media/qc_03_vstat_Full_Fstat*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-08_REML+tlrc.HEAD
+ Number of voxels in mask = 68948
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0++ Writing palette image to QC_sub-07/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-07/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 17.298092 -com SET_THRESHNEW 6.020756 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-07/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-07/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-07/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0
+* Removing temporary image directory 'QC_sub-07/media/__tmp_chauf_ssFqqJuqKz0'.
[1] Done Xvfb :300 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-07/media/qc_03_vstat_Full_Fstat*
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ++ Writing palette image to QC_sub-09/media/qc_00_vorig_EPI.pbar.jpg
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_00_vorig_EPI.sag.jpg'
++ Writing one 1816x212 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_00_vorig_EPI.cor.jpg'
++ Writing one 1816x256 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_00_vorig_EPI.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:7:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:8:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:5:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 gray_scale -com PBAR_SAVEIM QC_sub-09/media/qc_00_vorig_EPI.pbar.jpg dim=64x512H -com SET_SUBBRICKS -1 -1 -1 -com SET_ULAY_RANGE A.all 0 773.000000 -com SET_FUNC_RANGE 773.000000 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 7.283096 -0.054207 -13.95181 -com SAVE_JPEG sagittalimage QC_sub-09/media/qc_00_vorig_EPI.sag blowup=4 -com SAVE_JPEG coronalimage QC_sub-09/media/qc_00_vorig_EPI.cor blowup=4 -com SAVE_JPEG axialimage QC_sub-09/media/qc_00_vorig_EPI.axi blowup=4 -com QUITT QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ
+* Removing temporary image directory 'QC_sub-09/media/__tmp_chauf_GABbbCwHnBZ'.
++ DONE (good exit)
see: QC_sub-09/media/qc_00_vorig_EPI*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__workdir_EAC_jPRgefnngt1/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__workdir_EAC_jPRgefnngt1/eac_0_cp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__workdir_EAC_jPRgefnngt1/eac_0_cp.nii
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-07_REML+tlrc.HEAD
++ Output dataset QC_sub-09/media/__workdir_EAC_jPRgefnngt1/eac_1_grid2olay.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ Output dataset ./eac_2_ulay_shrp.nii
++ Just copy olay, bc ulay will get regridded
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+ Number of voxels in mask = 71768
++ Output dataset ./QC_sub-09/media/__workdir_EAC_jPRgefnngt1/eac_3_cp.nii
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_3_cp.nii
++ Forming automask
+ Fixed clip level = 104.504402
+ Used gradual clip level = 97.497398 .. 110.996803
+ Number voxels above clip level = 219051
+ Clustering voxels ...
+ Largest cluster has 218758 voxels
+ Clustering voxels ...
+ Largest cluster has 212824 voxels
+ Filled 13536 voxels in small holes; now have 226360 voxels
+ Filled 8720 voxels in large holes; now have 235080 voxels
+ Clustering voxels ...
+ Largest cluster has 235077 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 667231 voxels
+ Mask now has 235398 voxels
++ 235398 voxels in the mask [out of 902629: 26.08%]
++ first 10 x-planes are zero [from L]
++ last 8 x-planes are zero [from R]
++ first 10 y-planes are zero [from P]
++ last 8 y-planes are zero [from A]
++ first 2 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_4_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_5_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_2_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_6_edgy.nii -ulay_range_nz 2% 98% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 37.262657 -opacity 9 -prefix ../qc_01_ve2a_epi2anat -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_2_ulay_shrp.nii
++ Found input file: eac_6_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_D1cscnsnCsG
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[2%, 98%] -> [0.305168, 799.974182]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_D1cscnsnCsG/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_D1cscnsnCsG/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 89.243538
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_D1cscnsnCsG/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_D1cscnsnCsG/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Rd2I7QOksxLbBfU3_yUi-A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Rd2I7QOksxLbBfU3_yUi-A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :369
[1] 2594876
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+ 3dGrayplot: Elapsed = 8.9 s
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-08+tlrc.HEAD -olay full_mask.sub-08+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-08/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-08+tlrc.HEAD
++ Found input file: full_mask.sub-08+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_OGxKFoEaK9U
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_OGxKFoEaK9U/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_OGxKFoEaK9U/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_OGxKFoEaK9U/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_OGxKFoEaK9U/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_rkPVUPtmdkJB5mMGdnRGoQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_rkPVUPtmdkJB5mMGdnRGoQ+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :539
[1] 2595251
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_D1cscnsnCsG++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_01_ve2a_epi2anat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.305168 799.974182 -com SET_FUNC_RANGE 89.243538 -com SET_THRESHNEW 37.262657 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_01_ve2a_epi2anat.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_01_ve2a_epi2anat.cor blowup=2 -com SAVE_JPEG axialimage ../qc_01_ve2a_epi2anat.axi blowup=2 -com QUITT ../__tmp_chauf_D1cscnsnCsG
+* Removing temporary image directory '../__tmp_chauf_D1cscnsnCsG'.
[1] Done Xvfb :369 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_01_ve2a_epi2anat*
+* Removing temporary workdir 'QC_sub-09/media/__workdir_EAC_jPRgefnngt1*'
++ DONE! Image output:
QC_sub-09/media/qc_01_ve2a_epi2anat
++ Prepare for running @djunct_edgy_align_check (ver = 1.45)
++ Copy refbox (/opt/afni-latest/MNI_avg152T1+tlrc) to workdir
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__workdir_EAC_Ar7zOQhsHXL/REFBOX.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__workdir_EAC_Ar7zOQhsHXL/eac_0_cp.nii
++ Output dataset ./eac_1_ulay_shrp.nii
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: /opt/afni-latest/MNI_avg152T1+tlrc.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 57.780 69.336 57.780
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset QC_sub-09/media/__workdir_EAC_Ar7zOQhsHXL/eac_2_res.nii
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.2
++ ###########################################################
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset eac_2_res.nii
++ Forming automask
+ Fixed clip level = 0.298080
+ Used gradual clip level = 0.286276 .. 0.307885
+ Number voxels above clip level = 231114
+ Clustering voxels ...
+ Largest cluster has 231101 voxels
+ Clustering voxels ...
+ Largest cluster has 230543 voxels
+ Filled 1108 voxels in small holes; now have 231651 voxels
+ Filled 1424 voxels in large holes; now have 233075 voxels
+ Clustering voxels ...
+ Largest cluster has 233075 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 669554 voxels
+ Mask now has 233075 voxels
++ 233075 voxels in the mask [out of 902629: 25.82%]
++ first 10 x-planes are zero [from L]
++ last 9 x-planes are zero [from R]
++ first 11 y-planes are zero [from P]
++ last 9 y-planes are zero [from A]
++ first 3 z-planes are zero [from I]
++ last 13 z-planes are zero [from S]
++ Output dataset ./eac_3_mask.nii
++ CPU time = 0.000000 sec
++ 3dMedianFilter: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Output dataset ./eac_4_mfilt.nii
++ 3dedge3: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ olay_alpha has known value: No
++ My command:
@chauffeur_afni -ulay eac_1_ulay_shrp.nii -box_focus_slices REFBOX.nii -olay eac_5_edgy.nii -ulay_range 0% 120% -func_range_perc_nz 33 -cbar Reds_and_Blues_Inv -set_subbricks 0 0 0 -olay_alpha No -olay_boxed No -thr_olay 0.500561 -opacity 9 -prefix ../qc_02_va2t_anat2temp -montx 7 -monty 1 -montgap 1 -montcolor black -save_ftype JPEG -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: eac_1_ulay_shrp.nii
++ Found input file: eac_5_edgy.nii
++ Found focus refbox file: REFBOX.nii
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ../__tmp_chauf_vV8GwxB8wHE
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, 407.719703]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_vV8GwxB8wHE/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./../__tmp_chauf_vV8GwxB8wHE/tmp_olay.nii
++ For olay, the 33%ile value leads to
--> upper range value: 0.519532
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ 3dGrayplot: Elapsed = 9.3 s
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = ../__tmp_chauf_vV8GwxB8wHE/ulay_box_0.nii
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-07+tlrc.HEAD -olay full_mask.sub-07+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-07/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-07+tlrc.HEAD
++ Found input file: full_mask.sub-07+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_Z3PFzut2JZT
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=../__tmp_chauf_vV8GwxB8wHE/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_2hUMOw-Ojn74aTJOOz3AEQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_2hUMOw-Ojn74aTJOOz3AEQ+tlrc.BRIK
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :816
[1] 2596156
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Output dataset ././__tmp_chauf_Z3PFzut2JZT/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_Z3PFzut2JZT/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/__tmp_chauf_OGxKFoEaK9U++ Writing palette image to QC_sub-08/media/qc_06_mot_grayplot.pbar.jpg
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_Z3PFzut2JZT/ulay_box_0.nii
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-08/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_OGxKFoEaK9U
+* Removing temporary image directory './__tmp_chauf_OGxKFoEaK9U'.
[1] Done Xvfb :539 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_Z3PFzut2JZT/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_4FGtEU_J61q_fH2GMyGVhw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-08/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
++ Output dataset /tmp/3dcalc_XYZ_4FGtEU_J61q_fH2GMyGVhw+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :799
[1] 2596637
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-08/media/qc_09_regr_df.dat
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-08/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-08/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_0-JYz9zd4IlKsVmArUcPWQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_0-JYz9zd4IlKsVmArUcPWQ+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :19
[1] 2597522
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_vV8GwxB8wHE++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > ../qc_02_va2t_anat2temp.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com DO_NOTHING -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 407.719703 -com SET_FUNC_RANGE 0.519532 -com SET_THRESHNEW 0.500561 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage ../qc_02_va2t_anat2temp.sag blowup=2 -com SAVE_JPEG coronalimage ../qc_02_va2t_anat2temp.cor blowup=2 -com SAVE_JPEG axialimage ../qc_02_va2t_anat2temp.axi blowup=2 -com QUITT ../__tmp_chauf_vV8GwxB8wHE
+* Removing temporary image directory '../__tmp_chauf_vV8GwxB8wHE'.
[1] Done Xvfb :816 -screen 0 1024x768x24
++ DONE (good exit)
see: ../qc_02_va2t_anat2temp*
+* Removing temporary workdir 'QC_sub-09/media/__workdir_EAC_Ar7zOQhsHXL*'
++ DONE! Image output:
QC_sub-09/media/qc_02_va2t_anat2temp
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/__tmp_chauf_Z3PFzut2JZT++ Writing palette image to QC_sub-07/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-07/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_Z3PFzut2JZT
+* Removing temporary image directory './__tmp_chauf_Z3PFzut2JZT'.
[1] Done Xvfb :799 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-07/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay stats.sub-09_REML+tlrc.HEAD -cbar Plasma -pbar_posonly -ulay_range 0% 120% -func_range 19.882336 -thr_olay 5.898844 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-09/media/qc_03_vstat_Full_Fstat.pbar.jpg -pbar_comm_range 99%ile in mask -pbar_comm_thr 90%ile in mask, alpha+boxed on -prefix QC_sub-09/media/qc_03_vstat_Full_Fstat -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: stats.sub-09_REML+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
++ Using opacity: 9
ModuleNotFoundError: No module named 'matplotlib'
++ Making temporary work directory to copy vis files: QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Output dataset ./QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Output dataset ./QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu/tmp_olay.nii
++ User-entered function range value value (19.882336)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-07/media/qc_09_regr_df.dat
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_egKdnup5Lpb5ORD-OTCSMA argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ Output dataset /tmp/3dcalc_XYZ_egKdnup5Lpb5ORD-OTCSMA+tlrc.BRIK
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-07/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-07/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Using blowup factor: 2
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Using opacity: 9
++ Will have the ref box central gapord: 10 13 10
++ Making temporary work directory to copy vis files: QC_sub-07/media/__tmp_chauf_hQgFMVwfj18
------------------- end of optionizing -------------------
-- trying to start Xvfb :763
[1] 2599248
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94++ Writing palette image to QC_sub-08/media/qc_10_regr_corr_errts.pbar.jpg
++ Output dataset ./QC_sub-07/media/__tmp_chauf_hQgFMVwfj18/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_hQgFMVwfj18/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_10_regr_corr_errts.sag.jpg'
++ 3dAutobox: output dataset = QC_sub-07/media/__tmp_chauf_hQgFMVwfj18/ulay_box_0.nii
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-08/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-08/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-08/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-08/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94
+* Removing temporary image directory 'QC_sub-08/media/__tmp_chauf_mfXuAXlgZ94'.
[1] Done Xvfb :19 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-08/media/qc_10_regr_corr_errts*
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-07/media/__tmp_chauf_hQgFMVwfj18/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_zVRCZn6mSMCn2Hz6yu9pxQ argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_zVRCZn6mSMCn2Hz6yu9pxQ+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :447
[1] 2599712
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-08+tlrc.HEAD -ulay_range 0% 120% -func_range 189 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 189=red 168=oran-red 148=orange 128=oran-yell 107=yell-oran 87=yellow 67=lt-blue2 37=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-08/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 67 - 189 -prefix QC_sub-08/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-08+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7/tmp_olay.nii
++ User-entered function range value value (189)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_ERM-5Wi6ttmyq2VMTLX4_A argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_ERM-5Wi6ttmyq2VMTLX4_A+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :630
[1] 2600366
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu++ Writing palette image to QC_sub-09/media/qc_03_vstat_Full_Fstat.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_03_vstat_Full_Fstat.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_03_vstat_Full_Fstat.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_03_vstat_Full_Fstat.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +99 1 Plasma -com PBAR_SAVEIM QC_sub-09/media/qc_03_vstat_Full_Fstat.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 19.882336 -com SET_THRESHNEW 5.898844 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-09/media/qc_03_vstat_Full_Fstat.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-09/media/qc_03_vstat_Full_Fstat.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-09/media/qc_03_vstat_Full_Fstat.axi blowup=2 -com QUITT QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu
+* Removing temporary image directory 'QC_sub-09/media/__tmp_chauf_P6LO3ZKyUFu'.
[1] Done Xvfb :763 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-09/media/qc_03_vstat_Full_Fstat*
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_hQgFMVwfj18++ Writing palette image to QC_sub-07/media/qc_10_regr_corr_errts.pbar.jpg
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-07/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-07/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-07/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-07/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-07/media/__tmp_chauf_hQgFMVwfj18
+* Removing temporary image directory 'QC_sub-07/media/__tmp_chauf_hQgFMVwfj18'.
[1] Done Xvfb :447 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-07/media/qc_10_regr_corr_errts*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
++ 3dGrayplot: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+ Loading dataset ./errts.sub-09_REML+tlrc.HEAD
+ Number of voxels in mask = 71499
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-07+tlrc.HEAD -ulay_range 0% 120% -func_range 190 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 190=red 170=oran-red 150=orange 131=oran-yell 111=yell-oran 91=yellow 72=lt-blue2 40=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-07/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 72 - 190 -prefix QC_sub-07/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-07+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7++ Writing palette image to QC_sub-08/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Output dataset ./QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb/tmp_olay.nii
++ User-entered function range value value (190)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-08/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 189=red 168=oran-red 148=orange 128=oran-yell 107=yell-oran 87=yellow 67=lt-blue2 37=blue 0=none -com PBAR_SAVEIM QC_sub-08/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 189 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-08/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-08/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-08/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7
+* Removing temporary image directory 'QC_sub-08/media/__tmp_chauf_HEPIM8HSmR7'.
[1] Done Xvfb :630 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-08/media/qc_11_regr_tsnr_fin*
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_2oCskdXEH53rd0mL-Prkzg argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_2oCskdXEH53rd0mL-Prkzg+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :335
[1] 2601957
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-08/media/qc_12_warns_xmat.dat
++ Check for censor fraction warnings (general): QC_sub-08/media/qc_13_warns_cen_total.dat
++ Check for censor fraction warnings (per stim): QC_sub-08/media/qc_14_warns_cen_stim.dat
++ Check basic summary quants from proc in: QC_sub-08/media/qc_16_qsumm_ssrev.dat
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-08
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-08+tlrc.HEAD
final stats dset : stats.sub-08_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 2
average motion (per TR) : 0.0767002
average censored motion : 0.0744091
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 0.868807
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 0.868807
average outlier frac (TR) : 0.00122644
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 142
num TRs per run (censored): 0 4
fraction censored per run : 0 0.0273973
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 288
degrees of freedom used : 20
degrees of freedom left : 268
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 4
censor fraction : 0.013699
num regs of interest : 2
num TRs per stim (orig) : 120 110
num TRs censored per stim : 0 4
fraction TRs censored : 0.000 0.036
ave mot per sresp (orig) : 0.070899 0.079812
ave mot per sresp (cens) : 0.070899 0.073748
TSNR average : 139.227
global correlation (GCOR) : 0.0804356
anat/EPI mask Dice coef : 0.928854
anat/templ mask Dice coef : 0.960445
maximum F-stat (masked) : 55.4792
blur estimates (ACF) : 0.793667 3.35254 12.6561
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-08
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-08/index.html
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-08.results/QC_sub-08/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-08.results/QC_sub-08/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:19:33 UTC 2025
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb++ Writing palette image to QC_sub-07/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-07/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 190=red 170=oran-red 150=orange 131=oran-yell 111=yell-oran 91=yellow 72=lt-blue2 40=blue 0=none -com PBAR_SAVEIM QC_sub-07/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 190 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-07/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-07/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-07/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb
+* Removing temporary image directory 'QC_sub-07/media/__tmp_chauf_DjNaCwK97Bb'.
++ DONE (good exit)
see: QC_sub-07/media/qc_11_regr_tsnr_fin*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-07/media/qc_12_warns_xmat.dat
++ Check for censor fraction warnings (general): QC_sub-07/media/qc_13_warns_cen_total.dat
++ Check for censor fraction warnings (per stim): QC_sub-07/media/qc_14_warns_cen_stim.dat
++ Check basic summary quants from proc in: QC_sub-07/media/qc_16_qsumm_ssrev.dat
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-07
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-07+tlrc.HEAD
final stats dset : stats.sub-07_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 0
average motion (per TR) : 0.0422039
average censored motion : 0.0422039
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 1.18663
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 1.18663
average outlier frac (TR) : 0.000896575
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
censor fraction : 0.000000
num regs of interest : 2
num TRs per stim (orig) : 120 110
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
ave mot per sresp (orig) : 0.042234 0.041772
ave mot per sresp (cens) : 0.042234 0.041772
TSNR average : 144.539
global correlation (GCOR) : 0.0512396
anat/EPI mask Dice coef : 0.943059
anat/templ mask Dice coef : 0.96351
maximum F-stat (masked) : 54.439
blur estimates (ACF) : 0.880565 3.30767 13.8377
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-07
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-07/index.html
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-07.results/QC_sub-07/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-07.results/QC_sub-07/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:19:38 UTC 2025
+ 3dGrayplot: Elapsed = 9.2 s
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay full_mask.sub-09+tlrc.HEAD -olay full_mask.sub-09+tlrc.HEAD -box_focus_slices AMASK_FOCUS_OLAY -cbar gray_scale -func_range 3.29 -blowup 1 -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-09/media/qc_06_mot_grayplot.pbar.jpg -pbar_comm_range for normal distr, bounds of 0.001 prob tail -prefix __tmp_ZXCV_img -save_ftype JPEG -montx 1 -monty 1 -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: full_mask.sub-09+tlrc.HEAD
++ Found input file: full_mask.sub-09+tlrc.HEAD
++ Using blowup factor: 1
++ Using opacity: 9
++ Making temporary work directory to copy vis files: ./__tmp_chauf_Er2GePzVUhS
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay to be visualized within user range:
[0%, 98%] -> [0.000000, 1.000000]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_Er2GePzVUhS/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tmp_chauf_Er2GePzVUhS/tmp_olay.nii
++ User-entered function range value value (3.29)
++ Dimensions (xyzt): 61 73 61 1
++ (initial) Slice spacing ordered (x,y,z) is: 61 73 61
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=0..60 y=0..72 z=0..60
++ THD_zeropad: all pad values are zero - just copying dataset
++ 3dAutobox: output dataset = ./__tmp_chauf_Er2GePzVUhS/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=./__tmp_chauf_Er2GePzVUhS/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_kketkQPOiBtvWjz8NwOS0g argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0: no scale factor
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_kketkQPOiBtvWjz8NwOS0g+tlrc.BRIK
++ 271633 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 3 18 18
++ Will have the ref box central gapord: 61 73 61
------------------- end of optionizing -------------------
-- trying to start Xvfb :186
[1] 2603502
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/__tmp_chauf_Er2GePzVUhS++ Writing palette image to QC_sub-09/media/qc_06_mot_grayplot.pbar.jpg
++ Writing one 73x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.sag.jpg'
++ Writing one 61x61 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.cor.jpg'
++ Writing one 61x73 image to filter '/opt/afni-latest/cjpeg -quality 95 > ./__tmp_ZXCV_img.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=1x1:73:0:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=1x1:61:0:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 gray_scale -com PBAR_SAVEIM QC_sub-09/media/qc_06_mot_grayplot.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 1.000000 -com SET_FUNC_RANGE 3.29 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 3 18 18 -com SAVE_JPEG sagittalimage ./__tmp_ZXCV_img.sag blowup=1 -com SAVE_JPEG coronalimage ./__tmp_ZXCV_img.cor blowup=1 -com SAVE_JPEG axialimage ./__tmp_ZXCV_img.axi blowup=1 -com QUITT ./__tmp_chauf_Er2GePzVUhS
+* Removing temporary image directory './__tmp_chauf_Er2GePzVUhS'.
[1] Done Xvfb :186 -screen 0 1024x768x24
++ DONE (good exit)
see: ./__tmp_ZXCV_img*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
djpeg: can't open __tmp_img_enorm.jpg
pnmcat: Error reading first byte of what is expected to be a Netpbm magic number. Most often, this means your input file is empty
Empty input file
+* Removing temporary files '__tmp_gluing*'
++ DONE! Image output:
QC_sub-09/media/qc_06_mot_grayplot.jpg
rm: cannot remove '__tmp_img_enorm.jpg': No such file or directory
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
Traceback (most recent call last):
File "/opt/afni-latest/1dplot.py", line 50, in <module>
from afnipy import lib_plot_1D as lpod
File "/opt/afni-latest/afnipy/lib_plot_1D.py", line 39, in <module>
import matplotlib.pyplot as plt
ModuleNotFoundError: No module named 'matplotlib'
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check summary of degrees of freedom in: QC_sub-09/media/qc_09_regr_df.dat
++ olay_alpha has known value: Yes
++ pbar name has known extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -olay corr_brain+tlrc.HEAD -cbar Reds_and_Blues_Inv -ulay_range 0% 120% -func_range 0.6 -thr_olay 0.3 -olay_alpha Yes -olay_boxed Yes -set_subbricks 0 0 0 -opacity 9 -pbar_saveim QC_sub-09/media/qc_10_regr_corr_errts.pbar.jpg -pbar_comm_range Pearson r -pbar_comm_thr alpha+boxed on -prefix QC_sub-09/media/qc_10_regr_corr_errts -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: corr_brain+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 9
++ Making temporary work directory to copy vis files: QC_sub-09/media/__tmp_chauf_de0e1OAF3KW
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__tmp_chauf_de0e1OAF3KW/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__tmp_chauf_de0e1OAF3KW/tmp_olay.nii
++ User-entered function range value value (0.6)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-09/media/__tmp_chauf_de0e1OAF3KW/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-09/media/__tmp_chauf_de0e1OAF3KW/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_igUx4OKNiClPO6E8u6CYCw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_igUx4OKNiClPO6E8u6CYCw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :653
[1] 2604618
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_de0e1OAF3KW++ Writing palette image to QC_sub-09/media/qc_10_regr_corr_errts.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_10_regr_corr_errts.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_10_regr_corr_errts.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_10_regr_corr_errts.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=9 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=9 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL -99 1 Reds_and_Blues_Inv -com PBAR_SAVEIM QC_sub-09/media/qc_10_regr_corr_errts.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 0.6 -com SET_THRESHNEW 0.3 * -com SET_FUNC_ALPHA Yes -com SET_FUNC_BOXED Yes -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-09/media/qc_10_regr_corr_errts.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-09/media/qc_10_regr_corr_errts.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-09/media/qc_10_regr_corr_errts.axi blowup=2 -com QUITT QC_sub-09/media/__tmp_chauf_de0e1OAF3KW
+* Removing temporary image directory 'QC_sub-09/media/__tmp_chauf_de0e1OAF3KW'.
[1] Done Xvfb :653 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-09/media/qc_10_regr_corr_errts*
++ Prepare for running adjunct_apqc_tsnr_general (ver = 1.1)
++ MODE: 10
++ olay_alpha has known value: No
*+ Setting pbar file extension: jpg
++ My command:
@chauffeur_afni -ulay /opt/afni-latest/MNI_avg152T1+tlrc -olay TSNR.sub-09+tlrc.HEAD -ulay_range 0% 120% -func_range 174 -box_focus_slices /opt/afni-latest/MNI_avg152T1+tlrc -set_subbricks 0 0 0 -pbar_posonly -blowup 2 -cbar 174=red 156=oran-red 138=orange 121=oran-yell 103=yell-oran 85=yellow 68=lt-blue2 38=blue 0=none -cbar_ncolors 8 -cbar_topval EMPTY -opacity 4 -olay_alpha No -olay_boxed No -thr_olay 0 -pbar_saveim QC_sub-09/media/qc_11_regr_tsnr_fin.pbar -pbar_comm_range 95%ile in mask -pbar_comm_gen cbar: hot color range (5-95%ile TSNR in mask): 68 - 174 -prefix QC_sub-09/media/qc_11_regr_tsnr_fin -save_ftype JPEG -montx 7 -monty 1 -montgap 1 -montcolor black -set_xhairs OFF -label_mode 1 -label_size 3 -do_clean -pass -pass
++ Using AFNI ver : AFNI_21.2.00
++ chauffeur ver : 6.26
------------------ start of optionizing ------------------
++ Found input file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Found input file: TSNR.sub-09+tlrc.HEAD
++ Found focus refbox file: /opt/afni-latest/MNI_avg152T1+tlrc
++ Using blowup factor: 2
++ Using opacity: 4
++ Making temporary work directory to copy vis files: QC_sub-09/media/__tmp_chauf_piIaUSSkwId
++ Converted 0 to labels
++ Final subbrick indices: 0 0 0
++ Copy ulay to visualize (volumetric) within user's range:
++ Ulay range calc for >100%ile ulay max:
calculating 98%ile value, and then
multiplying it by a scale factor = 1.22448,
in order to produce the 98%ile value
++ Ulay to be visualized within user range:
[0%, 120%] -> [0.000000, .893150]
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__tmp_chauf_piIaUSSkwId/tmp_ulay.nii
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./QC_sub-09/media/__tmp_chauf_piIaUSSkwId/tmp_olay.nii
++ User-entered function range value value (174)
++ Dimensions (xyzt): 91 109 91 1
++ (initial) Slice spacing ordered (x,y,z) is: 13 15 13
++ 3dAutobox: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Auto bbox: x=10..81 y=10..101 z=3..78
++ 3dAutobox: output dataset = QC_sub-09/media/__tmp_chauf_piIaUSSkwId/ulay_box_0.nii
++ 3dmaskdump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Executing 3dcalc()
argv[0]=3dcalc argv[1]=-a argv[2]=QC_sub-09/media/__tmp_chauf_piIaUSSkwId/ulay_box_0.nii[0] argv[3]=-expr argv[4]=a argv[5]=-byte argv[6]=-session argv[7]=/tmp argv[8]=-prefix argv[9]=3dcalc_XYZ_Fs_BDE0UUY8j1iWYDJjWAw argv[10]=-verbose
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Computing sub-brick 0
++ Scaling output to type byte brick(s)
++ Sub-brick 0 scale factor = 314.130402
++ Computing output statistics
++ Output dataset /tmp/3dcalc_XYZ_Fs_BDE0UUY8j1iWYDJjWAw+tlrc.BRIK
++ 503424 voxels in the entire dataset (no mask)
++ 1 voxels in the boxes and/or balls
++ Using only the boxes+balls mask
++ How many coors? 3
++ Will have the ref box central coors : SET_DICOM_XYZ 0 16 10
++ Will have the ref box central gapord: 10 13 10
------------------- end of optionizing -------------------
-- trying to start Xvfb :601
[1] 2605450
_XSERVTransmkdir: Owner of /tmp/.X11-unix should be set to root
+/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/QC_sub-09/media/__tmp_chauf_piIaUSSkwId++ Writing palette image to QC_sub-09/media/qc_11_regr_tsnr_fin.pbar.jpg
++ Writing one 1538x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_11_regr_tsnr_fin.sag.jpg'
++ Writing one 1286x182 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_11_regr_tsnr_fin.cor.jpg'
++ Writing one 1286x218 image to filter '/opt/afni-latest/cjpeg -quality 95 > QC_sub-09/media/qc_11_regr_tsnr_fin.axi.jpg'
AFNI QUITTs!
+++ Command Echo:
afni -q -no1D -noplugins -no_detach -com SWITCH_UNDERLAY tmp_ulay.nii -com SWITCH_OVERLAY tmp_olay.nii -com SEE_OVERLAY + -com OPEN_WINDOW sagittalimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com OPEN_WINDOW coronalimage opacity=4 mont=7x1:13:1:black crop=0:0,0:0 -com OPEN_WINDOW axialimage opacity=4 mont=7x1:10:1:black crop=0:0,0:0 -com SET_PBAR_ALL +8 174=red 156=oran-red 138=orange 121=oran-yell 103=yell-oran 85=yellow 68=lt-blue2 38=blue 0=none -com PBAR_SAVEIM QC_sub-09/media/qc_11_regr_tsnr_fin.pbar.jpg dim=64x512H -com SET_SUBBRICKS 0 0 0 -com SET_ULAY_RANGE A.all 0.000000 .893150 -com SET_FUNC_RANGE 174 -com SET_THRESHNEW 0 * -com SET_FUNC_ALPHA No -com SET_FUNC_BOXED No -com SET_FUNC_RESAM NN.NN -com SET_XHAIRS OFF -com SET_XHAIR_GAP -1 -com SET_DICOM_XYZ 0 16 10 -com SAVE_JPEG sagittalimage QC_sub-09/media/qc_11_regr_tsnr_fin.sag blowup=2 -com SAVE_JPEG coronalimage QC_sub-09/media/qc_11_regr_tsnr_fin.cor blowup=2 -com SAVE_JPEG axialimage QC_sub-09/media/qc_11_regr_tsnr_fin.axi blowup=2 -com QUITT QC_sub-09/media/__tmp_chauf_piIaUSSkwId
+* Removing temporary image directory 'QC_sub-09/media/__tmp_chauf_piIaUSSkwId'.
[1] Done Xvfb :601 -screen 0 1024x768x24
++ DONE (good exit)
see: QC_sub-09/media/qc_11_regr_tsnr_fin*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Check for corr matrix warnings in: QC_sub-09/media/qc_12_warns_xmat.dat
++ Check for censor fraction warnings (general): QC_sub-09/media/qc_13_warns_cen_total.dat
++ Check for censor fraction warnings (per stim): QC_sub-09/media/qc_14_warns_cen_stim.dat
++ Check basic summary quants from proc in: QC_sub-09/media/qc_16_qsumm_ssrev.dat
# +++++++++++ Check output of @ss_review_basic +++++++++++ #
subject ID : sub-09
AFNI version : AFNI_21.2.00
AFNI package : linux_openmp_64
TR : 2.0
TRs removed (per run) : 0
num stim classes provided : 2
final anatomy dset : anat_final.sub-09+tlrc.HEAD
final stats dset : stats.sub-09_REML+tlrc.HEAD
final voxel resolution : 3.000000 3.000000 3.000000
motion limit : 0.3
num TRs above mot limit : 0
average motion (per TR) : 0.057909
average censored motion : 0.057909
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max motion displacement : 0.850377
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
max censored displacement : 0.850377
average outlier frac (TR) : 0.000870959
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num runs found : 2
num TRs per run : 146 146
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
num TRs per run (applied) : 146 146
num TRs per run (censored): 0 0
fraction censored per run : 0 0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs total (uncensored) : 292
TRs total : 292
degrees of freedom used : 20
degrees of freedom left : 272
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
TRs censored : 0
censor fraction : 0.000000
num regs of interest : 2
num TRs per stim (orig) : 120 120
num TRs censored per stim : 0 0
fraction TRs censored : 0.000 0.000
ave mot per sresp (orig) : 0.058195 0.056895
ave mot per sresp (cens) : 0.058195 0.056895
TSNR average : 133.674
global correlation (GCOR) : 0.037331
anat/EPI mask Dice coef : 0.924751
anat/templ mask Dice coef : 0.957488
maximum F-stat (masked) : 68.6935
blur estimates (ACF) : 0.907516 3.31665 16.4517
blur estimates (FWHM) : 0 0 0
apqc_make_html.py -qc_dir QC_sub-09
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
++ Done! Wrote QC HTML. To check, consider:
afni_open -b QC_sub-09/index.html
echo \nconsider running: \n\n afni_open -b ./afni_pro_glm/sub-09.results/QC_sub-09/index.html\n
consider running:
afni_open -b ./afni_pro_glm/sub-09.results/QC_sub-09/index.html
endif
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 05:20:02 UTC 2025
Summary of results:
sub-03 done
sub-02 done
sub-01 done
sub-04 done
sub-05 done
sub-06 done
sub-08 done
sub-07 done
sub-09 done
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3. Group - Analysis#
After estimating the first-level general linear model (GLM) for each subject, we will perform a group-level analysis to assess whether the Incongruent - Congruent contrast showed a consistent effect across subjects.
Creating a Group Mask#
First, we’ll create a group-level mask by combining each subject’s individual mask using a logical union. This ensures that only voxels present in all subjects’ data are included in the group analysis:
! 3dmask_tool -input afni_pro_glm/sub-*.results/mask_group+tlrc.HEAD -prefix ./afni_pro_glm/group_mask+tlrc -union
++ processing 9 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 9 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ writing result group_mask...
++ Output dataset ./afni_pro_glm/group_mask+tlrc.BRIK
3.1 Group-Level Analysis with 3dttest++#
AFNI’s 3dttest++ performs a voxelwise one-sample t-test across subjects. This tests whether the average contrast estimate (e.g., Incongruent - Congruent) is significantly different from zero at each voxel.oxelwise t-scoresariability.
Identifying Contrast Sub-Brick#
Before running the group-level test, we must determine which sub-brick contains the desired contrast. We use 3dinfo to inspect the subject-level stats+tlrc dataset and find the index of the contrast sub-brick corresponding to incongruent minus congruent (e.g., sub-brick #7, depending on how GLTs were defined in afni_proc.py):
! 3dinfo -verb ./afni_pro_glm/sub-08.results/stats.sub-08+tlrc.BRIK
++ 3dinfo: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
Dataset File: stats.sub-08+tlrc
Identifier Code: XYZ_DS49wumVRjnXZB8zRjA58A Creation Date: Tue Nov 4 05:15:56 2025
Template Space: MNI
Dataset Type: Func-Bucket (-fbuc)
Byte Order: LSB_FIRST [this CPU native = LSB_FIRST]
Storage Mode: BRIK
Storage Space: 14,124,916 (14 million) bytes
Geometry String: "MATRIX(-3,0,0,90,0,-3,0,126,0,0,3,-72):61,73,61"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -90.000 [R] -to- 90.000 [L] -step- 3.000 mm [ 61 voxels]
A-to-P extent: -90.000 [A] -to- 126.000 [P] -step- 3.000 mm [ 73 voxels]
I-to-S extent: -72.000 [I] -to- 108.000 [S] -step- 3.000 mm [ 61 voxels]
Number of values stored at each pixel = 13
-- At sub-brick #0 'Full_Fstat' datum type is float: 0 to 55.6688
statcode = fift; statpar = 2 268
-- At sub-brick #1 'congruent#0_Coef' datum type is float: -20.0682 to 19.7864
-- At sub-brick #2 'congruent#0_Tstat' datum type is float: -4.95626 to 7.66608
statcode = fitt; statpar = 268
-- At sub-brick #3 'congruent_Fstat' datum type is float: 0 to 58.7687
statcode = fift; statpar = 1 268
-- At sub-brick #4 'incongruent#0_Coef' datum type is float: -16.9107 to 19.8846
-- At sub-brick #5 'incongruent#0_Tstat' datum type is float: -5.66548 to 9.71671
statcode = fitt; statpar = 268
-- At sub-brick #6 'incongruent_Fstat' datum type is float: 0 to 94.4144
statcode = fift; statpar = 1 268
-- At sub-brick #7 'incongruent-congruent_GLT#0_Coef' datum type is float: -19.9548 to 21.7222
-- At sub-brick #8 'incongruent-congruent_GLT#0_Tstat' datum type is float: -4.39914 to 5.95293
statcode = fitt; statpar = 268
-- At sub-brick #9 'incongruent-congruent_GLT_Fstat' datum type is float: 0 to 35.4373
statcode = fift; statpar = 1 268
-- At sub-brick #10 'congruent-incongruent_GLT#0_Coef' datum type is float: -21.7222 to 19.9548
-- At sub-brick #11 'congruent-incongruent_GLT#0_Tstat' datum type is float: -5.95293 to 4.39914
statcode = fitt; statpar = 268
-- At sub-brick #12 'congruent-incongruent_GLT_Fstat' datum type is float: 0 to 35.4373
statcode = fift; statpar = 1 268
----- HISTORY -----
[jovyan@jupyter-monidoerig: Tue Nov 4 05:15:56 2025] {AFNI_21.2.00:linux_openmp_64} 3dDeconvolve -input pb03.sub-08.r01.scale+tlrc.HEAD pb03.sub-08.r02.scale+tlrc.HEAD -censor motion_sub-08_censor.1D -ortvec mot_demean.r01.1D mot_demean_r01 -ortvec mot_demean.r02.1D mot_demean_r02 -polort 2 -num_stimts 2 -stim_times 1 stimuli/congruent.1D GAM -stim_label 1 congruent -stim_times 2 stimuli/incongruent.1D GAM -stim_label 2 incongruent -gltsym 'SYM: +incongruent -congruent' -glt_label 1 incongruent-congruent -gltsym 'SYM: +congruent -incongruent' -glt_label 2 congruent-incongruent -jobs 8 -GOFORIT 0 -fout -tout -x1D X.xmat.1D -xjpeg X.jpg -x1D_uncensored X.nocensor.xmat.1D -fitts fitts.sub-08 -errts errts.sub-08 -bucket stats.sub-08
[jovyan@jupyter-monidoerig: Tue Nov 4 05:15:56 2025] Output prefix: stats.sub-08
This command lists all sub-bricks with their labels, allowing us to find the relevant contrast parameter estimate:
-- At sub-brick #7 'incongruent-congruent_GLT#0_Coef' datum type is float: -19.9548 to 21.7222
Sub-bricks labeled with “Coef” represent contrast (beta) estimates, while “Tstat” or “Fstat” indicate test statistics. For group analysis, we will extract the Coef sub-brick (here: index [7]).
We then automate the 3dttest++ execution with the following steps:
- Set the base directory containing all subjects’ AFNI result folders. **
Locate sub-brick
[**7]for each subject (the contrast of interest).**Verify dat**aset existence to avoid processing missing subjects.
Construct the
3dttest++command with:An output prefix for group results,
A group brain mask to constrain the analysis,
A label for the subject set (
Inc-Con),Subpaired with their corresponding contrast sub-brick pathssti**mates.
Run **the command using
subprocess.run()to capture and log output.
The resulting dataset (Flanker_Inc-Con_ttest+tlrc) contains:
Effect size map: average contrast values across subjects
T-statistic map: voxelwise t-values representing statistical significance
base_dir = os.path.abspath("./afni_pro_glm")
# Get all subject folders
subject_dirs = sorted(glob(os.path.join(base_dir, "sub-*.results")))
# Build paths to sub-brik 7 (e.g. Inc-Con contrast)
set_lines = []
for s in subject_dirs:
sub_id = os.path.basename(s).replace('.results', '')
stats_path = os.path.join(s, f"stats.{sub_id}+tlrc[7]")
if os.path.exists(stats_path.replace('[7]', '.BRIK')) or os.path.exists(stats_path): # extra check
set_lines.append(f"{sub_id} {stats_path}")
else:
print(f"⚠️ Missing file for subject {sub_id}: {stats_path}")
# Build 3dttest++ command
cmd_list = [
"3dttest++",
"-prefix", os.path.join(base_dir, "group_results", "Flanker-Inc-Con_ttest"),
"-mask", os.path.join(base_dir, "group_mask+tlrc"),
"-setA", "Inc-Con"
]
# Add subject lines
for line in set_lines:
cmd_list += line.split()
# Run the command
print("Running 3dttest++...")
print("\n")
print("Command:", " ".join(cmd_list))
result = subprocess.run(cmd_list, capture_output=True, text=True)
# Show output
print(result.stdout)
print(result.stderr)
Running 3dttest++...
Command: 3dttest++ -prefix /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_results/Flanker-Inc-Con_ttest -mask /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_mask+tlrc -setA Inc-Con sub-01 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/stats.sub-01+tlrc[7] sub-02 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/stats.sub-02+tlrc[7] sub-03 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/stats.sub-03+tlrc[7] sub-04 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/stats.sub-04+tlrc[7] sub-05 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/stats.sub-05+tlrc[7] sub-06 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/stats.sub-06+tlrc[7] sub-07 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/stats.sub-07+tlrc[7] sub-08 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/stats.sub-08+tlrc[7] sub-09 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/stats.sub-09+tlrc[7]
++ 3dttest++: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark++
++ 73409 voxels in -mask dataset
++ option -setA :: processing as LONG form (label label dset label dset ...)
++ have 9 volumes corresponding to option '-setA'
++ loading -setA datasets
++ t-testing:0123456789.0123456789.0123456789.0123456789.0123456789.!
++ ---------- End of analyses -- freeing workspaces ----------
++ Creating FDR curves in output dataset
*+ WARNING: Smallest FDR q [1 Inc-Con_Tstat] = 0.241757 ==> few true single voxel detections
+ Added 1 FDR curve to dataset
++ Output dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_results/Flanker-Inc-Con_ttest+tlrc.BRIK
++ ----- 3dttest++ says so long, farewell, and happy trails to you :) -----
This code snippet automates running AFNI’s 3dttest++ for a group analysis of the Incongruent - Congruent contrast:
Set the base directory where all subjects’ AFNI results are stored.
Find all subject result folders matching the pattern
sub-*.results.Build a list of dataset paths pointing to the 7th sub-brick
[7](the contrast of interest) in each subject’s stats dataset, verifying file existence and alerting if missing.Construct the
3dttest++command with:Output prefix for group results,
Group mask to restrict the test to voxels common across subjects,
Label for the group set (
Inc-Con),Subject IDs paired with their corresponding contrast sub-brick paths.
Run the command as a subprocess, capturing and printing AFNI’s output and errors for review.
The resulting output (Flanker_Inc-Con_ttest+tlrc) contains:
Effect size map: average contrast across subjects
T-statistic map: voxelwise t-sores
3.2 Performing Group Analysis with 3dMEMA#
To account for both the variability within subjects (parameter estimate differences) and the variability across subjects (contrast estimate precision), we will also perform a group-level analysis using AFNI’s 3dMEMA.
Unlike a simple t-test, 3dMEMA leverages each subject’s contrast estimate and corresponding t-statistic (or standard error), providing a more accurate mixed-effects model.
The steps we will take are:
Build a set list containing each subject’s ID along with the paths to their contrast estimate (
_REML+tlrc[7]) and t-statistic (_REML+tlrc[8]) sub-bricks.Verify that both files exist for each subject to avoid missing data errors.
Construct the
3dMEMAcommand with an output prefix, the group mask, and the full subject set list.Run the command as a subprocess, capturing output for review.
This process will produce a group-level statistical map that balances effect size and reliability, offering a robust inference about the Incongruent - Congruent contrast across subjects.
# Build set list: all subjects in one set block
set_list = ["-set", "IncCon"]
for s in subject_dirs:
sub_id = os.path.basename(s).replace(".results", "")
coef_path = os.path.join(s, f"stats.{sub_id}_REML+tlrc[7]")
tstat_path = os.path.join(s, f"stats.{sub_id}_REML+tlrc[8]")
# Sanity check (optional)
if os.path.exists(coef_path.replace("[7]", ".HEAD")) and os.path.exists(tstat_path.replace("[8]", ".HEAD")):
set_list += [sub_id, coef_path, tstat_path]
else:
print(f"⚠️ Missing for {sub_id}:\n {coef_path}\n {tstat_path}")
# Full 3dMEMA command
cmd_list = [
"3dMEMA",
"-prefix", os.path.join(base_dir, "group_results", "Flanker_Inc-Con_MEMA"),
"-mask", os.path.join(base_dir, "group_mask+tlrc"),
] + set_list
# Print and run
print("Running 3dMEMA...\n")
print("Command:", " ".join(cmd_list), "\n")
result = subprocess.run(cmd_list, capture_output=True, text=True)
print(result.stdout)
print(result.stderr)
Running 3dMEMA...
Command: 3dMEMA -prefix /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_results/Flanker_Inc-Con_MEMA -mask /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_mask+tlrc -set IncCon sub-01 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/stats.sub-01_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/stats.sub-01_REML+tlrc[8] sub-02 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/stats.sub-02_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/stats.sub-02_REML+tlrc[8] sub-03 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/stats.sub-03_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/stats.sub-03_REML+tlrc[8] sub-04 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/stats.sub-04_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/stats.sub-04_REML+tlrc[8] sub-05 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/stats.sub-05_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/stats.sub-05_REML+tlrc[8] sub-06 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/stats.sub-06_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/stats.sub-06_REML+tlrc[8] sub-07 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/stats.sub-07_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/stats.sub-07_REML+tlrc[8] sub-08 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/stats.sub-08_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/stats.sub-08_REML+tlrc[8] sub-09 /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/stats.sub-09_REML+tlrc[7] /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/stats.sub-09_REML+tlrc[8]
[1] "-----------------"
[1] "Totally 61 slices in the volume data."
[1] "Starting to analyze data slice-wise. Running progression shown below:"
[1] "-----------------"
Z slice # 1 done: 11/04/25 05:20:20.846
Z slice # 2 done: 11/04/25 05:20:21.102
Z slice # 3 done: 11/04/25 05:20:21.405
Z slice # 4 done: 11/04/25 05:20:21.744
Z slice # 5 done: 11/04/25 05:20:22.224
Z slice # 6 done: 11/04/25 05:20:22.743
Z slice # 7 done: 11/04/25 05:20:23.239
Z slice # 8 done: 11/04/25 05:20:23.805
Z slice # 9 done: 11/04/25 05:20:24.481
Z slice # 10 done: 11/04/25 05:20:25.288
Z slice # 11 done: 11/04/25 05:20:26.118
Z slice # 12 done: 11/04/25 05:20:28.067
Z slice # 13 done: 11/04/25 05:20:28.915
Z slice # 14 done: 11/04/25 05:20:29.912
Z slice # 15 done: 11/04/25 05:20:30.910
Z slice # 16 done: 11/04/25 05:20:31.913
Z slice # 17 done: 11/04/25 05:20:33.018
Z slice # 18 done: 11/04/25 05:20:34.212
Z slice # 19 done: 11/04/25 05:20:35.392
Z slice # 20 done: 11/04/25 05:20:36.512
Z slice # 21 done: 11/04/25 05:20:37.653
Z slice # 22 done: 11/04/25 05:20:38.770
Z slice # 23 done: 11/04/25 05:20:40.007
Z slice # 24 done: 11/04/25 05:20:41.220
Z slice # 25 done: 11/04/25 05:20:42.504
Z slice # 26 done: 11/04/25 05:20:43.786
Z slice # 27 done: 11/04/25 05:20:45.005
Z slice # 28 done: 11/04/25 05:20:46.204
Z slice # 29 done: 11/04/25 05:20:47.372
Z slice # 30 done: 11/04/25 05:20:48.487
Z slice # 31 done: 11/04/25 05:20:49.563
Z slice # 32 done: 11/04/25 05:20:50.668
Z slice # 33 done: 11/04/25 05:20:51.756
Z slice # 34 done: 11/04/25 05:20:52.830
Z slice # 35 done: 11/04/25 05:20:53.897
Z slice # 36 done: 11/04/25 05:20:54.934
Z slice # 37 done: 11/04/25 05:20:56.057
Z slice # 38 done: 11/04/25 05:20:57.130
Z slice # 39 done: 11/04/25 05:20:58.124
Z slice # 40 done: 11/04/25 05:20:59.131
Z slice # 41 done: 11/04/25 05:21:00.167
Z slice # 42 done: 11/04/25 05:21:01.132
Z slice # 43 done: 11/04/25 05:21:02.026
Z slice # 44 done: 11/04/25 05:21:02.877
Z slice # 45 done: 11/04/25 05:21:03.630
Z slice # 46 done: 11/04/25 05:21:04.346
Z slice # 47 done: 11/04/25 05:21:04.878
Z slice # 48 done: 11/04/25 05:21:05.357
Z slice # 49 done: 11/04/25 05:21:05.734
Z slice # 50 done: 11/04/25 05:21:06.109
Z slice # 51 done: 11/04/25 05:21:06.463
Z slice # 52 done: 11/04/25 05:21:06.781
Z slice # 53 done: 11/04/25 05:21:07.041
Z slice # 54 done: 11/04/25 05:21:07.290
Z slice # 55 done: 11/04/25 05:21:07.543
Z slice # 56 done: 11/04/25 05:21:07.802
Z slice # 57 done: 11/04/25 05:21:08.055
Z slice # 58 done: 11/04/25 05:21:08.309
Z slice # 59 done: 11/04/25 05:21:08.562
Z slice # 60 done: 11/04/25 05:21:08.820
Z slice # 61 done: 11/04/25 05:21:09.075
[1] "Analysis finished: 11/04/25 05:21:09.077"
[1] "#++++++++++++++++++++++++++++++++++++++++++++"
[1] "system is computationally singular: reciprocal condition number = 0"
There were 50 or more warnings (use warnings() to see the first 50)
++ Smallest FDR q [1 IncCon:t] = 0.0805103
*+ WARNING: Smallest FDR q [3 QE:Chisq] = 0.102273 ==> few true single voxel detections
3.3 Visualizing Group-Level Results#
Now that we have completed both group analyses — one with 3dttest++ and one with 3dMEMA — let’s visualize the resulting voxelwise t-statistic maps.
Steps:#
Convert AFNI datasets to NIfTI format using
3dcopy, to ensure compatibility with Python neuroimaging tools likenilearn.Extract the t-statistic sub-brick (sub-brick index
[1]) using3dTcat. This gives us:Inc-Con_Tstatfrom the3dttest++resultIncCon_Tstatfrom the3dMEMAresult
Load the statistical maps into Python using
nilearn.image.load_img().Apply voxelwise thresholding:
A two-tailed threshold of T > 3.3
A minimum cluster size of 40 contiguous voxels
This reduces noise and highlights clusters likely to reflect meaningful effects.
Note on Multiple Comparison Correction:
Cluster-size correction using3dClustSimrequires stable estimates of spatial smoothness, which generally requires at least 14 subjects.
Since our analysis includes only 9 subjects, we do not apply3dClustSimor other parametric corrections.
Instead, we apply an uncorrected voxelwise threshold (T > 3.3) with a minimum cluster size of 40 voxels.
This approach is intended for exploratory visualization only and should not be used for formal statistical inference.Plot the thresholded maps using
nilearn.plotting.plot_stat_map()to qualitatively compare the spatial activation patterns between the two group-level methods.
This approach allows us to visually inspect and compare the sensitivity and spatial extent of effects revealed by the simple one-sample t-test (3dttest++) and the mixed-effects model (3dMEMA) for the Incongruent - Congruent contrast.
! 3dcopy ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest+tlrc ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest.nii.gz
! 3dcopy ./afni_pro_glm/group_results/Flanker_Inc-Con_MEMA+tlrc ./afni_pro_glm/group_results/Flanker_Inc-Con_MEMA.nii.gz
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
! 3dinfo -verb ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest.nii.gz
++ 3dinfo: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
Dataset File: /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_results/Flanker-Inc-Con_ttest.nii.gz
Identifier Code: XYZ_e6jy6nwqCv_6fUYGSp5k8w Creation Date: Tue Nov 4 05:21:10 2025
Template Space: MNI
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST {assumed} [this CPU native = LSB_FIRST]
Storage Mode: NIFTI
Storage Space: 2,173,064 (2.2 million) bytes
Geometry String: "MATRIX(-3,0,0,90,0,-3,0,126,0,0,3,-72):61,73,61"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -90.000 [R] -to- 90.000 [L] -step- 3.000 mm [ 61 voxels]
A-to-P extent: -90.000 [A] -to- 126.000 [P] -step- 3.000 mm [ 73 voxels]
I-to-S extent: -72.000 [I] -to- 108.000 [S] -step- 3.000 mm [ 61 voxels]
Number of values stored at each pixel = 2
-- At sub-brick #0 'Inc-Con_mean' datum type is float: -2.86885 to 2.65067
-- At sub-brick #1 'Inc-Con_Tstat' datum type is float: -7.60381 to 9.81179
statcode = fitt; statpar = 8
----- HISTORY -----
[jovyan@jupyter-monidoerig: Tue Nov 4 05:20:07 2025] {AFNI_21.2.00:linux_openmp_64} 3dttest++ -prefix /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_results/Flanker-Inc-Con_ttest -mask /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/group_mask+tlrc -setA Inc-Con sub-01 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-01.results/stats.sub-01+tlrc[7]' sub-02 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-02.results/stats.sub-02+tlrc[7]' sub-03 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-03.results/stats.sub-03+tlrc[7]' sub-04 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-04.results/stats.sub-04+tlrc[7]' sub-05 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-05.results/stats.sub-05+tlrc[7]' sub-06 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-06.results/stats.sub-06+tlrc[7]' sub-07 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-07.results/stats.sub-07+tlrc[7]' sub-08 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-08.results/stats.sub-08+tlrc[7]' sub-09 '/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_pro_glm/sub-09.results/stats.sub-09+tlrc[7]'
[jovyan@jupyter-monidoerig: Tue Nov 4 05:21:10 2025] {AFNI_21.2.00:linux_openmp_64} 3dcopy ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest+tlrc ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest.nii.gz
# Copy sub-brik 1 (T-statistic) for ttest++
!3dTcat -prefix ./afni_pro_glm/group_results/Flanker-Inc-Con_ttest_Tstat.nii.gz './afni_pro_glm/group_results/Flanker-Inc-Con_ttest+tlrc[1]'
# Copy sub-brik 1 (T-statistic) for MEMA
!3dTcat -prefix ./afni_pro_glm/group_results/Flanker_Inc-Con_MEMA_Tstat.nii.gz './afni_pro_glm/group_results/Flanker_Inc-Con_MEMA+tlrc[1]'
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.1 s
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.1 s
# Load T-stat map (3dttest++ and 3dMEMA results)
img_ttest = image.load_img("./afni_pro_glm/group_results/Flanker-Inc-Con_ttest_Tstat.nii.gz")
img_mema = image.load_img("./afni_pro_glm/group_results/Flanker_Inc-Con_MEMA_Tstat.nii.gz")
threshold = 3.3
# Apply uncorrected voxel threshold (T > 3.3 ≈ p < 0.001)
# and minimum cluster size of 40 voxels
thresholded_img_ttest = image.threshold_img(
img_ttest,
threshold=threshold,
cluster_threshold=40,
two_sided=True,
copy_header=True
)
thresholded_img_mema = image.threshold_img(
img_mema,
threshold=threshold,
cluster_threshold=40,
two_sided=True,
copy_header=True
)
# Plot result
plotting.plot_stat_map(
thresholded_img_ttest,
title="3dttest++ (p<0.001, k≥40 vox)",
threshold=threshold,
colorbar=True
)
plotting.plot_stat_map(
thresholded_img_mema,
title="3dMEMA (p<0.001, k≥40 vox)",
threshold=threshold,
colorbar=True
)
plotting.show()
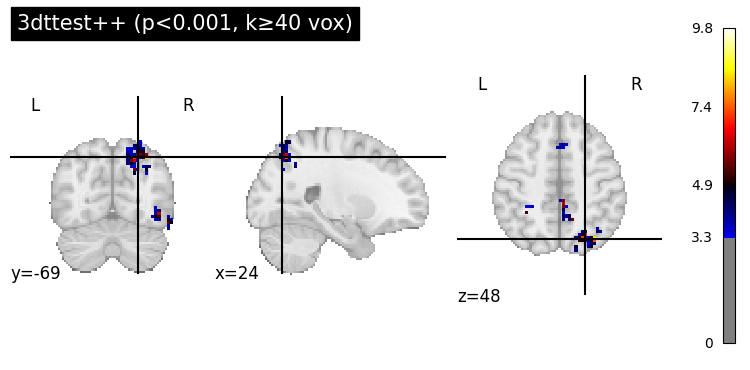
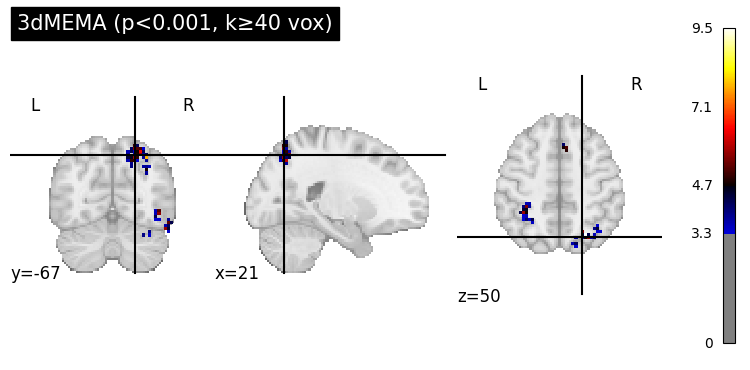
4. ROI Analysis#
4.1 ROI Analysis: Atlas-Based MidACC Mask#
To investigate group-level effects within a specific anatomical region, we use an atlas-based Region of Interest (ROI) mask focusing on the mid anterior cingulate cortex (midACC).
1. Identify Available Atlases:
We begin by confirming the available atlases using:
!whereami -show_atlas_code
This lists all atlases bundled with AFNI.
! whereami -show_atlas_code
++ Input coordinates orientation set by default rules to RAI
Atlas MNI_Glasser_HCP_v1.0, 360 regions
----------- Begin regions for MNI_Glasser_HCP_v1.0 atlas-----------
u:L_Primary_Visual_Cortex:1
u:R_Primary_Visual_Cortex:1001
u:L_Medial_Superior_Temporal_Area:2
u:R_Medial_Superior_Temporal_Area:1002
u:L_Sixth_Visual_Area:3
u:R_Sixth_Visual_Area:1003
u:L_Second_Visual_Area:4
u:R_Second_Visual_Area:1004
u:L_Third_Visual_Area:5
u:R_Third_Visual_Area:1005
u:L_Fourth_Visual_Area:6
u:R_Fourth_Visual_Area:1006
u:L_Eighth_Visual_Area:7
u:R_Eighth_Visual_Area:1007
u:L_Primary_Motor_Cortex:8
u:R_Primary_Motor_Cortex:1008
u:L_Primary_Sensory_Cortex:9
u:R_Primary_Sensory_Cortex:1009
u:L_Frontal_Eye_Fields:10
u:R_Frontal_Eye_Fields:1010
u:L_Premotor_Eye_Fields:11
u:R_Premotor_Eye_Fields:1011
u:L_Area_55b:12
u:R_Area_55b:1012
u:L_Area_V3A:13
u:R_Area_V3A:1013
u:L_RetroSplenial_Complex:14
u:R_RetroSplenial_Complex:1014
u:L_Parieto-Occipital_Sulcus_Area_2:15
u:R_Parieto-Occipital_Sulcus_Area_2:1015
u:L_Seventh_Visual_Area:16
u:R_Seventh_Visual_Area:1016
u:L_IntraParietal_Sulcus_Area_1:17
u:R_IntraParietal_Sulcus_Area_1:1017
u:L_Fusiform_Face_Complex:18
u:R_Fusiform_Face_Complex:1018
u:L_Area_V3B:19
u:R_Area_V3B:1019
u:L_Area_Lateral_Occipital_1:20
u:R_Area_Lateral_Occipital_1:1020
u:L_Area_Lateral_Occipital_2:21
u:R_Area_Lateral_Occipital_2:1021
u:L_Posterior_InferoTemporal:22
u:R_Posterior_InferoTemporal:1022
u:L_Middle_Temporal_Area:23
u:R_Middle_Temporal_Area:1023
u:L_Primary_Auditory_Cortex:24
u:R_Primary_Auditory_Cortex:1024
u:L_PeriSylvian_Language_Area:25
u:R_PeriSylvian_Language_Area:1025
u:L_Superior_Frontal_Language_Area:26
u:R_Superior_Frontal_Language_Area:1026
u:L_PreCuneus_Visual_Area:27
u:R_PreCuneus_Visual_Area:1027
u:L_Superior_Temporal_Visual_Area:28
u:R_Superior_Temporal_Visual_Area:1028
u:L_Medial_Area_7P:29
u:R_Medial_Area_7P:1029
u:L_Area_7m:30
u:R_Area_7m:1030
u:L_Parieto-Occipital_Sulcus_Area_1:31
u:R_Parieto-Occipital_Sulcus_Area_1:1031
u:L_Area_23d:32
u:R_Area_23d:1032
u:L_Area_ventral_23_a+b:33
u:R_Area_ventral_23_a+b:1033
u:L_Area_dorsal_23_a+b:34
u:R_Area_dorsal_23_a+b:1034
u:L_Area_31p_ventral:35
u:R_Area_31p_ventral:1035
u:L_Area_5m:36
u:R_Area_5m:1036
u:L_Area_5m_ventral:37
u:R_Area_5m_ventral:1037
u:L_Area_23c:38
u:R_Area_23c:1038
u:L_Area_5L:39
u:R_Area_5L:1039
u:L_Dorsal_Area_24d:40
u:R_Dorsal_Area_24d:1040
u:L_Ventral_Area_24d:41
u:R_Ventral_Area_24d:1041
u:L_Lateral_Area_7A:42
u:R_Lateral_Area_7A:1042
u:L_Supplementary_And_Cingulate_Eye:43
u:R_Supplementary_And_Cingulate_Eye:1043
u:L_Area_6m_anterior:44
u:R_Area_6m_anterior:1044
u:L_Medial_Area_7A:45
u:R_Medial_Area_7A:1045
u:L_Lateral_Area_7P:46
u:R_Lateral_Area_7P:1046
u:L_Area_7PC:47
u:R_Area_7PC:1047
u:L_Area_Lateral_IntraParietal_ventral:48
u:R_Area_Lateral_IntraParietal_ventral:1048
u:L_Ventral_IntraParietal_Complex:49
u:R_Ventral_IntraParietal_Complex:1049
u:L_Medial_IntraParietal_Area:50
u:R_Medial_IntraParietal_Area:1050
u:L_Area_1:51
u:R_Area_1:1051
u:L_Area_2:52
u:R_Area_2:1052
u:L_Area_3a:53
u:R_Area_3a:1053
u:L_Dorsal_area_6:54
u:R_Dorsal_area_6:1054
u:L_Area_6mp:55
u:R_Area_6mp:1055
u:L_Ventral_Area_6:56
u:R_Ventral_Area_6:1056
u:L_Area_Posterior_24_prime:57
u:R_Area_Posterior_24_prime:1057
u:L_Area_33_prime:58
u:R_Area_33_prime:1058
u:L_Anterior_24_prime:59
u:R_Anterior_24_prime:1059
u:L_Area_p32_prime:60
u:R_Area_p32_prime:1060
u:L_Area_a24:61
u:R_Area_a24:1061
u:L_Area_dorsal_32:62
u:R_Area_dorsal_32:1062
u:L_Area_8BM:63
u:R_Area_8BM:1063
u:L_Area_p32:64
u:R_Area_p32:1064
u:L_Area_10r:65
u:R_Area_10r:1065
u:L_Area_47m:66
u:R_Area_47m:1066
u:L_Area_8Av:67
u:R_Area_8Av:1067
u:L_Area_8Ad:68
u:R_Area_8Ad:1068
u:L_Area_9_Middle:69
u:R_Area_9_Middle:1069
u:L_Area_8B_Lateral:70
u:R_Area_8B_Lateral:1070
u:L_Area_9_Posterior:71
u:R_Area_9_Posterior:1071
u:L_Area_10d:72
u:R_Area_10d:1072
u:L_Area_8C:73
u:R_Area_8C:1073
u:L_Area_44:74
u:R_Area_44:1074
u:L_Area_45:75
u:R_Area_45:1075
u:L_Area_47l_(47_lateral):76
u:R_Area_47l_(47_lateral):1076
u:L_Area_anterior_47r:77
u:R_Area_anterior_47r:1077
u:L_Rostral_Area_6:78
u:R_Rostral_Area_6:1078
u:L_Area_IFJa:79
u:R_Area_IFJa:1079
u:L_Area_IFJp:80
u:R_Area_IFJp:1080
u:L_Area_IFSp:81
u:R_Area_IFSp:1081
u:L_Area__IFSa:82
u:R_Area__IFSa:1082
u:L_Area_posterior_9-46v:83
u:R_Area_posterior_9-46v:1083
u:L_Area_46:84
u:R_Area_46:1084
u:L_Area_anterior_9-46v:85
u:R_Area_anterior_9-46v:1085
u:L_Area_9-46d:86
u:R_Area_9-46d:1086
u:L_Area_9_anterior:87
u:R_Area_9_anterior:1087
u:L_Area_10v:88
u:R_Area_10v:1088
u:L_Area_anterior_10p:89
u:R_Area_anterior_10p:1089
u:L_Polar_10p:90
u:R_Polar_10p:1090
u:L_Area_11l:91
u:R_Area_11l:1091
u:L_Area_13l:92
u:R_Area_13l:1092
u:L_Orbital_Frontal_Complex:93
u:R_Orbital_Frontal_Complex:1093
u:L_Area_47s:94
u:R_Area_47s:1094
u:L_Area_Lateral_IntraParietal_dorsal:95
u:R_Area_Lateral_IntraParietal_dorsal:1095
u:L_Area_6_anterior:96
u:R_Area_6_anterior:1096
u:L_Inferior_6-8_Transitional_Area:97
u:R_Inferior_6-8_Transitional_Area:1097
u:L_Superior_6-8_Transitional_Area:98
u:R_Superior_6-8_Transitional_Area:1098
u:L_Area_43:99
u:R_Area_43:1099
u:L_Area_OP4/PV:100
u:R_Area_OP4/PV:1100
u:L_Area_OP1/SII:101
u:R_Area_OP1/SII:1101
u:L_Area_OP2-3/VS:102
u:R_Area_OP2-3/VS:1102
u:L_Area_52:103
u:R_Area_52:1103
u:L_RetroInsular_Cortex:104
u:R_RetroInsular_Cortex:1104
u:L_Area_PFcm:105
u:R_Area_PFcm:1105
u:L_Posterior_Insular_Area_2:106
u:R_Posterior_Insular_Area_2:1106
u:L_Area_TA2:107
u:R_Area_TA2:1107
u:L_Frontal_Opercular_Area_4:108
u:R_Frontal_Opercular_Area_4:1108
u:L_Middle_Insular_Area:109
u:R_Middle_Insular_Area:1109
u:L_Piriform_Cortex:110
u:R_Piriform_Cortex:1110
u:L_Anterior_Ventral_Insular_Area:111
u:R_Anterior_Ventral_Insular_Area:1111
u:L_Anterior_Agranular_Insula_Complex:112
u:R_Anterior_Agranular_Insula_Complex:1112
u:L_Frontal_Opercular_Area_1:113
u:R_Frontal_Opercular_Area_1:1113
u:L_Frontal_Opercular_Area_3:114
u:R_Frontal_Opercular_Area_3:1114
u:L_Frontal_Opercular_Area_2:115
u:R_Frontal_Opercular_Area_2:1115
u:L_Area_PFt:116
u:R_Area_PFt:1116
u:L_Anterior_IntraParietal_Area:117
u:R_Anterior_IntraParietal_Area:1117
u:L_Entorhinal_Cortex:118
u:R_Entorhinal_Cortex:1118
u:L_PreSubiculum:119
u:R_PreSubiculum:1119
u:L_Hippocampus:120
u:R_Hippocampus:1120
u:L_ProStriate_Area:121
u:R_ProStriate_Area:1121
u:L_Perirhinal_Ectorhinal_Cortex:122
u:R_Perirhinal_Ectorhinal_Cortex:1122
u:L_Area_STGa:123
u:R_Area_STGa:1123
u:L_ParaBelt_Complex:124
u:R_ParaBelt_Complex:1124
u:L_Auditory_5_Complex:125
u:R_Auditory_5_Complex:1125
u:L_ParaHippocampal_Area_1:126
u:R_ParaHippocampal_Area_1:1126
u:L_ParaHippocampal_Area_3:127
u:R_ParaHippocampal_Area_3:1127
u:L_Area_STSd_anterior:128
u:R_Area_STSd_anterior:1128
u:L_Area_STSd_posterior:129
u:R_Area_STSd_posterior:1129
u:L_Area_STSv_posterior:130
u:R_Area_STSv_posterior:1130
u:L_Area_TG_dorsal:131
u:R_Area_TG_dorsal:1131
u:L_Area_TE1_anterior:132
u:R_Area_TE1_anterior:1132
u:L_Area_TE1_posterior:133
u:R_Area_TE1_posterior:1133
u:L_Area_TE2_anterior:134
u:R_Area_TE2_anterior:1134
u:L_Area_TF:135
u:R_Area_TF:1135
u:L_Area_TE2_posterior:136
u:R_Area_TE2_posterior:1136
u:L_Area_PHT:137
u:R_Area_PHT:1137
u:L_Area_PH:138
u:R_Area_PH:1138
u:L_Area_TemporoParietoOccipital_Junction_1:139
u:R_Area_TemporoParietoOccipital_Junction_1:1139
u:L_Area_TemporoParietoOccipital_Junction_2:140
u:R_Area_TemporoParietoOccipital_Junction_2:1140
u:L_Area_TemporoParietoOccipital_Junction_3:141
u:R_Area_TemporoParietoOccipital_Junction_3:1141
u:L_Dorsal_Transitional_Visual_Area:142
u:R_Dorsal_Transitional_Visual_Area:1142
u:L_Area_PGp:143
u:R_Area_PGp:1143
u:L_Area_IntraParietal_2:144
u:R_Area_IntraParietal_2:1144
u:L_Area_IntraParietal_1:145
u:R_Area_IntraParietal_1:1145
u:L_Area_IntraParietal_0:146
u:R_Area_IntraParietal_0:1146
u:L_Area_PF_opercular:147
u:R_Area_PF_opercular:1147
u:L_Area_PF_Complex:148
u:R_Area_PF_Complex:1148
u:L_Area_PFm_Complex:149
u:R_Area_PFm_Complex:1149
u:L_Area_PGi:150
u:R_Area_PGi:1150
u:L_Area_PGs:151
u:R_Area_PGs:1151
u:L_Area_V6A:152
u:R_Area_V6A:1152
u:L_VentroMedial_Visual_Area_1:153
u:R_VentroMedial_Visual_Area_1:1153
u:L_VentroMedial_Visual_Area_3:154
u:R_VentroMedial_Visual_Area_3:1154
u:L_ParaHippocampal_Area_2:155
u:R_ParaHippocampal_Area_2:1155
u:L_Area_V4t:156
u:R_Area_V4t:1156
u:L_Area_FST:157
u:R_Area_FST:1157
u:L_Area_V3CD:158
u:R_Area_V3CD:1158
u:L_Area_Lateral_Occipital_3:159
u:R_Area_Lateral_Occipital_3:1159
u:L_VentroMedial_Visual_Area_2:160
u:R_VentroMedial_Visual_Area_2:1160
u:L_Area_31pd:161
u:R_Area_31pd:1161
u:L_Area_31a:162
u:R_Area_31a:1162
u:L_Ventral_Visual_Complex:163
u:R_Ventral_Visual_Complex:1163
u:L_Area_25:164
u:R_Area_25:1164
u:L_Area_s32:165
u:R_Area_s32:1165
u:L_posterior_OFC_Complex:166
u:R_posterior_OFC_Complex:1166
u:L_Area_Posterior_Insular_1:167
u:R_Area_Posterior_Insular_1:1167
u:L_Insular_Granular_Complex:168
u:R_Insular_Granular_Complex:1168
u:L_Area_Frontal_Opercular:169
u:R_Area_Frontal_Opercular:1169
u:L_Area_posterior_10p:170
u:R_Area_posterior_10p:1170
u:L_Area_posterior_47r:171
u:R_Area_posterior_47r:1171
u:L_Area_TG_Ventral:172
u:R_Area_TG_Ventral:1172
u:L_Medial_Belt_Complex:173
u:R_Medial_Belt_Complex:1173
u:L_Lateral_Belt_Complex:174
u:R_Lateral_Belt_Complex:1174
u:L_Auditory_4_Complex:175
u:R_Auditory_4_Complex:1175
u:L_Area_STSv_anterior:176
u:R_Area_STSv_anterior:1176
u:L_Area_TE1_Middle:177
u:R_Area_TE1_Middle:1177
u:L_Para-Insular_Area:178
u:R_Para-Insular_Area:1178
u:L_Area_anterior_32_prime:179
u:R_Area_anterior_32_prime:1179
u:L_Area_posterior_24:180
u:R_Area_posterior_24:1180
----------- End regions for MNI_Glasser_HCP_v1.0 atlas --------------
Atlas Brainnetome_1.0, 246 regions
----------- Begin regions for Brainnetome_1.0 atlas-----------
u:A8m_left:1
u:A8dl_left:3
u:A9l_left:5
u:A6dl_left:7
u:A6m_left:9
u:A9m_left:11
u:A10m_left:13
u:A9/46d_left:15
u:IFJ_left:17
u:A46_left:19
u:A9/46v_left:21
u:A8vl_left:23
u:A6vl_left:25
u:A10l_left:27
u:A44d_left:29
u:IFS_left:31
u:A45c_left:33
u:A45r_left:35
u:A44op_left:37
u:A44v_left:39
u:A14m_left:41
u:A12/47o_left:43
u:A11l_left:45
u:A11m_left:47
u:A13_left:49
u:A12/47l_left:51
u:A4hf_left:53
u:A6cdl_left:55
u:A4ul_left:57
u:A4t_left:59
u:A4tl_left:61
u:A6cvl_left:63
u:A1/2/3ll_left:65
u:A4ll_left:67
u:A38m_left:69
u:A41/42_left:71
u:TE1.0_1.2_left:73
u:A22c_left:75
u:A38l_left:77
u:A22r_left:79
u:A21c_left:81
u:A21r_left:83
u:A37dl_left:85
u:aSTS_left:87
u:A20iv_left:89
u:A37elv_left:91
u:A20r_left:93
u:A20il_left:95
u:A37vl_left:97
u:A20cl_left:99
u:A20cv_left:101
u:A20rv_left:103
u:A37mv_left:105
u:A37lv_left:107
u:A35/36r_left:109
u:A35/36c_left:111
u:TL_left:113
u:A28/34_left:115
u:TI_left:117
u:TH_left:119
u:rpSTS_left:121
u:cpSTS_left:123
u:A7r_left:125
u:A7c_left:127
u:A5l_left:129
u:A7pc_left:131
u:A7ip_left:133
u:A39c_left:135
u:A39rd_left:137
u:A40rd_left:139
u:A40c_left:141
u:A39rv_left:143
u:A40rv_left:145
u:A7m_left:147
u:A5m_left:149
u:dmPOS_left:151
u:A31_left:153
u:A1/2/3ulhf_left:155
u:A1/2/3tonIa_left:157
u:A2_left:159
u:A1/2/3tru_left:161
u:G_left:163
u:vIa_left:165
u:dIa_left:167
u:vId/vIg_left:169
u:dIg_left:171
u:dId_left:173
u:A23d_left:175
u:A24rv_left:177
u:A32p_left:179
u:A23v_left:181
u:A24cd_left:183
u:A23c_left:185
u:A32sg_left:187
u:cLinG_left:189
u:rCunG_left:191
u:cCunG_left:193
u:rLinG_left:195
u:vmPOS_left:197
u:mOccG_left:199
u:V5/MT+_left:201
u:OPC_left:203
u:iOccG_left:205
u:msOccG_left:207
u:lsOccG_left:209
u:mAmyg_left:211
u:lAmyg_left:213
u:rHipp_left:215
u:cHipp_left:217
u:vCa_left:219
u:GP_left:221
u:NAC_left:223
u:vmPu_left:225
u:dCa_left:227
u:dlPu_left:229
u:mPFtha_left:231
u:mPMtha_left:233
u:Stha_left:235
u:rTtha_left:237
u:PPtha_left:239
u:Otha_left:241
u:cTtha_left:243
u:lPFtha_left:245
u:A8m_right:2
u:A8dl_right:4
u:A9l_right:6
u:A6dl_right:8
u:A6m_right:10
u:A9m_right:12
u:A10m_right:14
u:A9/46d_right:16
u:IFJ_right:18
u:A46_right:20
u:A9/46v_right:22
u:A8vl_right:24
u:A6vl_right:26
u:A10l_right:28
u:A44d_right:30
u:IFS_right:32
u:A45c_right:34
u:A45r_right:36
u:A44op_right:38
u:A44v_right:40
u:A14m_right:42
u:A12/47o_right:44
u:A11l_right:46
u:A11m_right:48
u:A13_right:50
u:A12/47l_right:52
u:A4hf_right:54
u:A6cdl_right:56
u:A4ul_right:58
u:A4t_right:60
u:A4tl_right:62
u:A6cvl_right:64
u:A1/2/3ll_right:66
u:A4ll_right:68
u:A38m_right:70
u:A41/42_right:72
u:TE1.0_1.2_right:74
u:A22c_right:76
u:A38l_right:78
u:A22r_right:80
u:A21c_right:82
u:A21r_right:84
u:A37dl_right:86
u:aSTS_right:88
u:A20iv_right:90
u:A37elv_right:92
u:A20r_right:94
u:A20il_right:96
u:A37vl_right:98
u:A20cl_right:100
u:A20cv_right:102
u:A20rv_right:104
u:A37mv_right:106
u:A37lv_right:108
u:A35/36r_right:110
u:A35/36c_right:112
u:TL_right:114
u:A28/34_right:116
u:TI_right:118
u:TH_right:120
u:rpSTS_right:122
u:cpSTS_right:124
u:A7r_right:126
u:A7c_right:128
u:A5l_right:130
u:A7pc_right:132
u:A7ip_right:134
u:A39c_right:136
u:A39rd_right:138
u:A40rd_right:140
u:A40c_right:142
u:A39rv_right:144
u:A40rv_right:146
u:A7m_right:148
u:A5m_right:150
u:dmPOS_right:152
u:A31_right:154
u:A1/2/3ulhf_right:156
u:A1/2/3tonIa_right:158
u:A2_right:160
u:A1/2/3tru_right:162
u:G_right:164
u:vIa_right:166
u:dIa_right:168
u:vId/vIg_right:170
u:dIg_right:172
u:dId_right:174
u:A23d_right:176
u:A24rv_right:178
u:A32p_right:180
u:A23v_right:182
u:A24cd_right:184
u:A23c_right:186
u:A32sg_right:188
u:cLinG_right:190
u:rCunG_right:192
u:cCunG_right:194
u:rLinG_right:196
u:vmPOS_right:198
u:mOccG_right:200
u:V5/MT+_right:202
u:OPC_right:204
u:iOccG_right:206
u:msOccG_right:208
u:lsOccG_right:210
u:mAmyg_right:212
u:lAmyg_right:214
u:rHipp_right:216
u:cHipp_right:218
u:vCa_right:220
u:GP_right:222
u:NAC_right:224
u:vmPu_right:226
u:dCa_right:228
u:dlPu_right:230
u:mPFtha_right:232
u:mPMtha_right:234
u:Stha_right:236
u:rTtha_right:238
u:PPtha_right:240
u:Otha_right:242
u:cTtha_right:244
u:lPFtha_right:246
----------- End regions for Brainnetome_1.0 atlas --------------
Atlas CA_MPM_22_MNI, 100 regions
----------- Begin regions for CA_MPM_22_MNI atlas-----------
u:Interposed_Nucleus:111
u:Amygdala_(AStr):193
u:Amygdala_(CM):133
u:Amygdala_(LB):202
u:Amygdala_(SF):151
u:Area_FG3:117
u:Area_1:106
u:Area_2:243
u:Area_25:178
u:Area_33:119
u:Area_3a:250
u:Area_3b:187
u:Area_44:246
u:Area_45:129
u:Area_4a:160
u:Area_4p:191
u:Area_5Ci_(SPL):185
u:Area_5L_(SPL):159
u:Area_5M_(SPL):241
u:Area_7A_(SPL):165
u:Area_7M_(SPL):120
u:Area_7P_(SPL):224
u:Area_7PC_(SPL):167
u:Area_FG1:148
u:Area_FG2:171
u:Area_FG4:123
u:Area_Fo1:170
u:Area_Fo2:109
u:Area_Fo3:145
u:Area_Fp1:219
u:Area_Fp2:230
u:Area_Id1:126
u:Area_Ig1:236
u:Area_Ig2:204
u:Area_OP1_(SII):122
u:Area_OP2_(PIVC):139
u:Area_OP3_(VS):147
u:Area_OP4_(PV):173
u:Area_PF_(IPL):136
u:Area_PFcm_(IPL):153
u:Area_PFm_(IPL):128
u:Area_PFop_(IPL):125
u:Area_PFt_(IPL):240
u:Area_PGa_(IPL):201
u:Area_PGp_(IPL):198
u:Area_TE_1.0:210
u:Area_TE_1.1:213
u:Area_TE_1.2:218
u:Area_TE_3:181
u:Area_hIP1_(IPS):131
u:Area_hIP2_(IPS):226
u:Area_hIP3_(IPS):157
u:Area_hOc1_(V1):249
u:Area_hOc2_(V2):252
u:Area_hOc3d_(V3d):199
u:Area_hOc3v_(V3v):108
u:Area_hOc4d_(V3A):143
u:Area_hOc4la:162
u:Area_hOc4lp:233
u:Area_hOc4v_(V4(v)):103
u:Area_hOc5_(V5/MT):105
u:Area_s24:253
u:Area_s32:102
u:BF_(Ch_1-3):114
u:BF_(Ch_4):142
u:CA1_(Hippocampus):238
u:CA2_(Hippocampus):116
u:CA3_(Hippocampus):168
u:DG_(Hippocampus):150
u:Dorsal_Dentate_Nucleus:235
u:Entorhinal_Cortex:229
u:Fastigii_Nucleus:255
u:HATA_Region:174
u:Lobule_I_IV_(Hem):215
u:Lobule_IX_(Hem):134
u:Lobule_IX_(Verm):209
u:Lobule_V_(Hem):247
u:Lobule_VI_(Hem):222
u:Lobule_VI_(Verm):112
u:Lobule_VIIIa_(Hem):207
u:Lobule_VIIIa_(Verm):182
u:Lobule_VIIIb_(Hem):196
u:Lobule_VIIIb_(Verm):154
u:Lobule_VIIa_crusI_(Hem):212
u:Lobule_VIIa_crusI_(Verm):137
u:Lobule_VIIa_crusII_(Hem):216
u:Lobule_VIIa_crusII_(Verm):140
u:Lobule_VIIb_(Hem):164
u:Lobule_VIIb_(Verm):205
u:Lobule_X_(Hem):221
u:Lobule_X_(Verm):184
u:Subiculum:244
u:Thal:_Motor:176
u:Thal:_Parietal:179
u:Thal:_Prefrontal:156
u:Thal:_Premotor:188
u:Thal:_Somatosensory:190
u:Thal:_Temporal:232
u:Thal:_Visual:227
u:Ventral_Dentate_Nucleus:195
----------- End regions for CA_MPM_22_MNI atlas --------------
Atlas CA_MPM_22_TT, 100 regions
----------- Begin regions for CA_MPM_22_TT atlas-----------
u:Interposed_Nucleus :111
u:Amygdala_(AStr) :193
u:Amygdala_(CM) :133
u:Amygdala_(LB) :202
u:Amygdala_(SF) :151
u:Area_FG3 :117
u:Area_1 :106
u:Area_2 :243
u:Area_25 :178
u:Area_33 :119
u:Area_3a :250
u:Area_3b :187
u:Area_44 :246
u:Area_45 :129
u:Area_4a :160
u:Area_4p :191
u:Area_5Ci_(SPL) :185
u:Area_5L_(SPL) :159
u:Area_5M_(SPL) :241
u:Area_7A_(SPL) :165
u:Area_7M_(SPL) :120
u:Area_7P_(SPL) :224
u:Area_7PC_(SPL) :167
u:Area_FG1 :148
u:Area_FG2 :171
u:Area_FG4 :123
u:Area_Fo1 :170
u:Area_Fo2 :109
u:Area_Fo3 :145
u:Area_Fp1 :219
u:Area_Fp2 :230
u:Area_Id1 :126
u:Area_Ig1 :236
u:Area_Ig2 :204
u:Area_OP1_(SII) :122
u:Area_OP2_(PIVC) :139
u:Area_OP3_(VS) :147
u:Area_OP4_(PV) :173
u:Area_PF_(IPL) :136
u:Area_PFcm_(IPL) :153
u:Area_PFm_(IPL) :128
u:Area_PFop_(IPL) :125
u:Area_PFt_(IPL) :240
u:Area_PGa_(IPL) :201
u:Area_PGp_(IPL) :198
u:Area_TE_1.0 :210
u:Area_TE_1.1 :213
u:Area_TE_1.2 :218
u:Area_TE_3 :181
u:Area_hIP1_(IPS) :131
u:Area_hIP2_(IPS) :226
u:Area_hIP3_(IPS) :157
u:Area_hOc1_(V1) :249
u:Area_hOc2_(V2) :252
u:Area_hOc3d_(V3d) :199
u:Area_hOc3v_(V3v) :108
u:Area_hOc4d_(V3A) :143
u:Area_hOc4la :162
u:Area_hOc4lp :233
u:Area_hOc4v_(V4(v)) :103
u:Area_hOc5_(V5/MT) :105
u:Area_s24 :253
u:Area_s32 :102
u:BF_(Ch_1-3) :114
u:BF_(Ch_4) :142
u:CA1_(Hippocampus) :238
u:CA2_(Hippocampus) :116
u:CA3_(Hippocampus) :168
u:DG_(Hippocampus) :150
u:Dorsal_Dentate_Nucleus :235
u:Entorhinal_Cortex :229
u:Fastigii_Nucleus :255
u:HATA_Region :174
u:Lobule_I_IV_(Hem) :215
u:Lobule_IX_(Hem) :134
u:Lobule_IX_(Verm) :209
u:Lobule_V_(Hem) :247
u:Lobule_VI_(Hem) :222
u:Lobule_VI_(Verm) :112
u:Lobule_VIIIa_(Hem) :207
u:Lobule_VIIIa_(Verm) :182
u:Lobule_VIIIb_(Hem) :196
u:Lobule_VIIIb_(Verm) :154
u:Lobule_VIIa_crusI_(Hem) :212
u:Lobule_VIIa_crusI_(Verm) :137
u:Lobule_VIIa_crusII_(Hem) :216
u:Lobule_VIIa_crusII_(Verm) :140
u:Lobule_VIIb_(Hem) :164
u:Lobule_VIIb_(Verm) :205
u:Lobule_X_(Hem) :221
u:Lobule_X_(Verm) :184
u:Subiculum :244
u:Thal:_Motor :176
u:Thal:_Parietal :179
u:Thal:_Prefrontal :156
u:Thal:_Premotor :188
u:Thal:_Somatosensory :190
u:Thal:_Temporal :232
u:Thal:_Visual :227
u:Ventral_Dentate_Nucleus :195
----------- End regions for CA_MPM_22_TT atlas --------------
Atlas CA_N27_ML, 115 regions
----------- Begin regions for CA_N27_ML atlas-----------
l:Left Precentral Gyrus:1
r:Right Precentral Gyrus:2
l:Left Superior Frontal Gyrus:3
r:Right Superior Frontal Gyrus:4
l:Left Superior Orbital Gyrus:5
r:Right Superior Orbital Gyrus:6
l:Left Middle Frontal Gyrus:7
r:Right Middle Frontal Gyrus:8
l:Left Middle Orbital Gyrus:9
r:Right Middle Orbital Gyrus:10
l:Left Inferior Frontal Gyrus (p. Opercularis):11
r:Right Inferior Frontal Gyrus (p. Opercularis):12
l:Left Inferior Frontal Gyrus (p. Triangularis):13
r:Right Inferior Frontal Gyrus (p. Triangularis):14
l:Left Inferior Frontal Gyrus (p. Orbitalis):15
r:Right Inferior Frontal Gyrus (p. Orbitalis):16
l:Left Rolandic Operculum:17
r:Right Rolandic Operculum:18
l:Left SMA:19
r:Right SMA:20
l:Left Olfactory cortex:21
r:Right Olfactory cortex:22
l:Left Superior Medial Gyrus:23
r:Right Superior Medial Gyrus:24
l:Left Mid Orbital Gyrus:25
r:Right Mid Orbital Gyrus:26
l:Left Rectal Gyrus:27
r:Right Rectal Gyrus:28
l:Left Insula Lobe:29
r:Right Insula Lobe:30
l:Left Anterior Cingulate Cortex:31
r:Right Anterior Cingulate Cortex:32
l:Left Middle Cingulate Cortex:33
r:Right Middle Cingulate Cortex:34
l:Left Posterior Cingulate Cortex:35
r:Right Posterior Cingulate Cortex:36
l:Left Hippocampus:37
r:Right Hippocampus:38
l:Left ParaHippocampal Gyrus:39
r:Right ParaHippocampal Gyrus:40
l:Left Amygdala:41
r:Right Amygdala:42
l:Left Calcarine Gyrus:43
r:Right Calcarine Gyrus:44
l:Left Cuneus:45
r:Right Cuneus:46
l:Left Lingual Gyrus:47
r:Right Lingual Gyrus:48
l:Left Superior Occipital Gyrus:49
r:Right Superior Occipital Gyrus:50
l:Left Middle Occipital Gyrus:51
r:Right Middle Occipital Gyrus:52
l:Left Inferior Occipital Gyrus:53
r:Right Inferior Occipital Gyrus:54
l:Left Fusiform Gyrus:55
r:Right Fusiform Gyrus:56
l:Left Postcentral Gyrus:57
r:Right Postcentral Gyrus:58
l:Left Superior Parietal Lobule:59
r:Right Superior Parietal Lobule:60
l:Left Inferior Parietal Lobule:61
r:Right Inferior Parietal Lobule:62
l:Left SupraMarginal Gyrus:63
r:Right SupraMarginal Gyrus:64
l:Left Angular Gyrus:65
r:Right Angular Gyrus:66
l:Left Precuneus:67
r:Right Precuneus:68
l:Left Paracentral Lobule:69
r:Right Paracentral Lobule:70
l:Left Caudate Nucleus:71
r:Right Caudate Nucleus:72
l:Left Putamen:73
r:Right Putamen:74
l:Left Pallidum:75
r:Right Pallidum:76
l:Left Thalamus:77
r:Right Thalamus:78
l:Left Heschls Gyrus:79
r:Right Heschls Gyrus:80
l:Left Superior Temporal Gyrus:81
r:Right Superior Temporal Gyrus:82
l:Left Temporal Pole:83
r:Right Temporal Pole:84
l:Left Middle Temporal Gyrus:85
r:Right Middle Temporal Gyrus:86
l:Left Medial Temporal Pole:87
r:Right Medial Temporal Pole:88
l:Left Inferior Temporal Gyrus:89
r:Right Inferior Temporal Gyrus:90
l:Left Cerebellum (Crus 1):91
r:Right Cerebellum (Crus 1):92
l:Left Cerebellum (Crus 2):93
r:Right Cerebellum (Crus 2):94
l:Left Cerebellum (III):95
r:Right Cerebellum (III):96
l:Left Cerebellum (IV-V):97
r:Right Cerebellum (IV-V):98
l:Left Cerebellum (VI):99
r:Right Cerebellum (VI):100
l:Left Cerebellum (VII):101
r:Right Cerebellum (VII):102
l:Left Cerebellum (VIII):103
r:Right Cerebellum (VIII):104
l:Left Cerebellum (IX):105
r:Right Cerebellum (IX):106
l:Left Cerebellum (X):107
r:Right Cerebellum (X):108
u:Cerebellar Vermis (1/2):109
u:Cerebellar Vermis (3):110
u:Cerebellar Vermis (4/5):111
u:Cerebellar Vermis (6):112
u:Cerebellar Vermis (7):113
u:Cerebellar Vermis (8):114
u:Cerebellar Vermis (9):115
----------- End regions for CA_N27_ML atlas --------------
Atlas CA_N27_MPM, 75 regions
----------- Begin regions for CA_N27_MPM atlas-----------
u:hIP1:103
u:SPL (7M):105
u:Lobule X (Hem):107
u:Lobule V:109
u:Th-Parietal:111
u:Hipp (FD):113
u:Amyg (SF):115
u:TE 1.1:117
u:Area 4p:119
u:Insula (Id1):121
u:Hipp (CA):123
u:hOC3v (V3v):125
u:IPC (PGa):127
u:TE 1.2:129
u:Area 44:131
u:Area 45:133
u:Area 4a:135
u:Amyg (LB):137
u:Insula (Ig1):140
u:Lobule IX (Vermis):142
u:SPL (5L):144
u:Th-Premotor:146
u:Insula (Ig2):148
u:Lobules I-IV (Hem):150
u:SPL (7A):152
u:TE 1.0:154
u:Area 2:156
u:Lobule VIIa Crus I (Vermis):158
u:Amyg (CM):160
u:hOC4v (V4):162
u:Lobule IX (Hem):164
u:Th-Temporal:166
u:Lobule VIIIb (Hem):168
u:Lobule VI (Hem):170
u:Hipp (HATA):172
u:Th-Prefrontal:174
u:Area 6:176
u:TE 3:179
u:Area 17:181
u:OP 2:183
u:Lobule VIIIa (Vermis):185
u:Th-Somatosensory:187
u:IPC (PFm):189
u:Lobule X (Vermis):191
u:hOC5 (V5):193
u:Lobule VIIa Crus II (Vermis):195
u:Hipp (SUB):197
u:Lobule VI (Vermis):199
u:OP 1:201
u:SPL (5Ci):203
u:Lobule VIIa Crus II (Hem):205
u:hIP3:207
u:Lobule VIIIa (Hem):209
u:Th-Visual:211
u:Hipp (EC):213
u:IPC (PFt):215
u:Area 3b:218
u:Lobule VIIb (Vermis):220
u:Lobule VIIb (Hem):222
u:IPC (PFcm):224
u:IPC (PF):226
u:hIP2:228
u:SPL (7P):230
u:SPL (5M):232
u:IPC (PGp):234
u:Lobule VIIIb (Vermis):236
u:Th-Motor:238
u:Area 18:240
u:OP 4:242
u:IPC (PFop):244
u:OP 3:246
u:Lobule VIIa Crus I (Hem):248
u:Area 3a:250
u:Area 1:252
u:SPL (7PC):254
----------- End regions for CA_N27_MPM atlas --------------
** ERROR: Failed to get CA_N27_PM
*+ WARNING: NULL atlas
** ERROR: NULL atlas
Atlas CA_N27_GW, 2 regions
----------- Begin regions for CA_N27_GW atlas-----------
u:grey:0
u:white:1
----------- End regions for CA_N27_GW atlas --------------
Atlas CA_N27_LR, 2 regions
----------- Begin regions for CA_N27_LR atlas-----------
r:Right Brain:1
l:Left Brain:2
----------- End regions for CA_N27_LR atlas --------------
Atlas CA_ML_18_MNI, 116 regions
----------- Begin regions for CA_ML_18_MNI atlas-----------
l:Left Precentral Gyrus:1
r:Right Precentral Gyrus:2
l:Left Superior Frontal Gyrus:3
r:Right Superior Frontal Gyrus:4
l:Left Superior Orbital Gyrus:5
r:Right Superior Orbital Gyrus:6
l:Left Middle Frontal Gyrus:7
r:Right Middle Frontal Gyrus:8
l:Left Middle Orbital Gyrus:9
r:Right Middle Orbital Gyrus:10
l:Left Inferior Frontal Gyrus (p. Opercularis):11
r:Right Inferior Frontal Gyrus (p. Opercularis):12
l:Left Inferior Frontal Gyrus (p. Triangularis):13
r:Right Inferior Frontal Gyrus (p. Triangularis):14
l:Left Inferior Frontal Gyrus (p. Orbitalis):15
r:Right Inferior Frontal Gyrus (p. Orbitalis):16
l:Left Rolandic Operculum:17
r:Right Rolandic Operculum:18
l:Left SMA:19
r:Right SMA:20
l:Left Olfactory cortex:21
r:Right Olfactory cortex:22
l:Left Superior Medial Gyrus:23
r:Right Superior Medial Gyrus:24
l:Left Mid Orbital Gyrus:25
r:Right Mid Orbital Gyrus:26
l:Left Rectal Gyrus:27
r:Right Rectal Gyrus:28
l:Left Insula Lobe:29
r:Right Insula Lobe:30
l:Left Anterior Cingulate Cortex:31
r:Right Anterior Cingulate Cortex:32
l:Left Middle Cingulate Cortex:33
r:Right Middle Cingulate Cortex:34
l:Left Posterior Cingulate Cortex:35
r:Right Posterior Cingulate Cortex:36
l:Left Hippocampus:37
r:Right Hippocampus:38
l:Left ParaHippocampal Gyrus:39
r:Right ParaHippocampal Gyrus:40
l:Left Amygdala:41
r:Right Amygdala:42
l:Left Calcarine Gyrus:43
r:Right Calcarine Gyrus:44
l:Left Cuneus:45
r:Right Cuneus:46
l:Left Lingual Gyrus:47
r:Right Lingual Gyrus:48
l:Left Superior Occipital Gyrus:49
r:Right Superior Occipital Gyrus:50
l:Left Middle Occipital Gyrus:51
r:Right Middle Occipital Gyrus:52
l:Left Inferior Occipital Gyrus:53
r:Right Inferior Occipital Gyrus:54
l:Left Fusiform Gyrus:55
r:Right Fusiform Gyrus:56
l:Left Postcentral Gyrus:57
r:Right Postcentral Gyrus:58
l:Left Superior Parietal Lobule:59
r:Right Superior Parietal Lobule:60
l:Left Inferior Parietal Lobule:61
r:Right Inferior Parietal Lobule:62
l:Left SupraMarginal Gyrus:63
r:Right SupraMarginal Gyrus:64
l:Left Angular Gyrus:65
r:Right Angular Gyrus:66
l:Left Precuneus:67
r:Right Precuneus:68
l:Left Paracentral Lobule:69
r:Right Paracentral Lobule:70
l:Left Caudate Nucleus:71
r:Right Caudate Nucleus:72
l:Left Putamen:73
r:Right Putamen:74
l:Left Pallidum:75
r:Right Pallidum:76
l:Left Thalamus:77
r:Right Thalamus:78
l:Left Heschls Gyrus:79
r:Right Heschls Gyrus:80
l:Left Superior Temporal Gyrus:81
r:Right Superior Temporal Gyrus:82
l:Left Temporal Pole:83
r:Right Temporal Pole:84
l:Left Middle Temporal Gyrus:85
r:Right Middle Temporal Gyrus:86
l:Left Medial Temporal Pole:87
r:Right Medial Temporal Pole:88
l:Left Inferior Temporal Gyrus:89
r:Right Inferior Temporal Gyrus:90
l:Left Cerebellum (Crus 1):91
r:Right Cerebellum (Crus 1):92
l:Left Cerebellum (Crus 2):93
r:Right Cerebellum (Crus 2):94
l:Left Cerebellum (III):95
r:Right Cerebellum (III):96
l:Left Cerebellum (IV-V):97
r:Right Cerebellum (IV-V):98
l:Left Cerebellum (VI):99
r:Right Cerebellum (VI):100
l:Left Cerebellum (VII):101
r:Right Cerebellum (VII):102
l:Left Cerebellum (VIII):103
r:Right Cerebellum (VIII):104
l:Left Cerebellum (IX):105
r:Right Cerebellum (IX):106
l:Left Cerebellum (X):107
r:Right Cerebellum (X):108
u:Cerebellar Vermis (1/2):109
u:Cerebellar Vermis (3):110
u:Cerebellar Vermis (4/5):111
u:Cerebellar Vermis (6):112
u:Cerebellar Vermis (7):113
u:Cerebellar Vermis (8):114
u:Cerebellar Vermis (9):115
u:Cerebellar Vermis (10):116
----------- End regions for CA_ML_18_MNI atlas --------------
Atlas CA_MPM_18_MNI, 75 regions
----------- Begin regions for CA_MPM_18_MNI atlas-----------
u:hIP1:103
u:SPL (7M):105
u:Lobule X (Hem):107
u:Lobule V:109
u:Th-Parietal:111
u:Hipp (FD):113
u:Amyg (SF):115
u:TE 1.1:117
u:Area 4p:119
u:Insula (Id1):121
u:Hipp (CA):123
u:hOC3v (V3v):125
u:IPC (PGa):127
u:TE 1.2:129
u:Area 44:131
u:Area 45:133
u:Area 4a:135
u:Amyg (LB):137
u:Insula (Ig1):140
u:Lobule IX (Vermis):142
u:SPL (5L):144
u:Th-Premotor:146
u:Insula (Ig2):148
u:Lobules I-IV (Hem):150
u:SPL (7A):152
u:TE 1.0:154
u:Area 2:156
u:Lobule VIIa Crus I (Vermis):158
u:Amyg (CM):160
u:hOC4v (V4):162
u:Lobule IX (Hem):164
u:Th-Temporal:166
u:Lobule VIIIb (Hem):168
u:Lobule VI (Hem):170
u:Hipp (HATA):172
u:Th-Prefrontal:174
u:Area 6:176
u:TE 3:179
u:Area 17:181
u:OP 2:183
u:Lobule VIIIa (Vermis):185
u:Th-Somatosensory:187
u:IPC (PFm):189
u:Lobule X (Vermis):191
u:hOC5 (V5):193
u:Lobule VIIa Crus II (Vermis):195
u:Hipp (SUB):197
u:Lobule VI (Vermis):199
u:OP 1:201
u:SPL (5Ci):203
u:Lobule VIIa Crus II (Hem):205
u:hIP3:207
u:Lobule VIIIa (Hem):209
u:Th-Visual:211
u:Hipp (EC):213
u:IPC (PFt):215
u:Area 3b:218
u:Lobule VIIb (Vermis):220
u:Lobule VIIb (Hem):222
u:IPC (PFcm):224
u:IPC (PF):226
u:hIP2:228
u:SPL (7P):230
u:SPL (5M):232
u:IPC (PGp):234
u:Lobule VIIIb (Vermis):236
u:Th-Motor:238
u:Area 18:240
u:OP 4:242
u:IPC (PFop):244
u:OP 3:246
u:Lobule VIIa Crus I (Hem):248
u:Area 3a:250
u:Area 1:252
u:SPL (7PC):254
----------- End regions for CA_MPM_18_MNI atlas --------------
Atlas CA_LR_18_MNI, 2 regions
----------- Begin regions for CA_LR_18_MNI atlas-----------
r:Right Brain:1
l:Left Brain:2
----------- End regions for CA_LR_18_MNI atlas --------------
Atlas CA_ML_18_MNIA, 115 regions
----------- Begin regions for CA_ML_18_MNIA atlas-----------
l:Left Precentral Gyrus:1
r:Right Precentral Gyrus:2
l:Left Superior Frontal Gyrus:3
r:Right Superior Frontal Gyrus:4
l:Left Superior Orbital Gyrus:5
r:Right Superior Orbital Gyrus:6
l:Left Middle Frontal Gyrus:7
r:Right Middle Frontal Gyrus:8
l:Left Middle Orbital Gyrus:9
r:Right Middle Orbital Gyrus:10
l:Left Inferior Frontal Gyrus (p. Opercularis):11
r:Right Inferior Frontal Gyrus (p. Opercularis):12
l:Left Inferior Frontal Gyrus (p. Triangularis):13
r:Right Inferior Frontal Gyrus (p. Triangularis):14
l:Left Inferior Frontal Gyrus (p. Orbitalis):15
r:Right Inferior Frontal Gyrus (p. Orbitalis):16
l:Left Rolandic Operculum:17
r:Right Rolandic Operculum:18
l:Left SMA:19
r:Right SMA:20
l:Left Olfactory cortex:21
r:Right Olfactory cortex:22
l:Left Superior Medial Gyrus:23
r:Right Superior Medial Gyrus:24
l:Left Mid Orbital Gyrus:25
r:Right Mid Orbital Gyrus:26
l:Left Rectal Gyrus:27
r:Right Rectal Gyrus:28
l:Left Insula Lobe:29
r:Right Insula Lobe:30
l:Left Anterior Cingulate Cortex:31
r:Right Anterior Cingulate Cortex:32
l:Left Middle Cingulate Cortex:33
r:Right Middle Cingulate Cortex:34
l:Left Posterior Cingulate Cortex:35
r:Right Posterior Cingulate Cortex:36
l:Left Hippocampus:37
r:Right Hippocampus:38
l:Left ParaHippocampal Gyrus:39
r:Right ParaHippocampal Gyrus:40
l:Left Amygdala:41
r:Right Amygdala:42
l:Left Calcarine Gyrus:43
r:Right Calcarine Gyrus:44
l:Left Cuneus:45
r:Right Cuneus:46
l:Left Lingual Gyrus:47
r:Right Lingual Gyrus:48
l:Left Superior Occipital Gyrus:49
r:Right Superior Occipital Gyrus:50
l:Left Middle Occipital Gyrus:51
r:Right Middle Occipital Gyrus:52
l:Left Inferior Occipital Gyrus:53
r:Right Inferior Occipital Gyrus:54
l:Left Fusiform Gyrus:55
r:Right Fusiform Gyrus:56
l:Left Postcentral Gyrus:57
r:Right Postcentral Gyrus:58
l:Left Superior Parietal Lobule:59
r:Right Superior Parietal Lobule:60
l:Left Inferior Parietal Lobule:61
r:Right Inferior Parietal Lobule:62
l:Left SupraMarginal Gyrus:63
r:Right SupraMarginal Gyrus:64
l:Left Angular Gyrus:65
r:Right Angular Gyrus:66
l:Left Precuneus:67
r:Right Precuneus:68
l:Left Paracentral Lobule:69
r:Right Paracentral Lobule:70
l:Left Caudate Nucleus:71
r:Right Caudate Nucleus:72
l:Left Putamen:73
r:Right Putamen:74
l:Left Pallidum:75
r:Right Pallidum:76
l:Left Thalamus:77
r:Right Thalamus:78
l:Left Heschls Gyrus:79
r:Right Heschls Gyrus:80
l:Left Superior Temporal Gyrus:81
r:Right Superior Temporal Gyrus:82
l:Left Temporal Pole:83
r:Right Temporal Pole:84
l:Left Middle Temporal Gyrus:85
r:Right Middle Temporal Gyrus:86
l:Left Medial Temporal Pole:87
r:Right Medial Temporal Pole:88
l:Left Inferior Temporal Gyrus:89
r:Right Inferior Temporal Gyrus:90
l:Left Cerebellum (Crus 1):91
r:Right Cerebellum (Crus 1):92
l:Left Cerebellum (Crus 2):93
r:Right Cerebellum (Crus 2):94
l:Left Cerebellum (III):95
r:Right Cerebellum (III):96
l:Left Cerebellum (IV-V):97
r:Right Cerebellum (IV-V):98
l:Left Cerebellum (VI):99
r:Right Cerebellum (VI):100
l:Left Cerebellum (VII):101
r:Right Cerebellum (VII):102
l:Left Cerebellum (VIII):103
r:Right Cerebellum (VIII):104
l:Left Cerebellum (IX):105
r:Right Cerebellum (IX):106
l:Left Cerebellum (X):107
r:Right Cerebellum (X):108
u:Cerebellar Vermis (1/2):109
u:Cerebellar Vermis (3):110
u:Cerebellar Vermis (4/5):111
u:Cerebellar Vermis (6):112
u:Cerebellar Vermis (7):113
u:Cerebellar Vermis (8):114
u:Cerebellar Vermis (9):115
----------- End regions for CA_ML_18_MNIA atlas --------------
Atlas CA_MPM_18_MNIA, 75 regions
----------- Begin regions for CA_MPM_18_MNIA atlas-----------
u:hIP1:103
u:SPL (7M):105
u:Lobule X (Hem):107
u:Lobule V:109
u:Th-Parietal:111
u:Hipp (FD):113
u:Amyg (SF):115
u:TE 1.1:117
u:Area 4p:119
u:Insula (Id1):121
u:Hipp (CA):123
u:hOC3v (V3v):125
u:IPC (PGa):127
u:TE 1.2:129
u:Area 44:131
u:Area 45:133
u:Area 4a:135
u:Amyg (LB):137
u:Insula (Ig1):140
u:Lobule IX (Vermis):142
u:SPL (5L):144
u:Th-Premotor:146
u:Insula (Ig2):148
u:Lobules I-IV (Hem):150
u:SPL (7A):152
u:TE 1.0:154
u:Area 2:156
u:Lobule VIIa Crus I (Vermis):158
u:Amyg (CM):160
u:hOC4v (V4):162
u:Lobule IX (Hem):164
u:Th-Temporal:166
u:Lobule VIIIb (Hem):168
u:Lobule VI (Hem):170
u:Hipp (HATA):172
u:Th-Prefrontal:174
u:Area 6:176
u:TE 3:179
u:Area 17:181
u:OP 2:183
u:Lobule VIIIa (Vermis):185
u:Th-Somatosensory:187
u:IPC (PFm):189
u:Lobule X (Vermis):191
u:hOC5 (V5):193
u:Lobule VIIa Crus II (Vermis):195
u:Hipp (SUB):197
u:Lobule VI (Vermis):199
u:OP 1:201
u:SPL (5Ci):203
u:Lobule VIIa Crus II (Hem):205
u:hIP3:207
u:Lobule VIIIa (Hem):209
u:Th-Visual:211
u:Hipp (EC):213
u:IPC (PFt):215
u:Area 3b:218
u:Lobule VIIb (Vermis):220
u:Lobule VIIb (Hem):222
u:IPC (PFcm):224
u:IPC (PF):226
u:hIP2:228
u:SPL (7P):230
u:SPL (5M):232
u:IPC (PGp):234
u:Lobule VIIIb (Vermis):236
u:Th-Motor:238
u:Area 18:240
u:OP 4:242
u:IPC (PFop):244
u:OP 3:246
u:Lobule VIIa Crus I (Hem):248
u:Area 3a:250
u:Area 1:252
u:SPL (7PC):254
----------- End regions for CA_MPM_18_MNIA atlas --------------
** ERROR: Failed to get CA_PM_18_MNIA
*+ WARNING: NULL atlas
** ERROR: NULL atlas
Atlas CA_GW_18_MNIA, 2 regions
----------- Begin regions for CA_GW_18_MNIA atlas-----------
u:grey:0
u:white:1
----------- End regions for CA_GW_18_MNIA atlas --------------
Atlas CA_LR_18_MNIA, 2 regions
----------- Begin regions for CA_LR_18_MNIA atlas-----------
r:Right Brain:1
l:Left Brain:2
----------- End regions for CA_LR_18_MNIA atlas --------------
** ERROR: Failed to get DKD_Desai_PM
*+ WARNING: NULL atlas
** ERROR: NULL atlas
** ERROR: Failed to get DD_Desai_PM
*+ WARNING: NULL atlas
** ERROR: NULL atlas
** ERROR: Failed to get FS_Desai_PM
*+ WARNING: NULL atlas
** ERROR: NULL atlas
Atlas DD_Desai_MPM, 190 regions
----------- Begin regions for DD_Desai_MPM atlas-----------
u:ctx_lh_G_and_S_frontomargin:1
u:ctx_lh_G_and_S_occipital_inf:2
u:ctx_lh_G_and_S_paracentral:3
u:ctx_lh_G_and_S_subcentral:4
u:ctx_lh_G_and_S_transv_frontopol:5
u:ctx_lh_G_and_S_cingul-Ant:6
u:ctx_lh_G_and_S_cingul-Mid-Ant:7
u:ctx_lh_G_and_S_cingul-Mid-Post:8
u:ctx_lh_G_cingul-Post-dorsal:9
u:ctx_lh_G_cingul-Post-ventral:10
u:ctx_lh_G_cuneus:11
u:ctx_lh_G_front_inf-Opercular:12
u:ctx_lh_G_front_inf-Orbital:13
u:ctx_lh_G_front_inf-Triangul:14
u:ctx_lh_G_front_middle:15
u:ctx_lh_G_front_sup:16
u:ctx_lh_G_Ins_lg_and_S_cent_ins:17
u:ctx_lh_G_insular_short:18
u:ctx_lh_G_occipital_middle:19
u:ctx_lh_G_occipital_sup:20
u:ctx_lh_G_oc-temp_lat-fusifor:21
u:ctx_lh_G_oc-temp_med-Lingual:22
u:ctx_lh_G_oc-temp_med-Parahip:23
u:ctx_lh_G_orbital:24
u:ctx_lh_G_pariet_inf-Angular:25
u:ctx_lh_G_pariet_inf-Supramar:26
u:ctx_lh_G_parietal_sup:27
u:ctx_lh_G_postcentral:28
u:ctx_lh_G_precentral:29
u:ctx_lh_G_precuneus:30
u:ctx_lh_G_rectus:31
u:ctx_lh_G_subcallosal:32
u:ctx_lh_G_temp_sup-G_T_transv:33
u:ctx_lh_G_temp_sup-Lateral:34
u:ctx_lh_G_temp_sup-Plan_polar:35
u:ctx_lh_G_temp_sup-Plan_tempo:36
u:ctx_lh_G_temporal_inf:37
u:ctx_lh_G_temporal_middle:38
u:ctx_lh_Lat_Fis-ant-Horizont:39
u:ctx_lh_Lat_Fis-ant-Vertical:40
u:ctx_lh_Lat_Fis-post:41
u:ctx_lh_Medial_wall:42
u:ctx_lh_Pole_occipital:43
u:ctx_lh_Pole_temporal:44
u:ctx_lh_S_calcarine:45
u:ctx_lh_S_central:46
u:ctx_lh_S_cingul-Marginalis:47
u:ctx_lh_S_circular_insula_ant:48
u:ctx_lh_S_circular_insula_inf:49
u:ctx_lh_S_circular_insula_sup:50
u:ctx_lh_S_collat_transv_ant:51
u:ctx_lh_S_collat_transv_post:52
u:ctx_lh_S_front_inf:53
u:ctx_lh_S_front_middle:54
u:ctx_lh_S_front_sup:55
u:ctx_lh_S_interm_prim-Jensen:56
u:ctx_lh_S_intrapariet_and_P_trans:57
u:ctx_lh_S_oc_middle_and_Lunatus:58
u:ctx_lh_S_oc_sup_and_transversal:59
u:ctx_lh_S_occipital_ant:60
u:ctx_lh_S_oc-temp_lat:61
u:ctx_lh_S_oc-temp_med_and_Lingual:62
u:ctx_lh_S_orbital_lateral:63
u:ctx_lh_S_orbital_med-olfact:64
u:ctx_lh_S_orbital-H_Shaped:65
u:ctx_lh_S_parieto_occipital:66
u:ctx_lh_S_pericallosal:67
u:ctx_lh_S_postcentral:68
u:ctx_lh_S_precentral-inf-part:69
u:ctx_lh_S_precentral-sup-part:70
u:ctx_lh_S_suborbital:71
u:ctx_lh_S_subparietal:72
u:ctx_lh_S_temporal_inf:73
u:ctx_lh_S_temporal_sup:74
u:ctx_lh_S_temporal_transverse:75
u:ctx_rh_G_and_S_frontomargin:76
u:ctx_rh_G_and_S_occipital_inf:77
u:ctx_rh_G_and_S_paracentral:78
u:ctx_rh_G_and_S_subcentral:79
u:ctx_rh_G_and_S_transv_frontopol:80
u:ctx_rh_G_and_S_cingul-Ant:81
u:ctx_rh_G_and_S_cingul-Mid-Ant:82
u:ctx_rh_G_and_S_cingul-Mid-Post:83
u:ctx_rh_G_cingul-Post-dorsal:84
u:ctx_rh_G_cingul-Post-ventral:85
u:ctx_rh_G_cuneus:86
u:ctx_rh_G_front_inf-Opercular:87
u:ctx_rh_G_front_inf-Orbital:88
u:ctx_rh_G_front_inf-Triangul:89
u:ctx_rh_G_front_middle:90
u:ctx_rh_G_front_sup:91
u:ctx_rh_G_Ins_lg_and_S_cent_ins:92
u:ctx_rh_G_insular_short:93
u:ctx_rh_G_occipital_middle:94
u:ctx_rh_G_occipital_sup:95
u:ctx_rh_G_oc-temp_lat-fusifor:96
u:ctx_rh_G_oc-temp_med-Lingual:97
u:ctx_rh_G_oc-temp_med-Parahip:98
u:ctx_rh_G_orbital:99
u:ctx_rh_G_pariet_inf-Angular:100
u:ctx_rh_G_pariet_inf-Supramar:101
u:ctx_rh_G_parietal_sup:102
u:ctx_rh_G_postcentral:103
u:ctx_rh_G_precentral:104
u:ctx_rh_G_precuneus:105
u:ctx_rh_G_rectus:106
u:ctx_rh_G_subcallosal:107
u:ctx_rh_G_temp_sup-G_T_transv:108
u:ctx_rh_G_temp_sup-Lateral:109
u:ctx_rh_G_temp_sup-Plan_polar:110
u:ctx_rh_G_temp_sup-Plan_tempo:111
u:ctx_rh_G_temporal_inf:112
u:ctx_rh_G_temporal_middle:113
u:ctx_rh_Lat_Fis-ant-Horizont:114
u:ctx_rh_Lat_Fis-ant-Vertical:115
u:ctx_rh_Lat_Fis-post:116
u:ctx_rh_Medial_wall:117
u:ctx_rh_Pole_occipital:118
u:ctx_rh_Pole_temporal:119
u:ctx_rh_S_calcarine:120
u:ctx_rh_S_central:121
u:ctx_rh_S_cingul-Marginalis:122
u:ctx_rh_S_circular_insula_ant:123
u:ctx_rh_S_circular_insula_inf:124
u:ctx_rh_S_circular_insula_sup:125
u:ctx_rh_S_collat_transv_ant:126
u:ctx_rh_S_collat_transv_post:127
u:ctx_rh_S_front_inf:128
u:ctx_rh_S_front_middle:129
u:ctx_rh_S_front_sup:130
u:ctx_rh_S_interm_prim-Jensen:131
u:ctx_rh_S_intrapariet_and_P_trans:132
u:ctx_rh_S_oc_middle_and_Lunatus:133
u:ctx_rh_S_oc_sup_and_transversal:134
u:ctx_rh_S_occipital_ant:135
u:ctx_rh_S_oc-temp_lat:136
u:ctx_rh_S_oc-temp_med_and_Lingual:137
u:ctx_rh_S_orbital_lateral:138
u:ctx_rh_S_orbital_med-olfact:139
u:ctx_rh_S_orbital-H_Shaped:140
u:ctx_rh_S_parieto_occipital:141
u:ctx_rh_S_pericallosal:142
u:ctx_rh_S_postcentral:143
u:ctx_rh_S_precentral-inf-part:144
u:ctx_rh_S_precentral-sup-part:145
u:ctx_rh_S_suborbital:146
u:ctx_rh_S_subparietal:147
u:ctx_rh_S_temporal_inf:148
u:ctx_rh_S_temporal_sup:149
u:ctx_rh_S_temporal_transverse:150
l:Left-Cerebral-White-Matter:151
l:Left-Lateral-Ventricle:152
l:Left-Inf-Lat-Vent:153
l:Left-Cerebellum-White-Matter:154
l:Left-Cerebellum-Cortex:155
l:Left-Thalamus-Proper:156
l:Left-Caudate:157
l:Left-Putamen:158
l:Left-Pallidum:159
u:3rd-Ventricle:160
u:4th-Ventricle:161
u:Brain-Stem:162
l:Left-Hippocampus:163
l:Left-Amygdala:164
l:Left-Accumbens-area:165
l:Left-VentralDC:166
l:Left-vessel:167
l:Left-choroid-plexus:168
r:Right-Cerebral-White-Matter:169
r:Right-Lateral-Ventricle:170
r:Right-Inf-Lat-Vent:171
r:Right-Cerebellum-White-Matter:172
r:Right-Cerebellum-Cortex:173
r:Right-Thalamus-Proper:174
r:Right-Caudate:175
r:Right-Putamen:176
r:Right-Pallidum:177
r:Right-Hippocampus:178
r:Right-Amygdala:179
r:Right-Accumbens-area:180
r:Right-VentralDC:181
r:Right-vessel:182
r:Right-choroid-plexus:183
u:5th-Ventricle:184
u:Optic-Chiasm:185
u:CC_Posterior:186
u:CC_Mid_Posterior:187
u:CC_Central:188
u:CC_Mid_Anterior:189
u:CC_Anterior:190
----------- End regions for DD_Desai_MPM atlas --------------
Atlas DKD_Desai_MPM, 110 regions
----------- Begin regions for DKD_Desai_MPM atlas-----------
u:ctx-lh-bankssts:1
u:ctx-lh-caudalanteriorcingulate:2
u:ctx-lh-caudalmiddlefrontal:3
u:ctx-lh-corpuscallosum:4
u:ctx-lh-cuneus:5
u:ctx-lh-entorhinal:6
u:ctx-lh-fusiform:7
u:ctx-lh-inferiorparietal:8
u:ctx-lh-inferiortemporal:9
u:ctx-lh-isthmuscingulate:10
u:ctx-lh-lateraloccipital:11
u:ctx-lh-lateralorbitofrontal:12
u:ctx-lh-lingual:13
u:ctx-lh-medialorbitofrontal:14
u:ctx-lh-middletemporal:15
u:ctx-lh-parahippocampal:16
u:ctx-lh-paracentral:17
u:ctx-lh-parsopercularis:18
u:ctx-lh-parsorbitalis:19
u:ctx-lh-parstriangularis:20
u:ctx-lh-pericalcarine:21
u:ctx-lh-postcentral:22
u:ctx-lh-posteriorcingulate:23
u:ctx-lh-precentral:24
u:ctx-lh-precuneus:25
u:ctx-lh-rostralanteriorcingulate:26
u:ctx-lh-rostralmiddlefrontal:27
u:ctx-lh-superiorfrontal:28
u:ctx-lh-superiorparietal:29
u:ctx-lh-superiortemporal:30
u:ctx-lh-supramarginal:31
u:ctx-lh-frontalpole:32
u:ctx-lh-temporalpole:33
u:ctx-lh-transversetemporal:34
u:ctx-lh-insula:35
u:ctx-rh-bankssts:36
u:ctx-rh-caudalanteriorcingulate:37
u:ctx-rh-caudalmiddlefrontal:38
u:ctx-rh-corpuscallosum:39
u:ctx-rh-cuneus:40
u:ctx-rh-entorhinal:41
u:ctx-rh-fusiform:42
u:ctx-rh-inferiorparietal:43
u:ctx-rh-inferiortemporal:44
u:ctx-rh-isthmuscingulate:45
u:ctx-rh-lateraloccipital:46
u:ctx-rh-lateralorbitofrontal:47
u:ctx-rh-lingual:48
u:ctx-rh-medialorbitofrontal:49
u:ctx-rh-middletemporal:50
u:ctx-rh-parahippocampal:51
u:ctx-rh-paracentral:52
u:ctx-rh-parsopercularis:53
u:ctx-rh-parsorbitalis:54
u:ctx-rh-parstriangularis:55
u:ctx-rh-pericalcarine:56
u:ctx-rh-postcentral:57
u:ctx-rh-posteriorcingulate:58
u:ctx-rh-precentral:59
u:ctx-rh-precuneus:60
u:ctx-rh-rostralanteriorcingulate:61
u:ctx-rh-rostralmiddlefrontal:62
u:ctx-rh-superiorfrontal:63
u:ctx-rh-superiorparietal:64
u:ctx-rh-superiortemporal:65
u:ctx-rh-supramarginal:66
u:ctx-rh-frontalpole:67
u:ctx-rh-temporalpole:68
u:ctx-rh-transversetemporal:69
u:ctx-rh-insula:70
l:Left-Cerebral-White-Matter:71
l:Left-Lateral-Ventricle:72
l:Left-Inf-Lat-Vent:73
l:Left-Cerebellum-White-Matter:74
l:Left-Cerebellum-Cortex:75
l:Left-Thalamus-Proper:76
l:Left-Caudate:77
l:Left-Putamen:78
l:Left-Pallidum:79
u:3rd-Ventricle:80
u:4th-Ventricle:81
u:Brain-Stem:82
l:Left-Hippocampus:83
l:Left-Amygdala:84
l:Left-Accumbens-area:85
l:Left-VentralDC:86
l:Left-vessel:87
l:Left-choroid-plexus:88
r:Right-Cerebral-White-Matter:89
r:Right-Lateral-Ventricle:90
r:Right-Inf-Lat-Vent:91
r:Right-Cerebellum-White-Matter:92
r:Right-Cerebellum-Cortex:93
r:Right-Thalamus-Proper:94
r:Right-Caudate:95
r:Right-Putamen:96
r:Right-Pallidum:97
r:Right-Hippocampus:98
r:Right-Amygdala:99
r:Right-Accumbens-area:100
r:Right-VentralDC:101
r:Right-vessel:102
r:Right-choroid-plexus:103
u:5th-Ventricle:104
u:Optic-Chiasm:105
u:CC_Posterior:106
u:CC_Mid_Posterior:107
u:CC_Central:108
u:CC_Mid_Anterior:109
u:CC_Anterior:110
----------- End regions for DKD_Desai_MPM atlas --------------
Atlas MNI_VmPFC, 16 regions
----------- Begin regions for MNI_VmPFC atlas-----------
u:area11m_left:1
u:area11m_right:2
u:area14c_left:3
u:area14c_right:4
u:area14m_left:5
u:area14m_right:6
u:area14r_left:7
u:area14r_right:8
u:area14rr_left:9
u:area14rr_right:10
u:area24_left:11
u:area24_right:12
u:area25_left:13
u:area25_right:14
u:area32_left:15
u:area32_right:16
----------- End regions for MNI_VmPFC atlas --------------
Atlas TT_Daemon, 241 regions
----------- Begin regions for TT_Daemon atlas-----------
u:Anterior Commissure:1
u:Posterior Commissure:2
u:Corpus Callosum:3
l:Left Hippocampus:268
r:Right Hippocampus:68
l:Left Amygdala:271
r:Right Amygdala:71
l:Left Posterior Cingulate:220
r:Right Posterior Cingulate:20
l:Left Anterior Cingulate:221
r:Right Anterior Cingulate:21
l:Left Subcallosal Gyrus:222
r:Right Subcallosal Gyrus:22
l:Left Transverse Temporal Gyrus:224
r:Right Transverse Temporal Gyrus:24
l:Left Uncus:225
r:Right Uncus:25
l:Left Rectal Gyrus:226
r:Right Rectal Gyrus:26
l:Left Fusiform Gyrus:227
r:Right Fusiform Gyrus:27
l:Left Inferior Occipital Gyrus:228
r:Right Inferior Occipital Gyrus:28
l:Left Inferior Temporal Gyrus:229
r:Right Inferior Temporal Gyrus:29
l:Left Insula:230
r:Right Insula:30
l:Left Parahippocampal Gyrus:231
r:Right Parahippocampal Gyrus:31
l:Left Lingual Gyrus:232
r:Right Lingual Gyrus:32
l:Left Middle Occipital Gyrus:233
r:Right Middle Occipital Gyrus:33
l:Left Orbital Gyrus:234
r:Right Orbital Gyrus:34
l:Left Middle Temporal Gyrus:235
r:Right Middle Temporal Gyrus:35
l:Left Superior Temporal Gyrus:236
r:Right Superior Temporal Gyrus:36
l:Left Superior Occipital Gyrus:237
r:Right Superior Occipital Gyrus:37
l:Left Inferior Frontal Gyrus:239
r:Right Inferior Frontal Gyrus:39
l:Left Cuneus:240
r:Right Cuneus:40
l:Left Angular Gyrus:241
r:Right Angular Gyrus:41
l:Left Supramarginal Gyrus:242
r:Right Supramarginal Gyrus:42
l:Left Cingulate Gyrus:243
r:Right Cingulate Gyrus:43
l:Left Inferior Parietal Lobule:244
r:Right Inferior Parietal Lobule:44
l:Left Precuneus:245
r:Right Precuneus:45
l:Left Superior Parietal Lobule:246
r:Right Superior Parietal Lobule:46
l:Left Middle Frontal Gyrus:247
r:Right Middle Frontal Gyrus:47
l:Left Paracentral Lobule:248
r:Right Paracentral Lobule:48
l:Left Postcentral Gyrus:249
r:Right Postcentral Gyrus:49
l:Left Precentral Gyrus:250
r:Right Precentral Gyrus:50
l:Left Superior Frontal Gyrus:251
r:Right Superior Frontal Gyrus:51
l:Left Medial Frontal Gyrus:252
r:Right Medial Frontal Gyrus:52
l:Left Lentiform Nucleus:270
r:Right Lentiform Nucleus:70
l:Left Hypothalamus:272
r:Right Hypothalamus:72
l:Left Red Nucleus:273
r:Right Red Nucleus:73
l:Left Substantia Nigra:274
r:Right Substantia Nigra:74
l:Left Claustrum:275
r:Right Claustrum:75
l:Left Thalamus:276
r:Right Thalamus:76
l:Left Caudate:277
r:Right Caudate:77
l:Left Caudate Tail:324
r:Right Caudate Tail:124
l:Left Caudate Body:325
r:Right Caudate Body:125
l:Left Caudate Head:326
r:Right Caudate Head:126
l:Left Ventral Anterior Nucleus:328
r:Right Ventral Anterior Nucleus:128
l:Left Ventral Posterior Medial Nucleus:329
r:Right Ventral Posterior Medial Nucleus:129
l:Left Ventral Posterior Lateral Nucleus:330
r:Right Ventral Posterior Lateral Nucleus:130
l:Left Medial Dorsal Nucleus:331
r:Right Medial Dorsal Nucleus:131
l:Left Lateral Dorsal Nucleus:332
r:Right Lateral Dorsal Nucleus:132
l:Left Pulvinar:333
r:Right Pulvinar:133
l:Left Lateral Posterior Nucleus:334
r:Right Lateral Posterior Nucleus:134
l:Left Ventral Lateral Nucleus:335
r:Right Ventral Lateral Nucleus:135
l:Left Midline Nucleus:336
r:Right Midline Nucleus:136
l:Left Anterior Nucleus:337
r:Right Anterior Nucleus:137
l:Left Mammillary Body:338
r:Right Mammillary Body:138
l:Left Medial Globus Pallidus:344
r:Right Medial Globus Pallidus:144
l:Left Lateral Globus Pallidus:345
r:Right Lateral Globus Pallidus:145
l:Left Putamen:351
r:Right Putamen:151
l:Left Nucleus Accumbens:346
r:Right Nucleus Accumbens:146
l:Left Medial Geniculum Body:347
r:Right Medial Geniculum Body:147
l:Left Lateral Geniculum Body:348
r:Right Lateral Geniculum Body:148
l:Left Subthalamic Nucleus:349
r:Right Subthalamic Nucleus:149
l:Left Brodmann area 1:281
r:Right Brodmann area 1:81
l:Left Brodmann area 2:282
r:Right Brodmann area 2:82
l:Left Brodmann area 3:283
r:Right Brodmann area 3:83
l:Left Brodmann area 4:284
r:Right Brodmann area 4:84
l:Left Brodmann area 5:285
r:Right Brodmann area 5:85
l:Left Brodmann area 6:286
r:Right Brodmann area 6:86
l:Left Brodmann area 7:287
r:Right Brodmann area 7:87
l:Left Brodmann area 8:288
r:Right Brodmann area 8:88
l:Left Brodmann area 9:289
r:Right Brodmann area 9:89
l:Left Brodmann area 10:290
r:Right Brodmann area 10:90
l:Left Brodmann area 11:291
r:Right Brodmann area 11:91
l:Left Brodmann area 13:293
r:Right Brodmann area 13:93
l:Left Brodmann area 17:294
r:Right Brodmann area 17:94
l:Left Brodmann area 18:295
r:Right Brodmann area 18:95
l:Left Brodmann area 19:296
r:Right Brodmann area 19:96
l:Left Brodmann area 20:297
r:Right Brodmann area 20:97
l:Left Brodmann area 21:298
r:Right Brodmann area 21:98
l:Left Brodmann area 22:299
r:Right Brodmann area 22:99
l:Left Brodmann area 23:300
r:Right Brodmann area 23:100
l:Left Brodmann area 24:301
r:Right Brodmann area 24:101
l:Left Brodmann area 25:302
r:Right Brodmann area 25:102
l:Left Brodmann area 27:303
r:Right Brodmann area 27:103
l:Left Brodmann area 28:304
r:Right Brodmann area 28:104
l:Left Brodmann area 29:305
r:Right Brodmann area 29:105
l:Left Brodmann area 30:306
r:Right Brodmann area 30:106
l:Left Brodmann area 31:307
r:Right Brodmann area 31:107
l:Left Brodmann area 32:308
r:Right Brodmann area 32:108
l:Left Brodmann area 33:309
r:Right Brodmann area 33:109
l:Left Brodmann area 34:310
r:Right Brodmann area 34:110
l:Left Brodmann area 35:311
r:Right Brodmann area 35:111
l:Left Brodmann area 36:312
r:Right Brodmann area 36:112
l:Left Brodmann area 37:313
r:Right Brodmann area 37:113
l:Left Brodmann area 38:314
r:Right Brodmann area 38:114
l:Left Brodmann area 39:315
r:Right Brodmann area 39:115
l:Left Brodmann area 40:316
r:Right Brodmann area 40:116
l:Left Brodmann area 41:317
r:Right Brodmann area 41:117
l:Left Brodmann area 42:318
r:Right Brodmann area 42:118
l:Left Brodmann area 43:319
r:Right Brodmann area 43:119
l:Left Brodmann area 44:320
r:Right Brodmann area 44:120
l:Left Brodmann area 45:321
r:Right Brodmann area 45:121
l:Left Brodmann area 46:322
r:Right Brodmann area 46:122
l:Left Brodmann area 47:323
r:Right Brodmann area 47:123
l:Left Uvula of Vermis:253
r:Right Uvula of Vermis:53
l:Left Pyramis of Vermis:254
r:Right Pyramis of Vermis:54
l:Left Tuber of Vermis:255
r:Right Tuber of Vermis:55
l:Left Declive of Vermis:256
r:Right Declive of Vermis:56
l:Left Culmen of Vermis:257
r:Right Culmen of Vermis:57
l:Left Cerebellar Tonsil:258
r:Right Cerebellar Tonsil:58
l:Left Inferior Semi-Lunar Lobule:259
r:Right Inferior Semi-Lunar Lobule:59
l:Left Fastigium:260
r:Right Fastigium:60
l:Left Nodule:261
r:Right Nodule:61
l:Left Uvula:262
r:Right Uvula:62
l:Left Pyramis:263
r:Right Pyramis:63
l:Left Culmen:266
r:Right Culmen:66
l:Left Declive:265
r:Right Declive:65
l:Left Dentate:327
r:Right Dentate:127
l:Left Tuber:264
r:Right Tuber:64
l:Left Cerebellar Lingual:267
r:Right Cerebellar Lingual:67
----------- End regions for TT_Daemon atlas --------------
Atlas Haskins_Pediatric_Nonlinear_1.0, 107 regions
----------- Begin regions for Haskins_Pediatric_Nonlinear_1.0 atlas-----------
l:Left-Lateral-Ventricle:1
l:Left-Inf-Lat-Vent:2
l:Left-Cerebellum-White-Matter:3
l:Left-Cerebellum-Cortex:4
l:Left-Thalamus-Proper:5
l:Left-Caudate:6
l:Left-Putamen:7
l:Left-Pallidum:8
u:3rd-Ventricle:9
u:4th-Ventricle:10
u:Brain-Stem:11
l:Left-Hippocampus:12
l:Left-Amygdala:13
u:CSF:14
l:Left-Accumbens-area:15
l:Left-VentralDC:16
l:Left-vessel:17
l:Left-choroid-plexus:18
r:Right-Lateral-Ventricle:19
r:Right-Inf-Lat-Vent:20
r:Right-Cerebellum-White-Matter:21
r:Right-Cerebellum-Cortex:22
r:Right-Thalamus-Proper:23
r:Right-Caudate:24
r:Right-Putamen:25
r:Right-Pallidum:26
r:Right-Hippocampus:27
r:Right-Amygdala:28
r:Right-Accumbens-area:29
r:Right-VentralDC:30
r:Right-vessel:31
r:Right-choroid-plexus:32
u:5th-Ventricle:33
u:Optic-Chiasm:34
u:CC_Posterior:35
u:CC_Mid_Posterior:36
u:CC_Central:37
u:CC_Mid_Anterior:38
u:CC_Anterior:39
u:ctx-lh-bankssts:40
u:ctx-lh-caudalanteriorcingulate:41
u:ctx-lh-caudalmiddlefrontal:42
u:ctx-lh-cuneus:43
u:ctx-lh-entorhinal:44
u:ctx-lh-fusiform:45
u:ctx-lh-inferiorparietal:46
u:ctx-lh-inferiortemporal:47
u:ctx-lh-isthmuscingulate:48
u:ctx-lh-lateraloccipital:49
u:ctx-lh-lateralorbitofrontal:50
u:ctx-lh-lingual:51
u:ctx-lh-medialorbitofrontal:52
u:ctx-lh-middletemporal:53
u:ctx-lh-parahippocampal:54
u:ctx-lh-paracentral:55
u:ctx-lh-parsopercularis:56
u:ctx-lh-parsorbitalis:57
u:ctx-lh-parstriangularis:58
u:ctx-lh-pericalcarine:59
u:ctx-lh-postcentral:60
u:ctx-lh-posteriorcingulate:61
u:ctx-lh-precentral:62
u:ctx-lh-precuneus:63
u:ctx-lh-rostralanteriorcingulate:64
u:ctx-lh-rostralmiddlefrontal:65
u:ctx-lh-superiorfrontal:66
u:ctx-lh-superiorparietal:67
u:ctx-lh-superiortemporal:68
u:ctx-lh-supramarginal:69
u:ctx-lh-frontalpole:70
u:ctx-lh-temporalpole:71
u:ctx-lh-transversetemporal:72
u:ctx-lh-insula:73
u:ctx-rh-bankssts:74
u:ctx-rh-caudalanteriorcingulate:75
u:ctx-rh-caudalmiddlefrontal:76
u:ctx-rh-cuneus:77
u:ctx-rh-entorhinal:78
u:ctx-rh-fusiform:79
u:ctx-rh-inferiorparietal:80
u:ctx-rh-inferiortemporal:81
u:ctx-rh-isthmuscingulate:82
u:ctx-rh-lateraloccipital:83
u:ctx-rh-lateralorbitofrontal:84
u:ctx-rh-lingual:85
u:ctx-rh-medialorbitofrontal:86
u:ctx-rh-middletemporal:87
u:ctx-rh-parahippocampal:88
u:ctx-rh-paracentral:89
u:ctx-rh-parsopercularis:90
u:ctx-rh-parsorbitalis:91
u:ctx-rh-parstriangularis:92
u:ctx-rh-pericalcarine:93
u:ctx-rh-postcentral:94
u:ctx-rh-posteriorcingulate:95
u:ctx-rh-precentral:96
u:ctx-rh-precuneus:97
u:ctx-rh-rostralanteriorcingulate:98
u:ctx-rh-rostralmiddlefrontal:99
u:ctx-rh-superiorfrontal:100
u:ctx-rh-superiorparietal:101
u:ctx-rh-superiortemporal:102
u:ctx-rh-supramarginal:103
u:ctx-rh-frontalpole:104
u:ctx-rh-temporalpole:105
u:ctx-rh-transversetemporal:106
u:ctx-rh-insula:107
----------- End regions for Haskins_Pediatric_Nonlinear_1.0 atlas --------------
2. Generate ROI masks (midACC):
Using the DD_Desai_MPM atlas, we create separate masks for the left and right midACC based on their region labels:
!whereami -mask_atlas_region DD_Desai_MPM::ctx_lh_G_and_S_cingul-Mid-Ant -prefix ./afni_pro_glm/group_results/midACC_lh_mask
!whereami -mask_atlas_region DD_Desai_MPM::ctx_rh_G_and_S_cingul-Mid-Ant -prefix ./afni_pro_glm/group_results/midACC_rh_mask
++ Input coordinates orientation set by default rules to RAI
Best match for ctx_lh_G_and_S_cingul-Mid-Ant:
ctx_lh_G_and_S_cingul-Mid-Ant (code 7 )
++ Input coordinates orientation set by default rules to RAI
Best match for ctx_rh_G_and_S_cingul-Mid-Ant:
ctx_rh_G_and_S_cingul-Mid-Ant (code 82 )
We then combine both hemispheres into a single bilateral ROI mask:
!3dmask_tool \
-input ./afni_pro_glm/group_results/midACC_lh_mask+tlrc \
./afni_pro_glm/group_results/midACC_rh_mask+tlrc \
-union \
-prefix ./afni_pro_glm/group_results/midACC_mask
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 5546 survived, 4637855 were zero
++ writing result midACC_mask...
++ Output dataset ./afni_pro_glm/group_results/midACC_mask+tlrc.BRIK
To use the mask with Python tools like nilearn, we also convert it to NIfTI format:
! 3dAFNItoNIFTI -prefix ./afni_pro_glm/group_results/midACC_mask.nii.gz ./afni_pro_glm/group_results/midACC_mask+tlrc
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
# Load AFNI mask
roi_img = image.load_img('./afni_pro_glm/group_results/midACC_mask.nii.gz')
# Plot ROI over mni background
plotting.plot_roi(roi_img,
title="midACC ROI Mask",
display_mode='ortho',
cmap='autumn', draw_cross=True)
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7faed8d189d0>
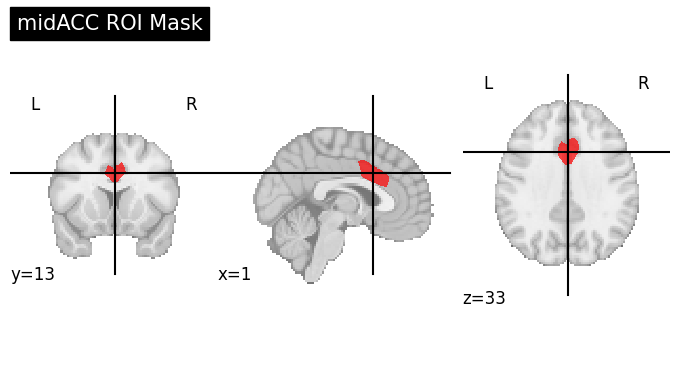
3. Resample the Mask to Match Functional Data:
To ensure compatibility with the functional/statistical datasets, we resample the ROI mask to the same voxel resolution as the subject-level stats+tlrc files:
! 3dresample -master ./afni_pro_glm/sub-01.results/stats.sub-01+tlrc -input ./afni_pro_glm/group_results/midACC_mask+tlrc -prefix ./afni_pro_glm/group_results/midACC_rs+tlrc
4. Extract Contrast Values from Each Subject:
We extract the contrast estimate for each subject by:
Collecting the congruent and incongruent beta sub-bricks across all subjects:
subjects = [f"sub-{i:02d}" for i in range(1, 10)]
for subj in subjects:
stats_path = f"./afni_pro_glm/{subj}.results/stats.{subj}+tlrc"
if os.path.exists(f"{stats_path}.HEAD"): # check AFNI header file
subprocess.run([
"3dbucket", "-aglueto", "./afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD", f"{stats_path}[1]"
])
subprocess.run([
"3dbucket", "-aglueto", "./afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD", f"{stats_path}[4]"
])
else:
print(f"Skipping {subj}: stats file not found")
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Congruent_betas+tlrc.HEAD
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: Over-writing dataset ././afni_pro_glm/group_results/Incongruent_betas+tlrc.HEAD
This will produce 4D images:
Congruent_betas+tlrc→ all congruent betas across subjectsIncongruent_betas+tlrc→ all incongruent betas across subjects
Computing the subject-wise contrast (Incongruent – Congruent) using
3dcalc:
# Create subject-wise contrast dataset (each sub-brick = Incongruent - Congruent)
! 3dcalc -a ./afni_pro_glm/group_results/Incongruent_betas+tlrc -b ./afni_pro_glm/group_results/Congruent_betas+tlrc -expr 'a-b' -prefix ./afni_pro_glm/group_results/incong_minus_cong+tlrc
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././afni_pro_glm/group_results/incong_minus_cong+tlrc.BRIK
Extracting the mean contrast value within the ROI for each subject using
3dmaskave:
# Extract average contrast value from ROI for each subject
! 3dmaskave -quiet -mask ./afni_pro_glm/group_results/midACC_rs+tlrc ./afni_pro_glm/group_results/incong_minus_cong+tlrc > ./afni_pro_glm/group_results/contrast_vals.txt
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 207 voxels survive the mask
5. Statistical Testing: Finally, we load the contrast values and run a one-sample t-test to determine whether the average contrast across subjects significantly differs from zero:
# Load contrast values (one per subject)
contrast_vals = np.loadtxt("./afni_pro_glm/group_results/contrast_vals.txt")
# Run a one-sample t-test against 0
t_stat, p_val = ttest_1samp(contrast_vals, 0)
# Print results
print(f"Mean contrast: {np.mean(contrast_vals):.4f}")
print(f"t = {t_stat:.4f}, p = {p_val:.4f}")
Mean contrast: 0.1277
t = 2.5196, p = 0.0358
✅ Interpretation#
This provides a simple but interpretable ROI-based statistical test focused on the Incongruent - Congruent contrast in the midACC. This result indicates that:
On average, the contrast Incongruent - Congruent is positive within the mid anterior cingulate cortex (midACC) ROI.
The difference is statistically significant at the α = 0.05 level (p = 0.036).
This suggests that the midACC shows greater activation during Incongruent trials compared to Congruent trials across hte 9 subjects.
4.2 ROI Analysis Using a Spherical Mask#
To complement the atlas-based ROI analysis, we can also define a spherical ROI centered on a coordinate of interest. This can be useful for hypothesis-driven analyses targeting specific anatomical or functional peaks.
1. Define the sphere:
A 5mm radius sphere is created around the coordinate (0, 20, 44) using AFNI’s
3dUndump.The sphere is aligned to the grid of the functional data via the
-masteroption.
echo "0 20 44" | 3dUndump -orient LPI -srad 5 -master Incongruent_betas+tlrc -prefix ConflictROI+tlrc -xyz -
# This script creates a 5mm sphere around a coordinate
# Change the x,y,z, coordinates on the left side to select a different peak
# Radius size can be changed with the -srad option
! echo "0 20 44" | 3dUndump -orient LPI -srad 5 -master ./afni_pro_glm/group_results/Incongruent_betas+tlrc -prefix ./afni_pro_glm/group_results/ConflictROI+tlrc -xyz -
++ 3dUndump: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Starting to fill via -xyz coordinates
++ Total number of voxels filled = 19
++ Wrote out dataset ././afni_pro_glm/group_results/ConflictROI+tlrc.BRIK
2. Extract ROI contrast values:
The average
Incongruent - Congruentcontrast value will be computed for each subject using3dmaskave:
! 3dmaskave -quiet -mask ./afni_pro_glm/group_results/ConflictROI+tlrc ./afni_pro_glm/group_results/incong_minus_cong+tlrc > ./afni_pro_glm/group_results/contrast_vals_sphere.txt
++ 3dmaskave: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
+++ 19 voxels survive the mask
3. Statistical test:
A one-sample t-test can be performed to determine whether the contrast values within the sphere significantly differ from zero.
# Load contrast values (one per subject)
contrast_vals_sphere = np.loadtxt("./afni_pro_glm/group_results/contrast_vals_sphere.txt")
# Run a one-sample t-test against 0
t_stat_sphere, p_val_sphere = ttest_1samp(contrast_vals_sphere, 0)
# Print results
print(f"Mean contrast: {np.mean(contrast_vals_sphere):.4f}")
print(f"t = {t_stat_sphere:.4f}, p = {p_val_sphere:.4f}")
Mean contrast: 0.2919
t = 3.3636, p = 0.0099
✅ Interpretation of Sphere ROI Results#
The mean contrast value for Incongruent-Congruent in the spherical ROI is 0.2919.
The one-sample t-test shows a significant effect at the α = 0.05 level:
t(8) = 3.36, p = 0.0099.This indicates that the ROI exhibits significantly greater activation during Incongruent trials compared to Congruent trials across subjects.
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-11-04T05:21:43.675155+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
ipyniivue : 2.3.2
tqdm : 4.66.1
numpy : 2.2.4
nibabel : 5.3.2
nilearn : 0.11.1
IPython : 8.16.1
scipy : 1.13.0
matplotlib: 3.8.4
