Introduction to Preprocessing#
Author: Marla Pinkert
Date: 10 Nov 2024
Citation and Resources:#
Tools included in this workflow#
FSL:
M. Jenkinson, C.F. Beckmann, T.E. Behrens, M.W. Woolrich, S.M. Smith. FSL. NeuroImage, 62:782-90, 2012
Nipype:
Esteban, O., Markiewicz, C. J., Burns, C., Goncalves, M., Jarecka, D., Ziegler, E., Berleant, S., Ellis, D. G., Pinsard, B., Madison, C., Waskom, M., Notter, M. P., Clark, D., Manhães-Savio, A., Clark, D., Jordan, K., Dayan, M., Halchenko, Y. O., Loney, F., … Ghosh, S. (2025). nipy/nipype: 1.8.6 (1.8.6). Zenodo https://doi.org/10.5281/zenodo.15054147
Dataset#
Flanker Task dataset from OpenNeuro:
Kelly AMC and Uddin LQ and Biswal BB and Castellanos FX and Milham MP (2018). Flanker task (event-related). OpenNeuro Dataset ds000102
Kelly, A.M., Uddin, L.Q., Biswal, B.B., Castellanos, F.X., Milham, M.P. (2008). Competition between functional brain networks mediates behavioral variability. Neuroimage, 39(1):527-37
Introduction#
This notebook is geared towards people who are taking their first steps preprocessing fMRI data and want to learn more, and also towards people who are interested in teaching using Neurodesk and want to check out an example to see whether this might be a fit for them. Specifically, we will look at how to deal with motion. There will be explanations on what we are doing along the way!
We will use the NYU Slow Flanker dataset from Openneuro. In this study, healthy adults underwent MRI scanning while performing an event-related Eriksen Flanker task.
Objectives:
Using Neurodesk to access neuroimaging analysis software within Colab
Understanding Nipype interfaces
Learning how to do quality checks regarding motion
There are many different packages and software used to preprocess MRI data. Installing them locally can be complicated and frustrating. Especially for teaching, when you have to make sure everyone is able install specific software, or as a student, when you are just starting out. We will therefore use Neurodesk to provide us with an environment which already comes with all we need - no complicated set-up.
Data preparation#
# download data
!datalad install https://github.com/OpenNeuroDatasets/ds000102.git
!cd ds000102 && datalad get sub-01
Cloning: 0%| | 0.00/2.00 [00:00<?, ? candidates/s]
Enumerating: 0.00 Objects [00:00, ? Objects/s]
Counting: 0%| | 0.00/27.0 [00:00<?, ? Objects/s]
Compressing: 0%| | 0.00/23.0 [00:00<?, ? Objects/s]
Receiving: 0%| | 0.00/2.15k [00:00<?, ? Objects/s]
Resolving: 0%| | 0.00/537 [00:00<?, ? Deltas/s]
[INFO ] scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
[INFO ] access to 1 dataset sibling s3-PRIVATE not auto-enabled, enable with:
| datalad siblings -d "/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102" enable -s s3-PRIVATE
install(ok): /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102 (dataset)
Total: 0%| | 0.00/66.8M [00:00<?, ? Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 0%| | 33.3k/10.6M [00:00<00:34, 305k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 1%| | 85.6k/10.6M [00:00<00:25, 412k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 2%| | 190k/10.6M [00:00<00:15, 668k Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 4%|▏ | 382k/10.6M [00:00<00:09, 1.11M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 8%|▎ | 799k/10.6M [00:00<00:04, 2.12M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 13%|▍ | 1.41M/10.6M [00:00<00:02, 3.34M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 18%|▌ | 1.91M/10.6M [00:00<00:02, 3.78M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 33%|█ | 3.53M/10.6M [00:00<00:01, 5.59M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 59%|█▊ | 6.23M/10.6M [00:01<00:00, 10.8M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 74%|██▏| 7.87M/10.6M [00:01<00:00, 12.2M Bytes/s]
Get sub-01/a .. 1_T1w.nii.gz: 94%|██▊| 9.97M/10.6M [00:01<00:00, 14.5M Bytes/s]
Total: 16%|████ | 10.6M/66.8M [00:02<00:10, 5.21M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 0%| | 0.00/28.1M [00:00<?, ? Bytes/s]
Get sub-01/f .. _bold.nii.gz: 9%|▎ | 2.46M/28.1M [00:00<00:01, 23.0M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 19%|▌ | 5.37M/28.1M [00:00<00:01, 19.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 29%|▉ | 8.19M/28.1M [00:00<00:01, 18.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 37%|█ | 10.3M/28.1M [00:00<00:00, 18.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 44%|█▎ | 12.2M/28.1M [00:00<00:00, 18.6M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 51%|█▌ | 14.3M/28.1M [00:00<00:00, 18.8M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 58%|█▋ | 16.3M/28.1M [00:00<00:00, 18.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 65%|█▉ | 18.3M/28.1M [00:00<00:00, 19.1M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 79%|██▍| 22.3M/28.1M [00:01<00:00, 18.6M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 87%|██▌| 24.3M/28.1M [00:01<00:00, 18.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 94%|██▊| 26.4M/28.1M [00:01<00:00, 18.9M Bytes/s]
Total: 58%|███████████████ | 38.6M/66.8M [00:03<00:02, 10.0M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 0%| | 0.00/28.1M [00:00<?, ? Bytes/s]
Get sub-01/f .. _bold.nii.gz: 7%|▏ | 2.02M/28.1M [00:00<00:01, 16.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 17%|▌ | 4.73M/28.1M [00:00<00:01, 14.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 24%|▋ | 6.68M/28.1M [00:00<00:01, 15.8M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 31%|▉ | 8.68M/28.1M [00:00<00:01, 16.8M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 38%|█▏ | 10.7M/28.1M [00:00<00:01, 17.4M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 45%|█▎ | 12.7M/28.1M [00:00<00:00, 18.0M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 52%|█▌ | 14.8M/28.1M [00:00<00:00, 18.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 60%|█▊ | 16.8M/28.1M [00:00<00:00, 18.5M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 67%|██ | 18.8M/28.1M [00:01<00:00, 18.7M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 74%|██▏| 20.9M/28.1M [00:01<00:00, 18.8M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 82%|██▍| 23.0M/28.1M [00:01<00:00, 19.1M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 89%|██▋| 25.1M/28.1M [00:01<00:00, 19.2M Bytes/s]
Get sub-01/f .. _bold.nii.gz: 97%|██▉| 27.2M/28.1M [00:01<00:00, 19.3M Bytes/s]
get(ok): sub-01/anat/sub-01_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-01/func/sub-01_task-flanker_run-2_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-01 (directory)
action summary:
get (ok: 4)
FSL and MCFLIRT#
FSL is a commonly used library of analysis tools for fMRI. It has a GUI, but can be used via the command line as well. We can use it for both preprocessing and first- and second-level analysis, however, we will use it for preprocessing only. Although you would often combine different tools, so you can cherry pick the best ones, it is completely possible to do every preprocessing step within FSL.
MCFLIRT is an FSL tool which we can use for realigning our functional data and calculation of motion parameters. We can not only run it using the command line, but also via a Nipype interface.
# This enables us to access FSL using Neurodesk
import module
await module.load('fsl/6.0.4')
# this makes it so FSL knows how we want our output files
import os
os.environ["FSLOUTPUTTYPE"]="NIFTI_GZ"
Above, the basic steps of preprocessing a functional file are demonstrated. There is an endless amount of steps to add, and variations in the ordering of the steps. For example, depending how your data was aqcuired, you might want to start slice-timing correction before realignment (see here for more information). However, smoothing usually is the last step.
Let’s first check which flags the command line tool MCLFLIRT needs:
!mcflirt -help
Usage: mcflirt -in <infile> [options]
Available options are:
-out, -o <outfile> (default is infile_mcf)
-cost {mutualinfo,woods,corratio,normcorr,normmi,leastsquares} (default is normcorr)
-bins <number of histogram bins> (default is 256)
-dof <number of transform dofs> (default is 6)
-refvol <number of reference volume> (default is no_vols/2)- registers to (n+1)th volume in series
-reffile, -r <filename> use a separate 3d image file as the target for registration (overrides refvol option)
-scaling <num> (6.0 is default)
-smooth <num> (1.0 is default - controls smoothing in cost function)
-rotation <num> specify scaling factor for rotation optimization tolerances
-verbose <num> (0 is least and default)
-stages <number of search levels> (default is 3 - specify 4 for final sinc interpolation)
-fov <num> (default is 20mm - specify size of field of view when padding 2d volume)
-2d Force padding of volume
-sinc_final (applies final transformations using sinc interpolation)
-spline_final (applies final transformations using spline interpolation)
-nn_final (applies final transformations using Nearest Neighbour interpolation)
-init <filename> (initial transform matrix to apply to all vols)
-gdt (run search on gradient images)
-meanvol register timeseries to mean volume (overrides refvol and reffile options)
-stats produce variance and std. dev. images
-mats save transformation matricies in subdirectory outfilename.mat
-plots save transformation parameters in file outputfilename.par
-report report progress to screen
-help
Now, we are ready to run MCFLIRT on the first run of our first subject. Apart from the inpute file (in_file), we specify we want to save the transformation parameters (-plots).
# run mcflirt
!mcflirt -in ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz -plots
In the /func folder of our first subject, two new files have appeared: sub-01_task-flanker_run-1_bold_mcf.nii.gz and sub-01_task-flanker_run-1_bold_mcf.par. The NIfTI file is our realigned functional file, the .par file contains our motion parameters!
Nipype Interfaces#
It is possible to do this exact same thing (using MCFLIRT) in Python using a Nipype interface. Nipype is an open-source project that allows for integrating a variety of different neuroimaging softwares into a single workflow. Nipype provides “wrappers” (= interfaces) for tools and software that is written in other languages than Python.
When we use Nipype, we use the interface classes provided by Nipype to instantiate objects we can use for analysis.
So, what are objects and classes? One way to think about classes is to view them as a sort of construction plan, or blueprint. Imagine a development area in a town where many very similar looking family homes are being built. Usually, these sorts of building projects still offer buyers certain customization options - such as the color, the doors, whether there is a garage. So, the construction plan (class) already contains all the information for building the house, and the custommized houses themselves would then correspond to different objects derived fromm the class.
To use MCFLIRT via Nipype, we have to import the MCFLIRT class that wraps the functionality of the tool within Python.
The nipype.interfaces module gives us access to many different external tools and software - of course these external tools have to be installed for this to work. You can find a list of all the available packages here.
For MCFLIRT, we will import the fsl module.
from nipype.interfaces import fsl
First, let’s have a look at the docstring of the MCFLIRT class:
help(fsl.MCFLIRT)
Help on class MCFLIRT in module nipype.interfaces.fsl.preprocess:
class MCFLIRT(nipype.interfaces.fsl.base.FSLCommand)
| MCFLIRT(**inputs)
|
| FSL MCFLIRT wrapper for within-modality motion correction
|
| For complete details, see the `MCFLIRT Documentation.
| <https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MCFLIRT>`_
|
| Examples
| --------
| >>> from nipype.interfaces import fsl
| >>> mcflt = fsl.MCFLIRT()
| >>> mcflt.inputs.in_file = 'functional.nii'
| >>> mcflt.inputs.cost = 'mutualinfo'
| >>> mcflt.inputs.out_file = 'moco.nii'
| >>> mcflt.cmdline
| 'mcflirt -in functional.nii -cost mutualinfo -out moco.nii'
| >>> res = mcflt.run() # doctest: +SKIP
|
| Method resolution order:
| MCFLIRT
| nipype.interfaces.fsl.base.FSLCommand
| nipype.interfaces.base.core.CommandLine
| nipype.interfaces.base.core.BaseInterface
| nipype.interfaces.base.core.Interface
| builtins.object
|
| Data and other attributes defined here:
|
| input_spec = <class 'nipype.interfaces.fsl.preprocess.MCFLIRTInputSpec...
|
| output_spec = <class 'nipype.interfaces.fsl.preprocess.MCFLIRTOutputSp...
|
| ----------------------------------------------------------------------
| Methods inherited from nipype.interfaces.fsl.base.FSLCommand:
|
| __init__(self, **inputs)
| Subclasses must implement __init__
|
| ----------------------------------------------------------------------
| Class methods inherited from nipype.interfaces.fsl.base.FSLCommand:
|
| set_default_output_type(output_type) from builtins.type
| Set the default output type for FSL classes.
|
| This method is used to set the default output type for all fSL
| subclasses. However, setting this will not update the output
| type for any existing instances. For these, assign the
| <instance>.inputs.output_type.
|
| ----------------------------------------------------------------------
| Readonly properties inherited from nipype.interfaces.fsl.base.FSLCommand:
|
| version
| interfaces should implement a version property
|
| ----------------------------------------------------------------------
| Methods inherited from nipype.interfaces.base.core.CommandLine:
|
| raise_exception(self, runtime)
|
| version_from_command(self, flag='-v', cmd=None)
|
| ----------------------------------------------------------------------
| Class methods inherited from nipype.interfaces.base.core.CommandLine:
|
| set_default_terminal_output(output_type) from builtins.type
| Set the default terminal output for CommandLine Interfaces.
|
| This method is used to set default terminal output for
| CommandLine Interfaces. However, setting this will not
| update the output type for any existing instances. For these,
| assign the <instance>.terminal_output.
|
| ----------------------------------------------------------------------
| Readonly properties inherited from nipype.interfaces.base.core.CommandLine:
|
| cmd
| sets base command, immutable
|
| cmdline
| `command` plus any arguments (args)
| validates arguments and generates command line
|
| ----------------------------------------------------------------------
| Data descriptors inherited from nipype.interfaces.base.core.CommandLine:
|
| terminal_output
|
| write_cmdline
|
| ----------------------------------------------------------------------
| Methods inherited from nipype.interfaces.base.core.BaseInterface:
|
| aggregate_outputs(self, runtime=None, needed_outputs=None)
| Collate expected outputs and apply output traits validation.
|
| load_inputs_from_json(self, json_file, overwrite=True)
| A convenient way to load pre-set inputs from a JSON file.
|
| run(self, cwd=None, ignore_exception=None, **inputs)
| Execute this interface.
|
| This interface will not raise an exception if runtime.returncode is
| non-zero.
|
| Parameters
| ----------
| cwd : specify a folder where the interface should be run
| inputs : allows the interface settings to be updated
|
| Returns
| -------
| results : :obj:`nipype.interfaces.base.support.InterfaceResult`
| A copy of the instance that was executed, provenance information and,
| if successful, results
|
| save_inputs_to_json(self, json_file)
| A convenient way to save current inputs to a JSON file.
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from nipype.interfaces.base.core.BaseInterface:
|
| resource_monitor = True
|
| ----------------------------------------------------------------------
| Class methods inherited from nipype.interfaces.base.core.Interface:
|
| help(returnhelp=False) from builtins.type
| Prints class help
|
| ----------------------------------------------------------------------
| Readonly properties inherited from nipype.interfaces.base.core.Interface:
|
| always_run
| Should the interface be always run even if the inputs were not changed?
| Only applies to interfaces being run within a workflow context.
|
| can_resume
| Defines if the interface can reuse partial results after interruption.
| Only applies to interfaces being run within a workflow context.
|
| ----------------------------------------------------------------------
| Data descriptors inherited from nipype.interfaces.base.core.Interface:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
The first step when using interfaces is always to create an object (or: instance) of the respective interface class (in this case the MCFLIRT class). We will also specify input and output file, and that we want to “save_plots” - remember how this relates to the -plots flag we used before.
moco = fsl.MCFLIRT(in_file = "ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz", out_file = "ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_mfc.nii.gz",save_plots=True)
One attribute that you can find for almost every Nipype interface is cmdline. It refers to the command that will be run on our computer. Nipype interfaces are just wrappers - “under the hood”, the external non-Python software will run.
If you run into errors with your code, you can try to run this command directly in the command line. Sometimes, that helps to figure out what the issue is.
print(moco.cmdline)
mcflirt -in ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz -out ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_mfc.nii.gz -plots
Now that we have instantiated our MCFLIRT object and provided the needed inputs, we can go ahead and run the interface using the run() method. This procedure is common to almost all Nipype interfaces:
instanstiate the object of a specific interface class
provide mandatory and optional inputs
run it by using the
runmethod.
moco_res = moco.run()
By saving the result of running our object, we can easily access the outputs attribute. This is helpful if we forget were we saved our files ;).
moco_res.outputs
mat_file = <undefined>
mean_img = <undefined>
out_file = /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_mfc.nii.gz
par_file = /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_mfc.nii.gz.par
rms_files = <undefined>
std_img = <undefined>
variance_img = <undefined>
Plotting Motion Parameters#
Now that we calculated our motion parameters, we can get to plotting them. First, we load them into memory using numpy, and have a look at their shape.
%%capture
!pip install numpy pandas
import numpy as np
motion_path = "ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_mcf.par"
motion_parameters = np.loadtxt(motion_path)
print(f"Shape of motion parameter matrix: {motion_parameters.shape}")
Shape of motion parameter matrix: (146, 6)
A quick look at the shape of our array reveals that there are 146 timepoints and 6 parameters per timepoint. These 6 parameters concern 3 directions (x, y, and z) - the translation, and 3 angles in which the head can rotate (pitch, yaw, and roll) - the rotation.
Let’s have a look at the first 10 timepoints, and how they are represented in an array.
motion_parameters[:10,:]
array([[-0.00273076, -0.00276311, -0.00237603, -0.154286 , 0.011919 ,
0.0328879 ],
[-0.00298859, -0.00262473, -0.00237603, -0.15885 , 0.0274672 ,
0.0875287 ],
[-0.0029029 , -0.00255196, -0.00224164, -0.166514 , 0.0269606 ,
0.0997821 ],
[-0.00252297, -0.00276311, -0.00237603, -0.166644 , 0.0142047 ,
0.0871495 ],
[-0.00240446, -0.00270943, -0.00237603, -0.168801 , -0.00752524,
0.0624465 ],
[-0.00274884, -0.00258874, -0.00237603, -0.172638 , 0.0512277 ,
0.116354 ],
[-0.00256525, -0.00251282, -0.00203235, -0.15235 , -0.0108426 ,
0.0804987 ],
[-0.00228808, -0.00276311, -0.00200398, -0.163281 , 0.00232863,
0.0529415 ],
[-0.00252837, -0.00250916, -0.00224777, -0.165916 , 0.00128811,
0.0960623 ],
[-0.00249458, -0.00276311, -0.00237603, -0.161301 , 0.00757243,
0.0870456 ]])
We now create two plots - the first one using the x, y, and z parameters, the second one using the pitch, yaw, and roll parameters. As you were able to see above, there are no labels to these arrays which tell us which parameter is which. This doesn’t matter, as their order is always x, y, z, pitch, yaw, and roll
%matplotlib inline
import matplotlib.pyplot as plt
# Create plot
fig, axes = plt.subplots(nrows=2, ncols=1, figsize=(15,10))
# Plot motion parameters
axes[0].plot(motion_parameters[:, 3:]) # x, y, z
axes[1].plot(motion_parameters[:, :3]) # pitch, roll, yaw
# Add labels
axes[0].set_ylabel("translation (mm)")
axes[1].set_ylabel("rotation (radians)")
axes[1].set_xlabel("time (TR)")
axes[0].legend(["x","y","z"])
axes[1].legend(["pitch","roll","yaw"])
# Add main figure title
fig.suptitle(f"Subject 1, Flanker-Task Run 1: Motion Parameters",fontsize = 18)
plt.show()
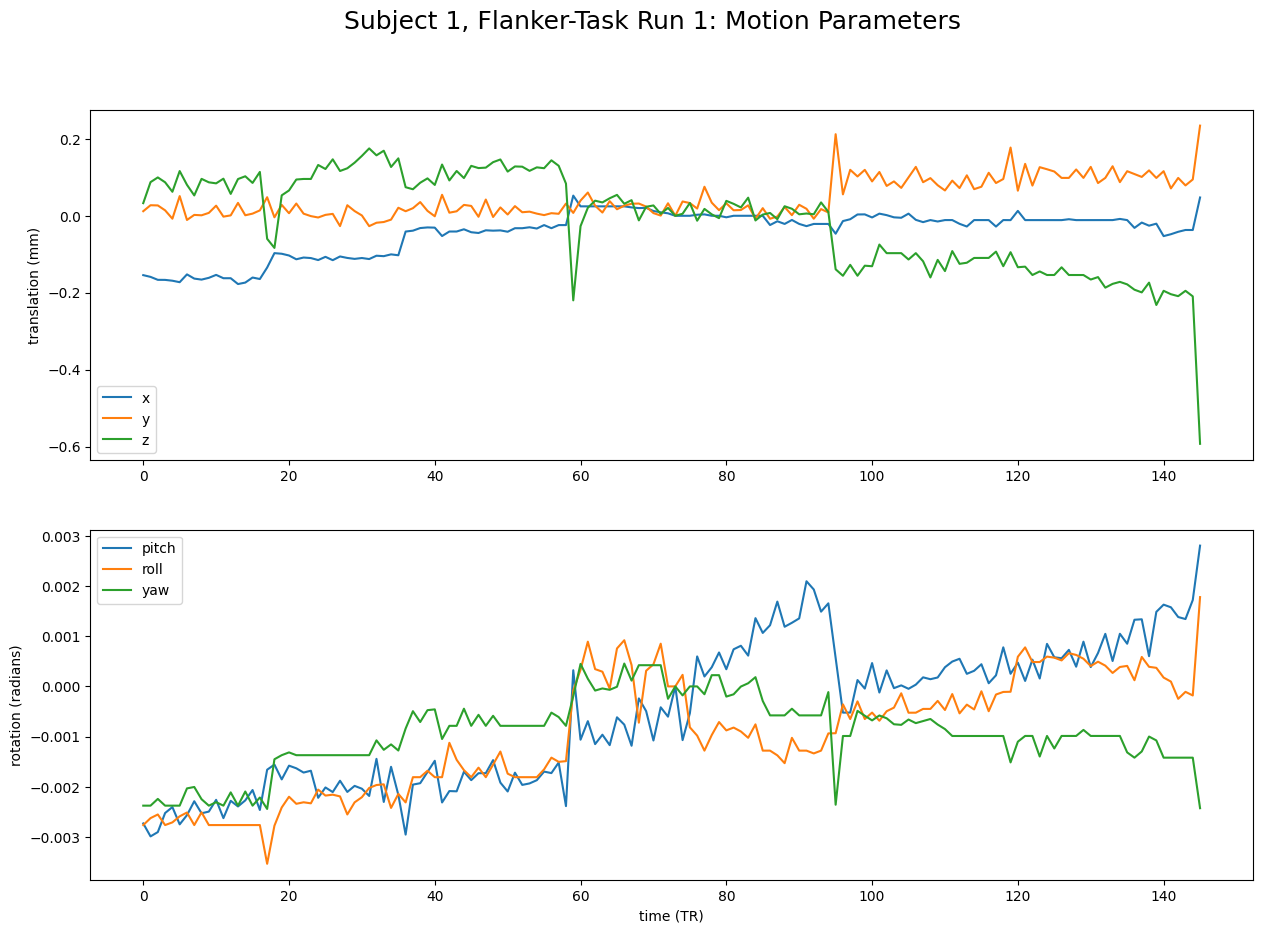
Sometimes, we might introduce motion due to our task. If you like to check whether this is true for your data, it may make sense to check by plotting the timings of the task.
# Read in events
import pandas as pd
events_path = "./ds000102/sub-01/func/sub-01_task-flanker_run-1_events.tsv"
events = pd.read_csv(events_path, sep = '\t')
# get timings of task onset and offset
task_on = events["onset"]
task_off = events["duration"]
# Divide by TR
task_on_tr = (task_on/2).tolist()
task_off_tr = ((task_on + task_off)/2).tolist()
# Create plot
fig, axes = plt.subplots(nrows=2, ncols=1, figsize=(15,10))
# Plot motion parameters
axes[0].plot(motion_parameters[:, 3:]) # x, y, z
axes[1].plot(motion_parameters[:, :3]) # pitch, roll, yaw
# Plot effort blocks
for motion_on, motion_off in zip(task_on_tr, task_off_tr):
axes[0].axvspan(motion_on, motion_off, color='grey', alpha=0.05, lw=0)
for motion_on, motion_off in zip(task_on_tr, task_off_tr):
axes[1].axvspan(motion_on, motion_off, color='grey', alpha=0.05, lw=0)
# Add labels to plots
axes[0].set_ylabel("translation (mm)")
axes[1].set_ylabel("rotation (radians)")
axes[1].set_xlabel("time (TR)")
axes[0].legend(["x","y","z"])
axes[1].legend(["pitch","roll","yaw"])
# Add main figure title
fig.suptitle(f"Subject 1, Flanker-Task Run 1: Motion Parameters",fontsize = 18)
Text(0.5, 0.98, 'Subject 1, Flanker-Task Run 1: Motion Parameters')
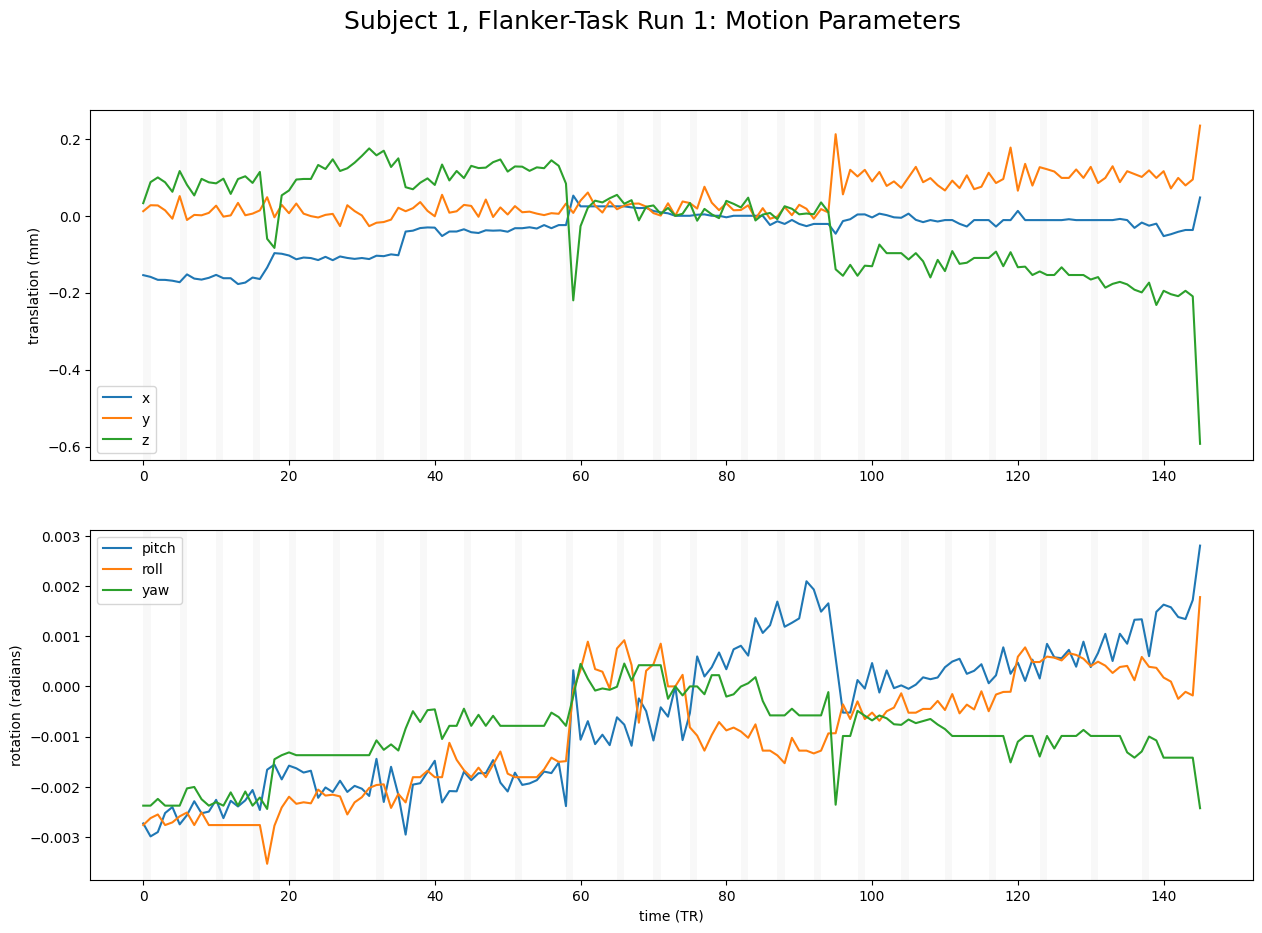
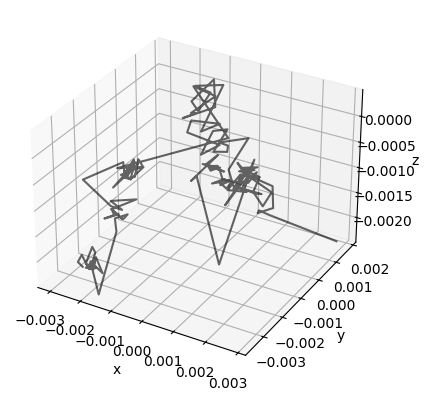
A student in my class had the idea to plot the motion parameters in 3D space, which I thought was fun. See below:
ax = plt.axes(projection="3d")
# Plot movement
ax.plot3D(motion_parameters[:, 0], motion_parameters[:, 1], motion_parameters[:, 2], color = "black", alpha = 0.6)
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_zlabel("z")
plt.show()
Additional Motion Parameters#
So far, we have inspected all motion parameters visually. That is an important part of figuring out whether our participants moved too much. However, we can also calculate additional parameters. We will use fsl.MotionOutliers, which is a wrapper for the FSL tool fsl_motion_outliers, to calculate mean & maximum framewise displacement, as well as outliers. See here for the documentation.
We start with instantiating our object with the necessary parameters.
mo = fsl.MotionOutliers(
in_file = "./ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz",
metric = "fdrms",
out_metric_values = "./ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_fdrms.par",
out_file = "./ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_outliers.txt"
)
We check what the command for the command line looks like:
print(mo.cmdline)
fsl_motion_outliers -i ds000102/sub-01/func/sub-01_task-flanker_run-1_bold.nii.gz --fdrms -o ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_outliers.txt -p sub-01_task-flanker_run-1_bold_metrics.png -s ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_fdrms.par
An we run our object!
res = mo.run()
Let’s check which outputs we got:
res.outputs
out_file = /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_outliers.txt
out_metric_plot = /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/sub-01_task-flanker_run-1_bold_metrics.png
out_metric_values = /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102/sub-01/func/sub-01_task-flanker_run-1_bold_fdrms.par
Now we can look at mean and maximum famewise displacement as well as outliers!
fdrms = np.loadtxt(res.outputs.out_metric_values )
outlier = np.loadtxt(res.outputs.out_file)
print(f"There is a maximum framewise displacement of {np.max(fdrms)} mm.")
print(f"There is a mean framewise displacement of {np.mean(fdrms)} mm.")
print(f"{round(np.sum(outlier)/len(outlier)*100,2)}% of timepoints are outliers.")
There is a maximum framewise displacement of 0.439103 mm.
There is a mean framewise displacement of 0.05534307294520548 mm.
6.16% of timepoints are outliers.
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-11-03T00:10:55.353324+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
nipype : 1.8.6
numpy : 2.2.6
matplotlib: 3.8.4
pandas : 2.2.3
