Brain Extraction Tools#
Author: Monika Doerig
Date: 10 June 2025
Citation and Resources:#
Educational resources#
Andy’s Brain Book:
This brain extraction example is based on the Advanced Normalization Tools (ANTs) chapter from Andy’s Brain Book (Jahn, 2022. doi:10.5281/zenodo.5879293)
Tools included in this workflow#
ANTs - antsBrainExtraction.sh:
Tustison, N.J., Cook, P.A., Holbrook, A.J. et al. The ANTsX ecosystem for quantitative biological and medical imaging. Sci Rep 11, 9068 (2021). https://doi.org/10.1038/s41598-021-87564-6
AFNI - 3dSkullStrip:
Cox RW (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29(3):162-173. doi:10.1006/cbmr.1996.0014 https://pubmed.ncbi.nlm.nih.gov/8812068/
RW Cox, JS Hyde (1997). Software tools for analysis and visualization of FMRI Data. NMR in Biomedicine, 10: 171-178. https://pubmed.ncbi.nlm.nih.gov/9430344/
FreeSurfer - SynthStrip:
SynthStrip: Skull-Stripping for Any Brain Image; Andrew Hoopes, Jocelyn S. Mora, Adrian V. Dalca, Bruce Fischl*, Malte Hoffmann* (*equal contribution); NeuroImage 260, 2022, 119474; https://doi.org/10.1016/j.neuroimage.2022.119474
Boosting skull-stripping performance for pediatric brain images; William Kelley, Nathan Ngo, Adrian V. Dalca, Bruce Fischl, Lilla Zöllei*, Malte Hoffmann* (*equal contribution); IEEE International Symposium on Biomedical Imaging (ISBI), 2024, forthcoming; https://arxiv.org/abs/2402.16634
FSL - BET:
M. Jenkinson, C.F. Beckmann, T.E. Behrens, M.W. Woolrich, S.M. Smith. FSL. NeuroImage, 62:782-90, 2012
Smith S. M. (2002). Fast robust automated brain extraction. Human brain mapping, 17(3), 143–155. https://doi.org/10.1002/hbm.10062
Dataset#
Opensource Data from OpenNeuro:
Kelly AMC and Uddin LQ and Biswal BB and Castellanos FX and Milham MP (2018). Flanker task (event-related). OpenNeuro Dataset ds000102
Kelly, A.M., Uddin, L.Q., Biswal, B.B., Castellanos, F.X., Milham, M.P. (2008). Competition between functional brain networks mediates behavioral variability. Neuroimage, 39(1):527-37
ANTs Brain Templates:
Avants, Brian; Tustison, Nick (2018). ANTs/ANTsR Brain Templates. figshare. Dataset. https://doi.org/10.6084/m9.figshare.915436.v2
Install and import python libraries#
%%capture
!pip install nilearn nibabel scipy numpy
import nibabel as nib
import numpy as np
from scipy import ndimage
from nilearn import plotting
from matplotlib.colors import ListedColormap
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
from ipyniivue import NiiVue
from IPython.display import display, Markdown, Image
Introduction#
Since fMRI studies focus on brain tissue, our first step is to remove the skull and non-brain areas from the image.
In order to analyze fMRI data, you will need to load an fMRI analysis package. In this example we will use the following packages and algorithms to skull-strip the anatomical image:
Advanced Normalization Tools (ANTs): antsBrainExtraction.sh
Analysis of Functional NeuroImages (AFNI): 3dSkullStrip
FreeSurfer: SynthStrip
FSL (FMRIB Software Library, created by the University of Oxford): BET - Brain Extraction Tool
Each package is maintained by a team of professionals, and each is updated at least every few years or so.
Advanced Normalization Tools (ANTs)#
ANTs is a software package for normalizing data to a template.
Templates for public neuroimaging datasets, such as those from IXI, Oasis, NKI-1, and Kirby/MMRR, are intended for use with ANTs and are available for download from figshare. These templates include an average T1 neuroimage of the head and various tissue priors for cortex, white matter, cerebrospinal fluid, deep gray matter, brainstem and the cerebellum.
import module
await module.load('ants/2.3.1')
await module.list()
['ants/2.3.1']
Analysis of Functional NeuroImages (AFNI)#
AFNI is a suite of programs designed to analyze fMRI data. Created in the mid-1990’s by Bob Cox, AFNI is now used by hundreds of imaging labs around the world.
await module.load('afni/24.1.02')
await module.list()
['ants/2.3.1', 'afni/24.1.02']
FreeSurfer#
FreeSurfer is a software package that enables you to analyze structural MRI images - in other words, you can use FreeSurfer to quantify the amount of grey matter and white matter in specific regions of the brain. You will also be able to calculate measurements such as the thickness, curvature, and volume of the different tissue types, and be able to correlate these with covariates; or, you can contrast these structural measurements between groups.
await module.load('freesurfer/7.3.2')
await module.list()
['ants/2.3.1', 'afni/24.1.02', 'freesurfer/7.3.2']
FMRIB Software Library (FSL)#
FSL is a comprehensive library of analysis tools for FMRI, MRI and diffusion brain imaging data. FSL has a tool to skull-strip an anatomical image called bet, or the Brain Extraction Tool.
await module.load('fsl/6.0.7.4')
await module.list()
['ants/2.3.1', 'afni/24.1.02', 'freesurfer/7.3.2', 'fsl/6.0.7.4']
Download Data#
T1 Image for brain extraction#
PATTERN = "sub-08/anat"
! datalad install https://github.com/OpenNeuroDatasets/ds000102.git
! cd ds000102 && datalad get $PATTERN
Cloning: 0%| | 0.00/2.00 [00:00<?, ? candidates/s]
Enumerating: 0.00 Objects [00:00, ? Objects/s]
Counting: 0%| | 0.00/27.0 [00:00<?, ? Objects/s]
Compressing: 0%| | 0.00/23.0 [00:00<?, ? Objects/s]
Receiving: 0%| | 0.00/2.15k [00:00<?, ? Objects/s]
Resolving: 0%| | 0.00/537 [00:00<?, ? Deltas/s]
[INFO ] scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
[INFO ] access to 1 dataset sibling s3-PRIVATE not auto-enabled, enable with:
| datalad siblings -d "/home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/ds000102" enable -s s3-PRIVATE
install(ok): /home/jovyan/Git_repositories/example-notebooks/books/structural_imaging/ds000102 (dataset)
Total: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 0%| | 50.8k/10.6M [00:00<00:22, 466k Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 1%| | 120k/10.6M [00:00<00:18, 570k Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 2%| | 242k/10.6M [00:00<00:12, 825k Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 4%|▏ | 416k/10.6M [00:00<00:08, 1.14M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 8%|▎ | 852k/10.6M [00:00<00:04, 2.18M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 13%|▍ | 1.39M/10.6M [00:00<00:02, 3.14M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 24%|▋ | 2.51M/10.6M [00:00<00:01, 5.49M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 44%|█▎ | 4.59M/10.6M [00:00<00:00, 7.48M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 67%|██ | 7.10M/10.6M [00:01<00:00, 11.7M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 84%|██▌| 8.85M/10.6M [00:01<00:00, 13.1M Bytes/s]
get(ok): sub-08/anat/sub-08_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-08/anat (directory)
action summary:
get (ok: 2)
input_image = 'ds000102/sub-08/anat/sub-08_T1w.nii.gz'
ANTs Brain Templates#
You will need templates to perform the brain extraction with ANTs. For this, we will use the OASIS brain templates, which are intended for use with ANTs medical image processing tools. However, it is up to you to determine which template works best for your data.
# Download the OASIS templates using wget
! wget -nc https://ndownloader.figshare.com/files/3133832 -O OASIS.zip
# Unzip the downloaded file
! unzip -n OASIS.zip -d OASIS
# Remove zip file
! rm -f OASIS.zip
# Delete templates that are not needed
! find OASIS/MICCAI2012-Multi-Atlas-Challenge-Data -type f ! -name 'T_template0.nii.gz' ! -name 'T_template0_BrainCerebellumProbabilityMask.nii.gz' -exec rm {} +
# Delete Priors2 subfolder
! rm -rf OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2
--2025-10-30 00:25:57-- https://ndownloader.figshare.com/files/3133832
Resolving ndownloader.figshare.com (ndownloader.figshare.com)... 52.49.230.183, 108.129.59.119, 34.251.62.251, ...
Connecting to ndownloader.figshare.com (ndownloader.figshare.com)|52.49.230.183|:443... connected.
HTTP request sent, awaiting response... 302 Found
Location: https://s3-eu-west-1.amazonaws.com/pfigshare-u-files/3133832/Oasis.zip?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIYCQYOYV5JSSROOA/20251030/eu-west-1/s3/aws4_request&X-Amz-Date=20251030T002557Z&X-Amz-Expires=10&X-Amz-SignedHeaders=host&X-Amz-Signature=7330301cb4c890c8d2c6fee73879cb41b88a663a80384d4cb0707b2b39d34f0d [following]
--2025-10-30 00:25:57-- https://s3-eu-west-1.amazonaws.com/pfigshare-u-files/3133832/Oasis.zip?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIYCQYOYV5JSSROOA/20251030/eu-west-1/s3/aws4_request&X-Amz-Date=20251030T002557Z&X-Amz-Expires=10&X-Amz-SignedHeaders=host&X-Amz-Signature=7330301cb4c890c8d2c6fee73879cb41b88a663a80384d4cb0707b2b39d34f0d
Resolving s3-eu-west-1.amazonaws.com (s3-eu-west-1.amazonaws.com)... 3.5.71.23, 3.5.68.185, 52.92.33.248, ...
Connecting to s3-eu-west-1.amazonaws.com (s3-eu-west-1.amazonaws.com)|3.5.71.23|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 55360609 (53M) [binary/octet-stream]
Saving to: ‘OASIS.zip’
OASIS.zip 100%[===================>] 52.80M 38.1MB/s in 1.4s
2025-10-30 00:25:59 (38.1 MB/s) - ‘OASIS.zip’ saved [55360609/55360609]
Archive: OASIS.zip
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellum.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumExtractionMask.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumMask.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumRegistrationMask.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_glm_4labelsJointFusion.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_glm_6labelsJointFusion.nii.gz
creating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors1.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors2.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors3.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors4.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors5.nii.gz
inflating: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/Priors2/priors6.nii.gz
brain_template = 'OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz'
brain_prior = 'OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz'
Brain Extraction#
1. ANTs#
First, we will perform brain extraction with this ANTs commands:
! antsBrainExtraction.sh
antsBrainExtraction.sh performs template-based brain extraction.
Usage:
antsBrainExtraction.sh -d imageDimension
-a anatomicalImage
-e brainExtractionTemplate
-m brainExtractionProbabilityMask
<OPT_ARGS>
-o outputPrefix
Example:
bash /opt/ants-2.3.1/antsBrainExtraction.sh -d 3 -a t1.nii.gz -e brainWithSkullTemplate.nii.gz -m brainPrior.nii.gz -o output
Required arguments:
-d: Image dimension 2 or 3 (for 2- or 3-dimensional image)
-a: Anatomical image Structural image, typically T1. If more than one
anatomical image is specified, subsequently specified
images are used during the segmentation process. However,
only the first image is used in the registration of priors.
Our suggestion would be to specify the T1 as the first image.
-e: Brain extraction template Anatomical template created using e.g. LPBA40 data set with
buildtemplateparallel.sh in ANTs.
-m: Brain extraction probability mask Brain probability mask created using e.g. LPBA40 data set which
have brain masks defined, and warped to anatomical template and
averaged resulting in a probability image.
-o: Output prefix Output directory + file prefix
Optional arguments:
-c: Tissue classification A k-means segmentation is run to find gray or white matter around
the edge of the initial brain mask warped from the template.
This produces a segmentation image with K classes, ordered by mean
intensity in increasing order. With this option, you can control
K and tell the script which classes represent CSF, gray and white matter.
Format (\"KxcsfLabelxgmLabelxwmLabel\")
Examples:
-c 3x1x2x3 for T1 with K=3, CSF=1, GM=2, WM=3 (default)
-c 3x3x2x1 for T2 with K=3, CSF=3, GM=2, WM=1
-c 3x1x3x2 for FLAIR with K=3, CSF=1 GM=3, WM=2
-c 4x4x2x3 uses K=4, CSF=4, GM=2, WM=3
-f: Brain extraction registration mask Mask used for registration to limit the metric computation to
a specific region.
-s: image file suffix Any of the standard ITK IO formats e.g. nrrd, nii.gz (default), mhd
-u: use random seeding Use random number generated from system clock in Atropos (default = 1)
-k: keep temporary files Keep brain extraction/segmentation warps, etc (default = false).
-q: use floating point precision Use antsRegistration with floating point precision.
-z: Test / debug mode If > 0, runs a faster version of the script. Only for debugging, results will not be good.
To run the command using the OASIS templates, you can follow this structure:
! antsBrainExtraction.sh -d 3 -a $input_image -e $brain_template -m $brain_prior -o ANTS_Stripped_
Will run Atropos segmentation with K=3. Classes labeled in order of mean intensity. Assuming CSF=1, GM=2, WM=3
The output directory "ANTS_Stripped_" does not exist. Making it.
Using antsBrainExtraction with the following arguments:
image dimension = 3
anatomical image = ds000102/sub-08/anat/sub-08_T1w.nii.gz
extraction template = OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz
extraction reg. mask =
extraction prior = OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz
output prefix = ANTS_Stripped_
output image suffix = nii.gz
N4 parameters (pre brain extraction):
convergence = [50x50x50x50,0.0000001]
shrink factor = 4
B-spline parameters = [200]
Atropos parameters (extraction):
convergence = [3,0.0]
likelihood = Gaussian
initialization = kmeans[3]
mrf = [0.1,1x1x1]
use clock random seed = 1
--------------------- Running antsBrainExtraction.sh on jupyter-monidoerig ---------------------
--------------------------------------------------------------------------------------
Bias correction of anatomical images (pre brain extraction)
1) pre-process by truncating the image intensities
2) run N4
--------------------------------------------------------------------------------------
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_N4Truncated0.nii.gz TruncateImageIntensity ds000102/sub-08/anat/sub-08_T1w.nii.gz 0.01 0.999 256
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//N4BiasFieldCorrection -d 3 -i ANTS_Stripped_N4Truncated0.nii.gz -s 4 -c [50x50x50x50,0.0000001] -b [200] -o ANTS_Stripped_N4Corrected0.nii.gz --verbose 1
Running N4 for 3-dimensional images.
Mask not read. Using the entire image as the mask.
Specified spline distance: 200mm
original image size: [176, 256, 256]
padded image size: [201, 401, 401]
number of control points: [4, 5, 5]
Current level = 1
Iteration 1 (of 50). Current convergence value = 0.000258536 (threshold = 1e-07)
Iteration 2 (of 50). Current convergence value = 0.000248828 (threshold = 1e-07)
Iteration 3 (of 50). Current convergence value = 0.000240245 (threshold = 1e-07)
Iteration 4 (of 50). Current convergence value = 0.000232668 (threshold = 1e-07)
Iteration 5 (of 50). Current convergence value = 0.00022608 (threshold = 1e-07)
Iteration 6 (of 50). Current convergence value = 0.000220297 (threshold = 1e-07)
Iteration 7 (of 50). Current convergence value = 0.000214742 (threshold = 1e-07)
Iteration 8 (of 50). Current convergence value = 0.000209961 (threshold = 1e-07)
Iteration 9 (of 50). Current convergence value = 0.000205729 (threshold = 1e-07)
Iteration 10 (of 50). Current convergence value = 0.000201525 (threshold = 1e-07)
Iteration 11 (of 50). Current convergence value = 0.000197393 (threshold = 1e-07)
Iteration 12 (of 50). Current convergence value = 0.000193677 (threshold = 1e-07)
Iteration 13 (of 50). Current convergence value = 0.00019035 (threshold = 1e-07)
Iteration 14 (of 50). Current convergence value = 0.000187294 (threshold = 1e-07)
Iteration 15 (of 50). Current convergence value = 0.000184611 (threshold = 1e-07)
Iteration 16 (of 50). Current convergence value = 0.000182274 (threshold = 1e-07)
Iteration 17 (of 50). Current convergence value = 0.000180015 (threshold = 1e-07)
Iteration 18 (of 50). Current convergence value = 0.000177968 (threshold = 1e-07)
Iteration 19 (of 50). Current convergence value = 0.000176021 (threshold = 1e-07)
Iteration 20 (of 50). Current convergence value = 0.000174094 (threshold = 1e-07)
Iteration 21 (of 50). Current convergence value = 0.000171951 (threshold = 1e-07)
Iteration 22 (of 50). Current convergence value = 0.000169746 (threshold = 1e-07)
Iteration 23 (of 50). Current convergence value = 0.000167645 (threshold = 1e-07)
Iteration 24 (of 50). Current convergence value = 0.000165552 (threshold = 1e-07)
Iteration 25 (of 50). Current convergence value = 0.000163502 (threshold = 1e-07)
Iteration 26 (of 50). Current convergence value = 0.000162474 (threshold = 1e-07)
Iteration 27 (of 50). Current convergence value = 0.00016215 (threshold = 1e-07)
Iteration 28 (of 50). Current convergence value = 0.000161625 (threshold = 1e-07)
Iteration 29 (of 50). Current convergence value = 0.000160964 (threshold = 1e-07)
Iteration 30 (of 50). Current convergence value = 0.000160438 (threshold = 1e-07)
Iteration 31 (of 50). Current convergence value = 0.000159709 (threshold = 1e-07)
Iteration 32 (of 50). Current convergence value = 0.000159056 (threshold = 1e-07)
Iteration 33 (of 50). Current convergence value = 0.000158104 (threshold = 1e-07)
Iteration 34 (of 50). Current convergence value = 0.000157114 (threshold = 1e-07)
Iteration 35 (of 50). Current convergence value = 0.000155974 (threshold = 1e-07)
Iteration 36 (of 50). Current convergence value = 0.000154861 (threshold = 1e-07)
Iteration 37 (of 50). Current convergence value = 0.000153546 (threshold = 1e-07)
Iteration 38 (of 50). Current convergence value = 0.000152239 (threshold = 1e-07)
Iteration 39 (of 50). Current convergence value = 0.000151147 (threshold = 1e-07)
Iteration 40 (of 50). Current convergence value = 0.000149921 (threshold = 1e-07)
Iteration 41 (of 50). Current convergence value = 0.000148829 (threshold = 1e-07)
Iteration 42 (of 50). Current convergence value = 0.000147763 (threshold = 1e-07)
Iteration 43 (of 50). Current convergence value = 0.000146349 (threshold = 1e-07)
Iteration 44 (of 50). Current convergence value = 0.00014523 (threshold = 1e-07)
Iteration 45 (of 50). Current convergence value = 0.000144099 (threshold = 1e-07)
Iteration 46 (of 50). Current convergence value = 0.000142761 (threshold = 1e-07)
Iteration 47 (of 50). Current convergence value = 0.000141342 (threshold = 1e-07)
Iteration 48 (of 50). Current convergence value = 0.000140069 (threshold = 1e-07)
Iteration 49 (of 50). Current convergence value = 0.000138855 (threshold = 1e-07)
Iteration 50 (of 50). Current convergence value = 0.000137506 (threshold = 1e-07)
Current level = 2
Iteration 1 (of 50). Current convergence value = 0.000342861 (threshold = 1e-07)
Iteration 2 (of 50). Current convergence value = 0.000341625 (threshold = 1e-07)
Iteration 3 (of 50). Current convergence value = 0.000340768 (threshold = 1e-07)
Iteration 4 (of 50). Current convergence value = 0.000340175 (threshold = 1e-07)
Iteration 5 (of 50). Current convergence value = 0.00033827 (threshold = 1e-07)
Iteration 6 (of 50). Current convergence value = 0.000336399 (threshold = 1e-07)
Iteration 7 (of 50). Current convergence value = 0.000334478 (threshold = 1e-07)
Iteration 8 (of 50). Current convergence value = 0.000331891 (threshold = 1e-07)
Iteration 9 (of 50). Current convergence value = 0.000329826 (threshold = 1e-07)
Iteration 10 (of 50). Current convergence value = 0.000327873 (threshold = 1e-07)
Iteration 11 (of 50). Current convergence value = 0.000326366 (threshold = 1e-07)
Iteration 12 (of 50). Current convergence value = 0.000325203 (threshold = 1e-07)
Iteration 13 (of 50). Current convergence value = 0.000324245 (threshold = 1e-07)
Iteration 14 (of 50). Current convergence value = 0.000323119 (threshold = 1e-07)
Iteration 15 (of 50). Current convergence value = 0.000322634 (threshold = 1e-07)
Iteration 16 (of 50). Current convergence value = 0.000322995 (threshold = 1e-07)
Iteration 17 (of 50). Current convergence value = 0.00032329 (threshold = 1e-07)
Iteration 18 (of 50). Current convergence value = 0.000323898 (threshold = 1e-07)
Iteration 19 (of 50). Current convergence value = 0.000324528 (threshold = 1e-07)
Iteration 20 (of 50). Current convergence value = 0.000321647 (threshold = 1e-07)
Iteration 21 (of 50). Current convergence value = 0.000318242 (threshold = 1e-07)
Iteration 22 (of 50). Current convergence value = 0.000315148 (threshold = 1e-07)
Iteration 23 (of 50). Current convergence value = 0.000312036 (threshold = 1e-07)
Iteration 24 (of 50). Current convergence value = 0.000309086 (threshold = 1e-07)
Iteration 25 (of 50). Current convergence value = 0.00030673 (threshold = 1e-07)
Iteration 26 (of 50). Current convergence value = 0.00030398 (threshold = 1e-07)
Iteration 27 (of 50). Current convergence value = 0.000301205 (threshold = 1e-07)
Iteration 28 (of 50). Current convergence value = 0.000298513 (threshold = 1e-07)
Iteration 29 (of 50). Current convergence value = 0.000295251 (threshold = 1e-07)
Iteration 30 (of 50). Current convergence value = 0.000288666 (threshold = 1e-07)
Iteration 31 (of 50). Current convergence value = 0.000270682 (threshold = 1e-07)
Iteration 32 (of 50). Current convergence value = 0.000259328 (threshold = 1e-07)
Iteration 33 (of 50). Current convergence value = 0.000247768 (threshold = 1e-07)
Iteration 34 (of 50). Current convergence value = 0.00023528 (threshold = 1e-07)
Iteration 35 (of 50). Current convergence value = 0.000224417 (threshold = 1e-07)
Iteration 36 (of 50). Current convergence value = 0.000218353 (threshold = 1e-07)
Iteration 37 (of 50). Current convergence value = 0.000213534 (threshold = 1e-07)
Iteration 38 (of 50). Current convergence value = 0.000210124 (threshold = 1e-07)
Iteration 39 (of 50). Current convergence value = 0.000207168 (threshold = 1e-07)
Iteration 40 (of 50). Current convergence value = 0.000205513 (threshold = 1e-07)
Iteration 41 (of 50). Current convergence value = 0.000205272 (threshold = 1e-07)
Iteration 42 (of 50). Current convergence value = 0.000204942 (threshold = 1e-07)
Iteration 43 (of 50). Current convergence value = 0.000204254 (threshold = 1e-07)
Iteration 44 (of 50). Current convergence value = 0.000202326 (threshold = 1e-07)
Iteration 45 (of 50). Current convergence value = 0.000199774 (threshold = 1e-07)
Iteration 46 (of 50). Current convergence value = 0.000197384 (threshold = 1e-07)
Iteration 47 (of 50). Current convergence value = 0.000195031 (threshold = 1e-07)
Iteration 48 (of 50). Current convergence value = 0.000193075 (threshold = 1e-07)
Iteration 49 (of 50). Current convergence value = 0.00019113 (threshold = 1e-07)
Iteration 50 (of 50). Current convergence value = 0.0001893 (threshold = 1e-07)
Current level = 3
Iteration 1 (of 50). Current convergence value = 0.000998225 (threshold = 1e-07)
Iteration 2 (of 50). Current convergence value = 0.000911791 (threshold = 1e-07)
Iteration 3 (of 50). Current convergence value = 0.000837971 (threshold = 1e-07)
Iteration 4 (of 50). Current convergence value = 0.000763517 (threshold = 1e-07)
Iteration 5 (of 50). Current convergence value = 0.000704851 (threshold = 1e-07)
Iteration 6 (of 50). Current convergence value = 0.00065979 (threshold = 1e-07)
Iteration 7 (of 50). Current convergence value = 0.000624687 (threshold = 1e-07)
Iteration 8 (of 50). Current convergence value = 0.000589346 (threshold = 1e-07)
Iteration 9 (of 50). Current convergence value = 0.000554393 (threshold = 1e-07)
Iteration 10 (of 50). Current convergence value = 0.000525235 (threshold = 1e-07)
Iteration 11 (of 50). Current convergence value = 0.000501895 (threshold = 1e-07)
Iteration 12 (of 50). Current convergence value = 0.000483087 (threshold = 1e-07)
Iteration 13 (of 50). Current convergence value = 0.000468356 (threshold = 1e-07)
Iteration 14 (of 50). Current convergence value = 0.000457002 (threshold = 1e-07)
Iteration 15 (of 50). Current convergence value = 0.000448627 (threshold = 1e-07)
Iteration 16 (of 50). Current convergence value = 0.000442205 (threshold = 1e-07)
Iteration 17 (of 50). Current convergence value = 0.000437965 (threshold = 1e-07)
Iteration 18 (of 50). Current convergence value = 0.000434013 (threshold = 1e-07)
Iteration 19 (of 50). Current convergence value = 0.000430327 (threshold = 1e-07)
Iteration 20 (of 50). Current convergence value = 0.000422352 (threshold = 1e-07)
Iteration 21 (of 50). Current convergence value = 0.000408076 (threshold = 1e-07)
Iteration 22 (of 50). Current convergence value = 0.000395871 (threshold = 1e-07)
Iteration 23 (of 50). Current convergence value = 0.0003853 (threshold = 1e-07)
Iteration 24 (of 50). Current convergence value = 0.000375409 (threshold = 1e-07)
Iteration 25 (of 50). Current convergence value = 0.000366843 (threshold = 1e-07)
Iteration 26 (of 50). Current convergence value = 0.000359712 (threshold = 1e-07)
Iteration 27 (of 50). Current convergence value = 0.000353946 (threshold = 1e-07)
Iteration 28 (of 50). Current convergence value = 0.000349232 (threshold = 1e-07)
Iteration 29 (of 50). Current convergence value = 0.000345705 (threshold = 1e-07)
Iteration 30 (of 50). Current convergence value = 0.000343062 (threshold = 1e-07)
Iteration 31 (of 50). Current convergence value = 0.000341917 (threshold = 1e-07)
Iteration 32 (of 50). Current convergence value = 0.000342084 (threshold = 1e-07)
Iteration 33 (of 50). Current convergence value = 0.000343344 (threshold = 1e-07)
Iteration 34 (of 50). Current convergence value = 0.00034619 (threshold = 1e-07)
Iteration 35 (of 50). Current convergence value = 0.000350448 (threshold = 1e-07)
Iteration 36 (of 50). Current convergence value = 0.000356079 (threshold = 1e-07)
Iteration 37 (of 50). Current convergence value = 0.000363458 (threshold = 1e-07)
Iteration 38 (of 50). Current convergence value = 0.000370693 (threshold = 1e-07)
Iteration 39 (of 50). Current convergence value = 0.000363499 (threshold = 1e-07)
Iteration 40 (of 50). Current convergence value = 0.000354278 (threshold = 1e-07)
Iteration 41 (of 50). Current convergence value = 0.000344335 (threshold = 1e-07)
Iteration 42 (of 50). Current convergence value = 0.000326012 (threshold = 1e-07)
Iteration 43 (of 50). Current convergence value = 0.000283165 (threshold = 1e-07)
Iteration 44 (of 50). Current convergence value = 0.000262942 (threshold = 1e-07)
Iteration 45 (of 50). Current convergence value = 0.000256401 (threshold = 1e-07)
Iteration 46 (of 50). Current convergence value = 0.000256516 (threshold = 1e-07)
Iteration 47 (of 50). Current convergence value = 0.000259121 (threshold = 1e-07)
Iteration 48 (of 50). Current convergence value = 0.000262287 (threshold = 1e-07)
Iteration 49 (of 50). Current convergence value = 0.000265913 (threshold = 1e-07)
Iteration 50 (of 50). Current convergence value = 0.000269532 (threshold = 1e-07)
Current level = 4
Iteration 1 (of 50). Current convergence value = 0.00146065 (threshold = 1e-07)
Iteration 2 (of 50). Current convergence value = 0.0014295 (threshold = 1e-07)
Iteration 3 (of 50). Current convergence value = 0.00134935 (threshold = 1e-07)
Iteration 4 (of 50). Current convergence value = 0.00127588 (threshold = 1e-07)
Iteration 5 (of 50). Current convergence value = 0.00122346 (threshold = 1e-07)
Iteration 6 (of 50). Current convergence value = 0.00117814 (threshold = 1e-07)
Iteration 7 (of 50). Current convergence value = 0.00114049 (threshold = 1e-07)
Iteration 8 (of 50). Current convergence value = 0.00109704 (threshold = 1e-07)
Iteration 9 (of 50). Current convergence value = 0.0010524 (threshold = 1e-07)
Iteration 10 (of 50). Current convergence value = 0.00101912 (threshold = 1e-07)
Iteration 11 (of 50). Current convergence value = 0.000998206 (threshold = 1e-07)
Iteration 12 (of 50). Current convergence value = 0.000984174 (threshold = 1e-07)
Iteration 13 (of 50). Current convergence value = 0.000974387 (threshold = 1e-07)
Iteration 14 (of 50). Current convergence value = 0.000967613 (threshold = 1e-07)
Iteration 15 (of 50). Current convergence value = 0.00096344 (threshold = 1e-07)
Iteration 16 (of 50). Current convergence value = 0.000962287 (threshold = 1e-07)
Iteration 17 (of 50). Current convergence value = 0.000964042 (threshold = 1e-07)
Iteration 18 (of 50). Current convergence value = 0.000968691 (threshold = 1e-07)
Iteration 19 (of 50). Current convergence value = 0.000974578 (threshold = 1e-07)
Iteration 20 (of 50). Current convergence value = 0.000980622 (threshold = 1e-07)
Iteration 21 (of 50). Current convergence value = 0.000988265 (threshold = 1e-07)
Iteration 22 (of 50). Current convergence value = 0.000996283 (threshold = 1e-07)
Iteration 23 (of 50). Current convergence value = 0.000980695 (threshold = 1e-07)
Iteration 24 (of 50). Current convergence value = 0.000977463 (threshold = 1e-07)
Iteration 25 (of 50). Current convergence value = 0.000974733 (threshold = 1e-07)
Iteration 26 (of 50). Current convergence value = 0.000967454 (threshold = 1e-07)
Iteration 27 (of 50). Current convergence value = 0.000956385 (threshold = 1e-07)
Iteration 28 (of 50). Current convergence value = 0.00094419 (threshold = 1e-07)
Iteration 29 (of 50). Current convergence value = 0.000928369 (threshold = 1e-07)
Iteration 30 (of 50). Current convergence value = 0.000910889 (threshold = 1e-07)
Iteration 31 (of 50). Current convergence value = 0.000896919 (threshold = 1e-07)
Iteration 32 (of 50). Current convergence value = 0.000881585 (threshold = 1e-07)
Iteration 33 (of 50). Current convergence value = 0.000863008 (threshold = 1e-07)
Iteration 34 (of 50). Current convergence value = 0.000846915 (threshold = 1e-07)
Iteration 35 (of 50). Current convergence value = 0.000831319 (threshold = 1e-07)
Iteration 36 (of 50). Current convergence value = 0.00081534 (threshold = 1e-07)
Iteration 37 (of 50). Current convergence value = 0.000798769 (threshold = 1e-07)
Iteration 38 (of 50). Current convergence value = 0.000781479 (threshold = 1e-07)
Iteration 39 (of 50). Current convergence value = 0.00076681 (threshold = 1e-07)
Iteration 40 (of 50). Current convergence value = 0.000758623 (threshold = 1e-07)
Iteration 41 (of 50). Current convergence value = 0.000749481 (threshold = 1e-07)
Iteration 42 (of 50). Current convergence value = 0.000746784 (threshold = 1e-07)
Iteration 43 (of 50). Current convergence value = 0.000746007 (threshold = 1e-07)
Iteration 44 (of 50). Current convergence value = 0.000749525 (threshold = 1e-07)
Iteration 45 (of 50). Current convergence value = 0.000754025 (threshold = 1e-07)
Iteration 46 (of 50). Current convergence value = 0.000755228 (threshold = 1e-07)
Iteration 47 (of 50). Current convergence value = 0.000758238 (threshold = 1e-07)
Iteration 48 (of 50). Current convergence value = 0.000775884 (threshold = 1e-07)
Iteration 49 (of 50). Current convergence value = 0.000795187 (threshold = 1e-07)
Iteration 50 (of 50). Current convergence value = 0.000814762 (threshold = 1e-07)
N4BiasFieldCorrectionImageFilter (0x1b1c36e0)
RTTI typeinfo: itk::N4BiasFieldCorrectionImageFilter<itk::Image<float, 3u>, itk::Image<float, 3u>, itk::Image<float, 3u> >
Reference Count: 1
Modified Time: 474
Debug: Off
Object Name:
Observers:
IterationEvent(Command)
Inputs:
Primary: (0x1b1cfa70) *
_1: (0x1b1d3410)
Indexed Inputs:
0: Primary (0x1b1cfa70)
1: _1 (0x1b1d3410)
Required Input Names: Primary
NumberOfRequiredInputs: 1
Outputs:
Primary: (0x1b1cae80)
Indexed Outputs:
0: Primary (0x1b1cae80)
NumberOfRequiredOutputs: 1
Number Of Threads: 32
ReleaseDataFlag: Off
ReleaseDataBeforeUpdateFlag: Off
AbortGenerateData: Off
Progress: 0
Multithreader:
RTTI typeinfo: itk::PoolMultiThreader
Reference Count: 1
Modified Time: 138
Debug: Off
Object Name:
Observers:
none
DynamicMultiThreading: On
CoordinateTolerance: 1e-06
DirectionTolerance: 1e-06
Use Mask Label: 0
Mask label: 1
Number of histogram bins: 200
Wiener filter noise: 0.01
Bias field FWHM: 0.15
Maximum number of iterations: [50, 50, 50, 50]
Convergence threshold: 1e-07
Spline order: 3
Number of fitting levels: [4, 4, 4]
Number of control points: [4, 5, 5]
CurrentConvergenceMeasurement: 0.000814762
CurrentLevel: 4
ElapsedIterations: 51
LogBiasFieldControlPointLattice:
Image (0x1b1d2dd0)
RTTI typeinfo: itk::Image<itk::Vector<float, 1u>, 3u>
Reference Count: 1
Modified Time: 396864244
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 0
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [11, 19, 19]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [11, 19, 19]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [11, 19, 19]
Spacing: [24.5, 24.75, 24.75]
Origin: [-24.5, 24.75, -24.75]
Direction:
1 0 0
0 -1 0
0 0 1
IndexToPointMatrix:
24.5 0 0
0 -24.75 0
0 0 24.75
PointToIndexMatrix:
0.0408163 0 0
0 -0.040404 0
0 0 0.040404
Inverse Direction:
1 0 0
0 -1 0
0 0 1
PixelContainer:
ImportImageContainer (0x1b1d3060)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, itk::Vector<float, 1u> >
Reference Count: 1
Modified Time: 396864245
Debug: Off
Object Name:
Observers:
none
Pointer: 0x1b97b520
Container manages memory: true
Size: 3971
Capacity: 3971
Elapsed time: 28.795
END <<<<<<<<<<<<<<<<<<<<
--------------------------------------------------------------------------------------
Done with N4 correction (pre brain extraction): 0h 0m 44s
--------------------------------------------------------------------------------------
--------------------------------------------------------------------------------------
Brain extraction using the following steps:
1) Register OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz to ANTS_Stripped_N4Corrected0.nii.gz
2) Warp OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz to ds000102/sub-08/anat/sub-08_T1w.nii.gz using, from 1),
ANTS_Stripped_BrainExtractionWarp/Affine
3) Refine segmentation results using Atropos
--------------------------------------------------------------------------------------
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ResampleImageBySpacing 3 OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz ANTS_Stripped_BrainExtractionInitialAffineFixed.nii.gz 4 4 4 1
spacing [1, 1, 1] dim 3
spacing2 [4, 4, 4]
smoothing by : 3 dir 0
smoothing by : 3 dir 1
smoothing by : 3 dir 2
out space [4, 4, 4]
output size [54, 64, 72] spc [4, 4, 4]
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ResampleImageBySpacing 3 ANTS_Stripped_N4Corrected0.nii.gz ANTS_Stripped_BrainExtractionInitialAffineMoving.nii.gz 4 4 4 1
spacing [1, 1, 1] dim 3
spacing2 [4, 4, 4]
smoothing by : 3 dir 0
smoothing by : 3 dir 1
smoothing by : 3 dir 2
out space [4, 4, 4]
output size [44, 64, 64] spc [4, 4, 4]
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionLaplacian.nii.gz Laplacian ANTS_Stripped_N4Corrected0.nii.gz 1.5 1
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionTemplateLaplacian.nii.gz Laplacian OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz 1.5 1
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//antsAI -d 3 -v 1 -m Mattes[ANTS_Stripped_BrainExtractionInitialAffineFixed.nii.gz,ANTS_Stripped_BrainExtractionInitialAffineMoving.nii.gz,32,Regular,0.2] -t Affine[0.1] -s [20,0.12] -g [40,0x40x40] -p 0 -c 10 -o ANTS_Stripped_BrainExtractionInitialAffine.mat
Using the Mattes MI metric (number of bins = 32)
Starting optimizer with 243 starting points
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//antsRegistration -d 3 -u 1 -w [0.025,0.975] -o ANTS_Stripped_BrainExtractionPrior -r ANTS_Stripped_BrainExtractionInitialAffine.mat -z 1 --float 0 --verbose 1 -m MI[OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz,ANTS_Stripped_N4Corrected0.nii.gz,1,32,Regular,0.25] -c [1000x500x250x100,1e-8,10] -t Rigid[0.1] -f 8x4x2x1 -s 4x2x1x0 -m MI[OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz,ANTS_Stripped_N4Corrected0.nii.gz,1,32,Regular,0.25] -c [1000x500x250x100,1e-8,10] -t Affine[0.1] -f 8x4x2x1 -s 4x2x1x0 -m CC[OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz,ANTS_Stripped_N4Corrected0.nii.gz,0.5,4] -m CC[ANTS_Stripped_BrainExtractionTemplateLaplacian.nii.gz,ANTS_Stripped_BrainExtractionLaplacian.nii.gz,0.5,4] -c [50x10x0,1e-9,15] -t SyN[0.1,3,0] -f 4x2x1 -s 2x1x0
All_Command_lines_OK
Using double precision for computations.
=============================================================================
The composite transform comprises the following transforms (in order):
1. ANTS_Stripped_BrainExtractionInitialAffine.mat (type = AffineTransform)
=============================================================================
number of levels = 4
number of levels = 4
number of levels = 3
fixed image: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz
moving image: ANTS_Stripped_N4Corrected0.nii.gz
fixed image: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz
moving image: ANTS_Stripped_N4Corrected0.nii.gz
fixed image: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0.nii.gz
moving image: ANTS_Stripped_N4Corrected0.nii.gz
fixed image: ANTS_Stripped_BrainExtractionTemplateLaplacian.nii.gz
moving image: ANTS_Stripped_BrainExtractionLaplacian.nii.gz
Dimension = 3
Number of stages = 3
Use Histogram Matching true
Winsorize image intensities true
Lower quantile = 0.025
Upper quantile = 0.975
Stage 1 State
Image metric = Mattes
Fixed image = Image (0xa9f33d0)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1254
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1065
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
Spacing: [1, 1, 1]
Origin: [0, 293, 0]
Direction:
1 0 0
0 -0 -1
0 -1 -0
IndexToPointMatrix:
1 0 0
0 0 -1
0 -1 0
PointToIndexMatrix:
1 0 0
0 0 -1
0 -1 0
Inverse Direction:
1 0 0
0 0 -1
0 -1 0
PixelContainer:
ImportImageContainer (0xa9e78d0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1062
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f35970010
Container manages memory: true
Size: 16091136
Capacity: 16091136
Moving image = Image (0xa9f68a0)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1255
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1252
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
Spacing: [1, 1, 1]
Origin: [0, 126.054, -133.283]
Direction:
1 0 0
0 -1 0
0 0 1
IndexToPointMatrix:
1 0 0
0 -1 0
0 0 1
PointToIndexMatrix:
1 0 0
0 -1 0
0 0 1
Inverse Direction:
1 0 0
0 -1 0
0 0 1
PixelContainer:
ImportImageContainer (0xa9e4390)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1249
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f3016f010
Container manages memory: true
Size: 11534336
Capacity: 11534336
Weighting = 1
Sampling strategy = regular
Number of bins = 32
Radius = 4
Sampling percentage = 0.25
Transform = Rigid
Gradient step = 0.1
Update field sigma (voxel space) = 0
Total field sigma (voxel space) = 0
Update field time sigma = 0
Total field time sigma = 0
Number of time indices = 0
Number of time point samples = 0
Stage 2 State
Image metric = Mattes
Fixed image = Image (0xa9fc2c0)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1630
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1441
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
Spacing: [1, 1, 1]
Origin: [0, 293, 0]
Direction:
1 0 0
0 -0 -1
0 -1 -0
IndexToPointMatrix:
1 0 0
0 0 -1
0 -1 0
PointToIndexMatrix:
1 0 0
0 0 -1
0 -1 0
Inverse Direction:
1 0 0
0 0 -1
0 -1 0
PixelContainer:
ImportImageContainer (0xa9fc550)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1438
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f286aa010
Container manages memory: true
Size: 16091136
Capacity: 16091136
Moving image = Image (0xa9fea00)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 1631
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1628
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
Spacing: [1, 1, 1]
Origin: [0, 126.054, -133.283]
Direction:
1 0 0
0 -1 0
0 0 1
IndexToPointMatrix:
1 0 0
0 -1 0
0 0 1
PointToIndexMatrix:
1 0 0
0 -1 0
0 0 1
Inverse Direction:
1 0 0
0 -1 0
0 0 1
PixelContainer:
ImportImageContainer (0xa9f71d0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1625
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f22ea9010
Container manages memory: true
Size: 11534336
Capacity: 11534336
Weighting = 1
Sampling strategy = regular
Number of bins = 32
Radius = 4
Sampling percentage = 0.25
Transform = Affine
Gradient step = 0.1
Update field sigma (voxel space) = 0
Total field sigma (voxel space) = 0
Update field time sigma = 0
Total field time sigma = 0
Number of time indices = 0
Number of time point samples = 0
Stage 3 State
Image metric = CC
Fixed image = Image (0xaa01b20)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 2006
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 1817
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [216, 256, 291]
Spacing: [1, 1, 1]
Origin: [0, 293, 0]
Direction:
1 0 0
0 -0 -1
0 -1 -0
IndexToPointMatrix:
1 0 0
0 0 -1
0 -1 0
PointToIndexMatrix:
1 0 0
0 0 -1
0 -1 0
Inverse Direction:
1 0 0
0 0 -1
0 -1 0
PixelContainer:
ImportImageContainer (0xaa01db0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 1814
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f1b3e4010
Container manages memory: true
Size: 16091136
Capacity: 16091136
Moving image = Image (0xaa041b0)
RTTI typeinfo: itk::Image<double, 3u>
Reference Count: 2
Modified Time: 2007
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 0
UpdateMTime: 2004
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [176, 256, 256]
Spacing: [1, 1, 1]
Origin: [0, 126.054, -133.283]
Direction:
1 0 0
0 -1 0
0 0 1
IndexToPointMatrix:
1 0 0
0 -1 0
0 0 1
PointToIndexMatrix:
1 0 0
0 -1 0
0 0 1
Inverse Direction:
1 0 0
0 -1 0
0 0 1
PixelContainer:
ImportImageContainer (0xaa04440)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, double>
Reference Count: 1
Modified Time: 2001
Debug: Off
Object Name:
Observers:
none
Pointer: 0x7f6f15be3010
Container manages memory: true
Size: 11534336
Capacity: 11534336
Weighting = 0.5
Sampling strategy = none
Number of bins = 32
Radius = 4
Sampling percentage = 1
Transform = SyN
Gradient step = 0.1
Update field sigma (voxel space) = 3
Total field sigma (voxel space) = 0
Update field time sigma = 0
Total field time sigma = 0
Number of time indices = 0
Number of time point samples = 0
Registration using 3 total stages.
Stage 0
iterations = 1000x500x250x100
convergence threshold = 1e-08
convergence window size = 10
number of levels = 4
using the Mattes MI metric (number of bins = 32, weight = 1)
preprocessing: winsorizing the image intensities
preprocessing: histogram matching the images
Shrink factors (level 1 out of 4): [8, 8, 8]
Shrink factors (level 2 out of 4): [4, 4, 4]
Shrink factors (level 3 out of 4): [2, 2, 2]
Shrink factors (level 4 out of 4): [1, 1, 1]
smoothing sigmas per level: [4, 2, 1, 0]
regular sampling (percentage = 0.25)
*** Running Euler3DTransform registration ***
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -7.536517046677e-01, 1.797693134862e+308, 1.5495e+00, 1.5495e+00,
2DIAGNOSTIC, 2, -7.537473866575e-01, 1.797693134862e+308, 1.5531e+00, 3.6302e-03,
2DIAGNOSTIC, 3, -7.537209862946e-01, 1.797693134862e+308, 1.5566e+00, 3.4928e-03,
2DIAGNOSTIC, 4, -7.543671684293e-01, 1.797693134862e+308, 1.5600e+00, 3.3751e-03,
2DIAGNOSTIC, 5, -7.551942106114e-01, 1.797693134862e+308, 1.5634e+00, 3.3939e-03,
2DIAGNOSTIC, 6, -7.558229085333e-01, 1.797693134862e+308, 1.5668e+00, 3.4618e-03,
2DIAGNOSTIC, 7, -7.574886200738e-01, 1.797693134862e+308, 1.5702e+00, 3.3751e-03,
2DIAGNOSTIC, 8, -7.609174688167e-01, 1.797693134862e+308, 1.5770e+00, 6.8331e-03,
2DIAGNOSTIC, 9, -7.611451814184e-01, 1.797693134862e+308, 1.5811e+00, 4.0920e-03,
2DIAGNOSTIC, 10, -7.613712387168e-01, 7.842351055372e-04, 1.5868e+00, 5.6369e-03,
2DIAGNOSTIC, 11, -7.611822160651e-01, 7.476310528839e-04, 1.5912e+00, 4.4429e-03,
2DIAGNOSTIC, 12, -7.617449459927e-01, 6.706595207591e-04, 1.5963e+00, 5.0919e-03,
2DIAGNOSTIC, 13, -7.621736295462e-01, 5.546344774527e-04, 1.6007e+00, 4.4358e-03,
2DIAGNOSTIC, 14, -7.637601636169e-01, 4.644438310239e-04, 1.6080e+00, 7.2691e-03,
2DIAGNOSTIC, 15, -7.644171923465e-01, 3.818023973478e-04, 1.6138e+00, 5.7831e-03,
2DIAGNOSTIC, 16, -7.643304010101e-01, 2.875599361090e-04, 1.6181e+00, 4.3011e-03,
2DIAGNOSTIC, 17, -7.643382585631e-01, 2.135897005037e-04, 1.6238e+00, 5.7070e-03,
2DIAGNOSTIC, 18, -7.643701985532e-01, 2.000056275030e-04, 1.6289e+00, 5.0490e-03,
2DIAGNOSTIC, 19, -7.643347106448e-01, 1.726381457220e-04, 1.6339e+00, 5.0359e-03,
2DIAGNOSTIC, 20, -7.641538548379e-01, 1.330630861163e-04, 1.6383e+00, 4.4289e-03,
2DIAGNOSTIC, 21, -7.641190795319e-01, 8.138953688107e-05, 1.6429e+00, 4.6091e-03,
2DIAGNOSTIC, 22, -7.641719306244e-01, 3.770725866920e-05, 1.6480e+00, 5.0559e-03,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -5.935296626196e-01, 1.797693134862e+308, 3.1932e+00, 1.5453e+00,
2DIAGNOSTIC, 2, -5.936448624567e-01, 1.797693134862e+308, 3.2162e+00, 2.2935e-02,
2DIAGNOSTIC, 3, -5.935640133563e-01, 1.797693134862e+308, 3.2428e+00, 2.6624e-02,
2DIAGNOSTIC, 4, -5.936385409286e-01, 1.797693134862e+308, 3.2829e+00, 4.0090e-02,
2DIAGNOSTIC, 5, -5.936264283812e-01, 1.797693134862e+308, 3.3052e+00, 2.2314e-02,
2DIAGNOSTIC, 6, -5.936225364109e-01, 1.797693134862e+308, 3.3308e+00, 2.5596e-02,
2DIAGNOSTIC, 7, -5.936219262068e-01, 1.797693134862e+308, 3.3523e+00, 2.1468e-02,
2DIAGNOSTIC, 8, -5.936192768262e-01, 1.797693134862e+308, 3.3751e+00, 2.2843e-02,
2DIAGNOSTIC, 9, -5.936140842558e-01, 1.797693134862e+308, 3.3990e+00, 2.3932e-02,
2DIAGNOSTIC, 10, -5.936186882205e-01, 7.612452384329e-06, 3.4210e+00, 2.1995e-02,
2DIAGNOSTIC, 11, -5.936214230857e-01, 3.053105598602e-06, 3.4388e+00, 1.7790e-02,
2DIAGNOSTIC, 12, -5.936295663513e-01, 4.277984220771e-06, 3.4622e+00, 2.3356e-02,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -5.200706672309e-01, 1.797693134862e+308, 4.5364e+00, 1.0742e+00,
2DIAGNOSTIC, 2, -5.204283649917e-01, 1.797693134862e+308, 4.6723e+00, 1.3592e-01,
2DIAGNOSTIC, 3, -5.208455852462e-01, 1.797693134862e+308, 4.7986e+00, 1.2629e-01,
2DIAGNOSTIC, 4, -5.211450614565e-01, 1.797693134862e+308, 4.9244e+00, 1.2583e-01,
2DIAGNOSTIC, 5, -5.213348714731e-01, 1.797693134862e+308, 5.0626e+00, 1.3822e-01,
2DIAGNOSTIC, 6, -5.216572235098e-01, 1.797693134862e+308, 5.2888e+00, 2.2616e-01,
2DIAGNOSTIC, 7, -5.216576403991e-01, 1.797693134862e+308, 5.4647e+00, 1.7593e-01,
2DIAGNOSTIC, 8, -5.216498868667e-01, 1.797693134862e+308, 5.6863e+00, 2.2154e-01,
2DIAGNOSTIC, 9, -5.216507468874e-01, 1.797693134862e+308, 5.8329e+00, 1.4661e-01,
2DIAGNOSTIC, 10, -5.216505093220e-01, 1.943054229684e-04, 6.0053e+00, 1.7244e-01,
2DIAGNOSTIC, 11, -5.216417328325e-01, 1.227842089593e-04, 6.1417e+00, 1.3634e-01,
2DIAGNOSTIC, 12, -5.216363447978e-01, 6.816831474181e-05, 6.2911e+00, 1.4940e-01,
2DIAGNOSTIC, 13, -5.216375479921e-01, 3.355604128388e-05, 6.4669e+00, 1.7585e-01,
2DIAGNOSTIC, 14, -5.216379865711e-01, 1.267550016035e-05, 6.6344e+00, 1.6745e-01,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -4.551781313907e-01, 1.797693134862e+308, 1.0452e+01, 3.8175e+00,
2DIAGNOSTIC, 2, -4.553831823223e-01, 1.797693134862e+308, 1.1234e+01, 7.8184e-01,
2DIAGNOSTIC, 3, -4.554872191605e-01, 1.797693134862e+308, 1.2036e+01, 8.0192e-01,
2DIAGNOSTIC, 4, -4.555303779194e-01, 1.797693134862e+308, 1.2810e+01, 7.7417e-01,
2DIAGNOSTIC, 5, -4.556419290902e-01, 1.797693134862e+308, 1.3787e+01, 9.7687e-01,
2DIAGNOSTIC, 6, -4.557008254494e-01, 1.797693134862e+308, 1.4571e+01, 7.8429e-01,
2DIAGNOSTIC, 7, -4.558494998319e-01, 1.797693134862e+308, 1.5365e+01, 7.9406e-01,
2DIAGNOSTIC, 8, -4.565904019778e-01, 1.797693134862e+308, 1.6228e+01, 8.6343e-01,
2DIAGNOSTIC, 9, -4.573815029853e-01, 1.797693134862e+308, 1.7839e+01, 1.6104e+00,
2DIAGNOSTIC, 10, -4.573753332103e-01, 3.070908486995e-04, 1.8583e+01, 7.4418e-01,
2DIAGNOSTIC, 11, -4.573887918572e-01, 3.100256639842e-04, 1.9352e+01, 7.6919e-01,
2DIAGNOSTIC, 12, -4.574035418242e-01, 2.966730753228e-04, 2.0080e+01, 7.2748e-01,
2DIAGNOSTIC, 13, -4.574286069804e-01, 2.642874463690e-04, 2.0848e+01, 7.6821e-01,
2DIAGNOSTIC, 14, -4.574581839944e-01, 2.152614702367e-04, 2.1631e+01, 7.8354e-01,
2DIAGNOSTIC, 15, -4.575078979323e-01, 1.604094209176e-04, 2.2368e+01, 7.3685e-01,
2DIAGNOSTIC, 16, -4.575741886508e-01, 1.034330662832e-04, 2.3121e+01, 7.5295e-01,
2DIAGNOSTIC, 17, -4.576520477389e-01, 5.165319176648e-05, 2.3885e+01, 7.6341e-01,
2DIAGNOSTIC, 18, -4.578041353705e-01, 3.109381026074e-05, 2.4748e+01, 8.6382e-01,
2DIAGNOSTIC, 19, -4.581065617596e-01, 4.571496912069e-05, 2.6011e+01, 1.2623e+00,
2DIAGNOSTIC, 20, -4.581102844447e-01, 5.449864857585e-05, 2.6972e+01, 9.6133e-01,
2DIAGNOSTIC, 21, -4.581115398587e-01, 5.809318081304e-05, 2.7910e+01, 9.3750e-01,
2DIAGNOSTIC, 22, -4.581117665335e-01, 5.659376937230e-05, 2.8847e+01, 9.3787e-01,
2DIAGNOSTIC, 23, -4.581111558556e-01, 5.064375224961e-05, 2.9671e+01, 8.2389e-01,
2DIAGNOSTIC, 24, -4.581105160366e-01, 4.119401262637e-05, 3.0505e+01, 8.3377e-01,
2DIAGNOSTIC, 25, -4.581099037140e-01, 2.998822225364e-05, 3.1584e+01, 1.0786e+00,
2DIAGNOSTIC, 26, -4.581092744206e-01, 1.862367380751e-05, 3.2751e+01, 1.1669e+00,
2DIAGNOSTIC, 27, -4.581092273406e-01, 8.240548398791e-06, 3.3601e+01, 8.5033e-01,
2DIAGNOSTIC, 28, -4.581091750594e-01, 1.118891451803e-06, 3.4343e+01, 7.4216e-01,
2DIAGNOSTIC, 29, -4.581091070281e-01, 9.702879958384e-07, 3.5311e+01, 9.6801e-01,
2DIAGNOSTIC, 30, -4.581089501725e-01, 9.170884486959e-07, 3.6057e+01, 7.4575e-01,
2DIAGNOSTIC, 31, -4.581089094266e-01, 9.052579336630e-07, 3.6899e+01, 8.4219e-01,
2DIAGNOSTIC, 32, -4.581088409796e-01, 9.088955359032e-07, 3.7747e+01, 8.4813e-01,
2DIAGNOSTIC, 33, -4.581090023567e-01, 9.121109990837e-07, 3.8602e+01, 8.5534e-01,
2DIAGNOSTIC, 34, -4.581089550822e-01, 9.077463881159e-07, 3.9461e+01, 8.5880e-01,
2DIAGNOSTIC, 35, -4.581088923050e-01, 8.948256594275e-07, 4.0304e+01, 8.4287e-01,
2DIAGNOSTIC, 36, -4.581089436467e-01, 8.742070163083e-07, 4.1356e+01, 1.0518e+00,
2DIAGNOSTIC, 37, -4.581089188142e-01, 8.544524457317e-07, 4.2229e+01, 8.7362e-01,
2DIAGNOSTIC, 38, -4.581088810271e-01, 8.348505437412e-07, 4.3078e+01, 8.4865e-01,
2DIAGNOSTIC, 39, -4.581087884893e-01, 8.141083124337e-07, 4.3828e+01, 7.4973e-01,
2DIAGNOSTIC, 40, -4.581088105867e-01, 7.926594123159e-07, 4.4595e+01, 7.6763e-01,
2DIAGNOSTIC, 41, -4.581087995302e-01, 7.718301750954e-07, 4.5372e+01, 7.7667e-01,
2DIAGNOSTIC, 42, -4.581088629049e-01, 7.523039190077e-07, 4.6222e+01, 8.4999e-01,
2DIAGNOSTIC, 43, -4.581089264879e-01, 7.375316825077e-07, 4.7143e+01, 9.2089e-01,
2DIAGNOSTIC, 44, -4.581089129429e-01, 7.229126020927e-07, 4.7906e+01, 7.6270e-01,
2DIAGNOSTIC, 45, -4.581089736488e-01, 7.090046949508e-07, 4.8707e+01, 8.0153e-01,
2DIAGNOSTIC, 46, -4.581090667274e-01, 6.975034535957e-07, 4.9597e+01, 8.8938e-01,
2DIAGNOSTIC, 47, -4.581091530624e-01, 6.867719001755e-07, 5.0455e+01, 8.5822e-01,
2DIAGNOSTIC, 48, -4.581091881347e-01, 6.755902119161e-07, 5.1211e+01, 7.5623e-01,
2DIAGNOSTIC, 49, -4.581091695985e-01, 6.623171814116e-07, 5.2091e+01, 8.8008e-01,
2DIAGNOSTIC, 50, -4.581092604611e-01, 6.498300094138e-07, 5.2947e+01, 8.5617e-01,
2DIAGNOSTIC, 51, -4.581091309985e-01, 6.348142804723e-07, 5.3700e+01, 7.5254e-01,
2DIAGNOSTIC, 52, -4.581090619524e-01, 6.193294131950e-07, 5.4589e+01, 8.8878e-01,
2DIAGNOSTIC, 53, -4.581093726155e-01, 6.081918261826e-07, 5.5342e+01, 7.5333e-01,
2DIAGNOSTIC, 54, -4.581081906988e-01, 5.828826813298e-07, 5.6124e+01, 7.8206e-01,
2DIAGNOSTIC, 55, -4.581084230176e-01, 5.628446773515e-07, 5.6992e+01, 8.6771e-01,
2DIAGNOSTIC, 56, -4.581088728443e-01, 5.509124610809e-07, 5.7778e+01, 7.8616e-01,
2DIAGNOSTIC, 57, -4.581090783868e-01, 5.439675498931e-07, 5.8507e+01, 7.2860e-01,
2DIAGNOSTIC, 58, -4.581102606643e-01, 5.517287561194e-07, 5.9333e+01, 8.2646e-01,
2DIAGNOSTIC, 59, -4.581098004247e-01, 5.538029330156e-07, 6.0071e+01, 7.3818e-01,
2DIAGNOSTIC, 60, -4.581102018096e-01, 5.599307363081e-07, 6.0919e+01, 8.4823e-01,
2DIAGNOSTIC, 61, -4.581098938697e-01, 5.583797963275e-07, 6.1645e+01, 7.2589e-01,
2DIAGNOSTIC, 62, -4.581094775310e-01, 5.482834270695e-07, 6.2505e+01, 8.5997e-01,
2DIAGNOSTIC, 63, -4.581099524938e-01, 5.432976657692e-07, 6.3229e+01, 7.2358e-01,
2DIAGNOSTIC, 64, -4.581108146268e-01, 5.315217349415e-07, 6.3978e+01, 7.4871e-01,
2DIAGNOSTIC, 65, -4.581131987729e-01, 5.416240811303e-07, 6.4951e+01, 9.7365e-01,
2DIAGNOSTIC, 66, -4.581127366389e-01, 5.458993811505e-07, 6.5706e+01, 7.5493e-01,
2DIAGNOSTIC, 67, -4.581138355036e-01, 5.562876634300e-07, 6.6602e+01, 8.9616e-01,
2DIAGNOSTIC, 68, -4.581153786469e-01, 5.834855755348e-07, 6.7501e+01, 8.9887e-01,
2DIAGNOSTIC, 69, -4.581153344096e-01, 5.934685166249e-07, 6.8407e+01, 9.0630e-01,
2DIAGNOSTIC, 70, -4.581170355447e-01, 6.091476977051e-07, 6.9261e+01, 8.5338e-01,
2DIAGNOSTIC, 71, -4.581197469994e-01, 6.305381149256e-07, 7.0121e+01, 8.5988e-01,
2DIAGNOSTIC, 72, -4.581192299490e-01, 6.253301255994e-07, 7.1045e+01, 9.2402e-01,
2DIAGNOSTIC, 73, -4.581193373371e-01, 6.079006328128e-07, 7.1964e+01, 9.1922e-01,
2DIAGNOSTIC, 74, -4.581237690857e-01, 6.185056045886e-07, 7.2728e+01, 7.6413e-01,
2DIAGNOSTIC, 75, -4.581289962498e-01, 6.726585455811e-07, 7.3470e+01, 7.4204e-01,
2DIAGNOSTIC, 76, -4.581403554416e-01, 7.891507849151e-07, 7.4240e+01, 7.6938e-01,
2DIAGNOSTIC, 77, -4.581552884344e-01, 9.944440992494e-07, 7.5014e+01, 7.7416e-01,
2DIAGNOSTIC, 78, -4.582171900555e-01, 1.642093364439e-06, 7.5783e+01, 7.6933e-01,
2DIAGNOSTIC, 79, -4.583540728479e-01, 3.217488072801e-06, 7.7306e+01, 1.5232e+00,
2DIAGNOSTIC, 80, -4.583555183646e-01, 4.481533818698e-06, 7.8196e+01, 8.8937e-01,
2DIAGNOSTIC, 81, -4.583571030637e-01, 5.371447017672e-06, 7.9082e+01, 8.8649e-01,
2DIAGNOSTIC, 82, -4.583579873664e-01, 5.775113150846e-06, 7.9972e+01, 8.8948e-01,
2DIAGNOSTIC, 83, -4.583617527142e-01, 5.666407015693e-06, 8.0742e+01, 7.7061e-01,
2DIAGNOSTIC, 84, -4.583683405698e-01, 5.113515294667e-06, 8.1446e+01, 7.0334e-01,
2DIAGNOSTIC, 85, -4.583817820313e-01, 4.253732693580e-06, 8.2228e+01, 7.8255e-01,
2DIAGNOSTIC, 86, -4.584106418264e-01, 3.345650175592e-06, 8.2997e+01, 7.6935e-01,
2DIAGNOSTIC, 87, -4.584945403565e-01, 2.870990937594e-06, 8.3803e+01, 8.0577e-01,
2DIAGNOSTIC, 88, -4.586332852785e-01, 3.539237075450e-06, 8.5297e+01, 1.4940e+00,
2DIAGNOSTIC, 89, -4.586335441757e-01, 4.823026878651e-06, 8.6512e+01, 1.2150e+00,
2DIAGNOSTIC, 90, -4.586341586745e-01, 5.679162264096e-06, 8.7364e+01, 8.5142e-01,
2DIAGNOSTIC, 91, -4.586338392259e-01, 6.020308437830e-06, 8.8213e+01, 8.4993e-01,
2DIAGNOSTIC, 92, -4.586330267344e-01, 5.793541105057e-06, 8.9065e+01, 8.5135e-01,
2DIAGNOSTIC, 93, -4.586335876298e-01, 5.058879835292e-06, 8.9914e+01, 8.4927e-01,
2DIAGNOSTIC, 94, -4.586334117849e-01, 3.930897319843e-06, 9.0655e+01, 7.4047e-01,
2DIAGNOSTIC, 95, -4.586302449078e-01, 2.562095125964e-06, 9.1521e+01, 8.6651e-01,
2DIAGNOSTIC, 96, -4.586292782469e-01, 1.182609673380e-06, 9.2568e+01, 1.0465e+00,
2DIAGNOSTIC, 97, -4.586296705683e-01, 2.632010438486e-07, 9.3457e+01, 8.8968e-01,
2DIAGNOSTIC, 98, -4.586295214467e-01, 2.460893804603e-07, 9.4180e+01, 7.2265e-01,
2DIAGNOSTIC, 99, -4.586290754818e-01, 2.356200834424e-07, 9.5055e+01, 8.7547e-01,
2DIAGNOSTIC, 100, -4.586284618825e-01, 2.342297151688e-07, 9.5980e+01, 9.2422e-01,
Elapsed time (stage 0): 9.7386e+01
Stage 1
iterations = 1000x500x250x100
convergence threshold = 1.0000e-08
convergence window size = 10
number of levels = 4
using the Mattes MI metric (number of bins = 32, weight = 1.0000e+00)
preprocessing: winsorizing the image intensities
preprocessing: histogram matching the images
Shrink factors (level 1 out of 4): [8, 8, 8]
Shrink factors (level 2 out of 4): [4, 4, 4]
Shrink factors (level 3 out of 4): [2, 2, 2]
Shrink factors (level 4 out of 4): [1, 1, 1]
smoothing sigmas per level: [4, 2, 1, 0]
regular sampling (percentage = 2.5000e-01)
*** Running AffineTransform registration ***
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -7.663299704249e-01, 1.797693134862e+308, 1.4018e+00, 1.4018e+00,
2DIAGNOSTIC, 2, -7.668271207238e-01, 1.797693134862e+308, 1.4061e+00, 4.2200e-03,
2DIAGNOSTIC, 3, -7.675272344377e-01, 1.797693134862e+308, 1.4102e+00, 4.1089e-03,
2DIAGNOSTIC, 4, -7.684427758400e-01, 1.797693134862e+308, 1.4149e+00, 4.7371e-03,
2DIAGNOSTIC, 5, -7.682876731730e-01, 1.797693134862e+308, 1.4195e+00, 4.6170e-03,
2DIAGNOSTIC, 6, -7.682926313184e-01, 1.797693134862e+308, 1.4235e+00, 3.9630e-03,
2DIAGNOSTIC, 7, -7.684786105269e-01, 1.797693134862e+308, 1.4277e+00, 4.2639e-03,
2DIAGNOSTIC, 8, -7.689981167963e-01, 1.797693134862e+308, 1.4324e+00, 4.6530e-03,
2DIAGNOSTIC, 9, -7.687463718841e-01, 1.797693134862e+308, 1.4381e+00, 5.7030e-03,
2DIAGNOSTIC, 10, -7.687222075653e-01, 1.926136796496e-04, 1.4439e+00, 5.7812e-03,
2DIAGNOSTIC, 11, -7.689013808683e-01, 1.271314056380e-04, 1.4497e+00, 5.8100e-03,
2DIAGNOSTIC, 12, -7.686460483609e-01, 6.808392262279e-05, 1.4561e+00, 6.4259e-03,
2DIAGNOSTIC, 13, -7.688319394900e-01, 3.664917030083e-05, 1.4634e+00, 7.2560e-03,
2DIAGNOSTIC, 14, -7.688091134873e-01, 3.030882787090e-05, 1.4691e+00, 5.7180e-03,
2DIAGNOSTIC, 15, -7.686345217063e-01, 1.437105278386e-05, 1.4754e+00, 6.3219e-03,
2DIAGNOSTIC, 16, -7.686687738123e-01, 2.697459019069e-07, 1.4804e+00, 5.0039e-03,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -5.984477576755e-01, 1.797693134862e+308, 2.7513e+00, 1.2708e+00,
2DIAGNOSTIC, 2, -5.987948328806e-01, 1.797693134862e+308, 2.7801e+00, 2.8876e-02,
2DIAGNOSTIC, 3, -5.992165375894e-01, 1.797693134862e+308, 2.8071e+00, 2.7019e-02,
2DIAGNOSTIC, 4, -5.996469328830e-01, 1.797693134862e+308, 2.8348e+00, 2.7618e-02,
2DIAGNOSTIC, 5, -6.004586643947e-01, 1.797693134862e+308, 2.8629e+00, 2.8108e-02,
2DIAGNOSTIC, 6, -6.025779673625e-01, 1.797693134862e+308, 2.8954e+00, 3.2557e-02,
2DIAGNOSTIC, 7, -6.030436354456e-01, 1.797693134862e+308, 2.9386e+00, 4.3148e-02,
2DIAGNOSTIC, 8, -6.033435386129e-01, 1.797693134862e+308, 2.9646e+00, 2.6059e-02,
2DIAGNOSTIC, 9, -6.048329212415e-01, 1.797693134862e+308, 2.9889e+00, 2.4278e-02,
2DIAGNOSTIC, 10, -6.072012030345e-01, 8.940196774672e-04, 3.0129e+00, 2.3936e-02,
2DIAGNOSTIC, 11, -6.102566727849e-01, 1.010687389260e-03, 3.0434e+00, 3.0524e-02,
2DIAGNOSTIC, 12, -6.118892276803e-01, 1.093126008877e-03, 3.0720e+00, 2.8653e-02,
2DIAGNOSTIC, 13, -6.136007053070e-01, 1.143078588685e-03, 3.1020e+00, 2.9934e-02,
2DIAGNOSTIC, 14, -6.168216759362e-01, 1.204251122920e-03, 3.1350e+00, 3.3004e-02,
2DIAGNOSTIC, 15, -6.187575013239e-01, 1.228440431501e-03, 3.1708e+00, 3.5878e-02,
2DIAGNOSTIC, 16, -6.187446765278e-01, 1.197723654199e-03, 3.1954e+00, 2.4596e-02,
2DIAGNOSTIC, 17, -6.191564611795e-01, 1.087010183137e-03, 3.2280e+00, 3.2570e-02,
2DIAGNOSTIC, 18, -6.192072564539e-01, 8.979992272779e-04, 3.2533e+00, 2.5289e-02,
2DIAGNOSTIC, 19, -6.192186811323e-01, 6.814396413875e-04, 3.2840e+00, 3.0664e-02,
2DIAGNOSTIC, 20, -6.191832395227e-01, 4.763848630064e-04, 3.3076e+00, 2.3619e-02,
2DIAGNOSTIC, 21, -6.191820463021e-01, 3.152049722327e-04, 3.3312e+00, 2.3667e-02,
2DIAGNOSTIC, 22, -6.191651256055e-01, 1.741356960888e-04, 3.3563e+00, 2.5006e-02,
2DIAGNOSTIC, 23, -6.191521547098e-01, 6.228702200889e-05, 3.3812e+00, 2.4927e-02,
2DIAGNOSTIC, 24, -6.191475552868e-01, 1.558103523664e-05, 3.4026e+00, 2.1413e-02,
2DIAGNOSTIC, 25, -6.191460427717e-01, 7.421340157875e-06, 3.4239e+00, 2.1338e-02,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -5.384348832009e-01, 1.797693134862e+308, 4.5035e+00, 1.0795e+00,
2DIAGNOSTIC, 2, -5.385250196881e-01, 1.797693134862e+308, 4.7082e+00, 2.0470e-01,
2DIAGNOSTIC, 3, -5.385463983108e-01, 1.797693134862e+308, 4.9172e+00, 2.0907e-01,
2DIAGNOSTIC, 4, -5.385488780616e-01, 1.797693134862e+308, 5.0800e+00, 1.6275e-01,
2DIAGNOSTIC, 5, -5.385450665576e-01, 1.797693134862e+308, 5.2786e+00, 1.9865e-01,
2DIAGNOSTIC, 6, -5.385503757993e-01, 1.797693134862e+308, 5.4399e+00, 1.6122e-01,
2DIAGNOSTIC, 7, -5.385633688005e-01, 1.797693134862e+308, 5.6000e+00, 1.6019e-01,
2DIAGNOSTIC, 8, -5.385924525570e-01, 1.797693134862e+308, 5.7756e+00, 1.7555e-01,
2DIAGNOSTIC, 9, -5.386559883748e-01, 1.797693134862e+308, 5.9393e+00, 1.6369e-01,
2DIAGNOSTIC, 10, -5.387764297080e-01, 2.935154679322e-05, 6.1130e+00, 1.7367e-01,
2DIAGNOSTIC, 11, -5.389449732466e-01, 3.744819680784e-05, 6.2847e+00, 1.7178e-01,
2DIAGNOSTIC, 12, -5.392957423741e-01, 6.033310367885e-05, 6.4904e+00, 2.0564e-01,
2DIAGNOSTIC, 13, -5.395134559517e-01, 8.390798897473e-05, 6.8398e+00, 3.4945e-01,
2DIAGNOSTIC, 14, -5.396266554473e-01, 1.010504483858e-04, 7.1707e+00, 3.3091e-01,
2DIAGNOSTIC, 15, -5.396172746330e-01, 1.065175820998e-04, 7.3716e+00, 2.0089e-01,
2DIAGNOSTIC, 16, -5.396170721013e-01, 1.019484539601e-04, 7.5724e+00, 2.0074e-01,
2DIAGNOSTIC, 17, -5.396163922343e-01, 8.898211927776e-05, 7.7879e+00, 2.1550e-01,
2DIAGNOSTIC, 18, -5.396137973109e-01, 7.009461299233e-05, 8.0164e+00, 2.2850e-01,
2DIAGNOSTIC, 19, -5.396126556048e-01, 4.864426620033e-05, 8.2256e+00, 2.0920e-01,
2DIAGNOSTIC, 20, -5.396121109466e-01, 2.820817445372e-05, 8.4105e+00, 1.8491e-01,
2DIAGNOSTIC, 21, -5.396115700907e-01, 1.131858158511e-05, 8.5713e+00, 1.6085e-01,
2DIAGNOSTIC, 22, -5.396116364028e-01, 3.337724552237e-06, 8.7692e+00, 1.9789e-01,
2DIAGNOSTIC, 23, -5.396116962539e-01, 7.898639296997e-07, 8.9492e+00, 1.7995e-01,
2DIAGNOSTIC, 24, -5.396115229136e-01, 9.989327680385e-07, 9.1321e+00, 1.8294e-01,
2DIAGNOSTIC, 25, -5.396110176546e-01, 1.013101277723e-06, 9.3375e+00, 2.0541e-01,
2DIAGNOSTIC, 26, -5.396110731122e-01, 1.047560471527e-06, 9.5239e+00, 1.8636e-01,
2DIAGNOSTIC, 27, -5.396104896252e-01, 1.072282722600e-06, 9.6982e+00, 1.7438e-01,
2DIAGNOSTIC, 28, -5.396092907415e-01, 1.034380217091e-06, 9.8620e+00, 1.6378e-01,
2DIAGNOSTIC, 29, -5.396086702018e-01, 9.775298770581e-07, 1.0030e+01, 1.6847e-01,
2DIAGNOSTIC, 30, -5.396087150693e-01, 9.274051934113e-07, 1.0231e+01, 2.0100e-01,
2DIAGNOSTIC, 31, -5.396040913648e-01, 8.048709510128e-07, 1.0411e+01, 1.7989e-01,
2DIAGNOSTIC, 32, -5.396041730758e-01, 7.155237033221e-07, 1.0597e+01, 1.8543e-01,
2DIAGNOSTIC, 33, -5.395999569444e-01, 5.911764485236e-07, 1.0858e+01, 2.6132e-01,
2DIAGNOSTIC, 34, -5.396000691833e-01, 5.096799690144e-07, 1.1031e+01, 1.7287e-01,
2DIAGNOSTIC, 35, -5.395996519988e-01, 4.585456640470e-07, 1.1214e+01, 1.8275e-01,
2DIAGNOSTIC, 36, -5.396030114903e-01, 5.033250347654e-07, 1.1409e+01, 1.9531e-01,
2DIAGNOSTIC, 37, -5.396025494442e-01, 5.664650359304e-07, 1.1597e+01, 1.8751e-01,
2DIAGNOSTIC, 38, -5.396020846378e-01, 6.320183503460e-07, 1.1789e+01, 1.9221e-01,
2DIAGNOSTIC, 39, -5.396009703661e-01, 6.906580571342e-07, 1.1955e+01, 1.6651e-01,
2DIAGNOSTIC, 40, -5.396004898572e-01, 7.536069083491e-07, 1.2170e+01, 2.1477e-01,
2DIAGNOSTIC, 41, -5.396005510795e-01, 7.574162709557e-07, 1.2348e+01, 1.7758e-01,
2DIAGNOSTIC, 42, -5.395993649863e-01, 7.500734914428e-07, 1.2509e+01, 1.6166e-01,
2DIAGNOSTIC, 43, -5.395988294297e-01, 6.866429119665e-07, 1.2707e+01, 1.9738e-01,
2DIAGNOSTIC, 44, -5.395989372012e-01, 6.346735973821e-07, 1.2892e+01, 1.8582e-01,
2DIAGNOSTIC, 45, -5.395977810990e-01, 5.735888331283e-07, 1.3079e+01, 1.8637e-01,
2DIAGNOSTIC, 46, -5.395846579412e-01, 4.163414429291e-07, 1.3272e+01, 1.9318e-01,
2DIAGNOSTIC, 47, -5.395840700686e-01, 2.846159978739e-07, 1.3427e+01, 1.5474e-01,
2DIAGNOSTIC, 48, -5.395834629653e-01, 1.828887088162e-07, 1.3604e+01, 1.7772e-01,
2DIAGNOSTIC, 49, -5.395847112425e-01, 1.305640752389e-07, 1.3770e+01, 1.6584e-01,
DIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
2DIAGNOSTIC, 1, -4.712809184955e-01, 1.797693134862e+308, 1.7499e+01, 3.7285e+00,
2DIAGNOSTIC, 2, -4.714535292766e-01, 1.797693134862e+308, 1.8543e+01, 1.0438e+00,
2DIAGNOSTIC, 3, -4.715541847974e-01, 1.797693134862e+308, 1.9470e+01, 9.2744e-01,
2DIAGNOSTIC, 4, -4.716316261965e-01, 1.797693134862e+308, 2.0350e+01, 8.8005e-01,
2DIAGNOSTIC, 5, -4.717309019140e-01, 1.797693134862e+308, 2.1389e+01, 1.0387e+00,
2DIAGNOSTIC, 6, -4.717705420960e-01, 1.797693134862e+308, 2.2777e+01, 1.3883e+00,
2DIAGNOSTIC, 7, -4.717758043260e-01, 1.797693134862e+308, 2.3657e+01, 8.8036e-01,
2DIAGNOSTIC, 8, -4.717969469237e-01, 1.797693134862e+308, 2.4519e+01, 8.6107e-01,
2DIAGNOSTIC, 9, -4.718106553715e-01, 1.797693134862e+308, 2.5578e+01, 1.0595e+00,
2DIAGNOSTIC, 10, -4.718217350084e-01, 6.928966714513e-05, 2.6430e+01, 8.5199e-01,
2DIAGNOSTIC, 11, -4.718520462095e-01, 4.614713762989e-05, 2.7312e+01, 8.8226e-01,
2DIAGNOSTIC, 12, -4.719260388716e-01, 3.530800662825e-05, 2.8467e+01, 1.1545e+00,
2DIAGNOSTIC, 13, -4.719378419907e-01, 2.829057096279e-05, 2.9408e+01, 9.4076e-01,
2DIAGNOSTIC, 14, -4.719604025796e-01, 2.416392492162e-05, 3.0997e+01, 1.5898e+00,
2DIAGNOSTIC, 15, -4.719596478160e-01, 2.253109955135e-05, 3.2213e+01, 1.2156e+00,
2DIAGNOSTIC, 16, -4.719596909608e-01, 2.072441188343e-05, 3.3179e+01, 9.6636e-01,
2DIAGNOSTIC, 17, -4.719601588132e-01, 1.756224565014e-05, 3.4235e+01, 1.0555e+00,
2DIAGNOSTIC, 18, -4.719592053693e-01, 1.396598251481e-05, 3.5462e+01, 1.2272e+00,
2DIAGNOSTIC, 19, -4.719590829447e-01, 1.007627868990e-05, 3.8437e+01, 2.9746e+00,
2DIAGNOSTIC, 20, -4.719590829411e-01, 6.132998020693e-06, 4.1317e+01, 2.8802e+00,
2DIAGNOSTIC, 21, -4.719590829411e-01, 2.965034572054e-06, 4.2364e+01, 1.0477e+00,
2DIAGNOSTIC, 22, -4.719590829411e-01, 1.973422965949e-06, 4.3395e+01, 1.0303e+00,
2DIAGNOSTIC, 23, -4.719590829411e-01, 1.311091641414e-06, 4.4530e+01, 1.1350e+00,
2DIAGNOSTIC, 24, -4.719590829411e-01, 1.277873539804e-06, 4.5549e+01, 1.0188e+00,
2DIAGNOSTIC, 25, -4.719590829411e-01, 1.233369128530e-06, 4.6606e+01, 1.0571e+00,
2DIAGNOSTIC, 26, -4.719590829411e-01, 1.196367388157e-06, 4.7616e+01, 1.0107e+00,
2DIAGNOSTIC, 27, -4.719590829411e-01, 1.175702662622e-06, 4.8662e+01, 1.0455e+00,
2DIAGNOSTIC, 28, -4.719590829411e-01, 1.136337023380e-06, 5.0071e+01, 1.4089e+00,
2DIAGNOSTIC, 29, -4.719590829411e-01, 1.097144124544e-06, 5.1087e+01, 1.0164e+00,
2DIAGNOSTIC, 30, -4.719590829411e-01, 1.060564589005e-06, 5.2114e+01, 1.0272e+00,
2DIAGNOSTIC, 31, -4.719590829411e-01, 1.026345527101e-06, 5.3112e+01, 9.9741e-01,
2DIAGNOSTIC, 32, -4.719590829411e-01, 9.942655987277e-07, 5.4135e+01, 1.0234e+00,
2DIAGNOSTIC, 33, -4.719590829411e-01, 9.641302981672e-07, 5.5203e+01, 1.0674e+00,
2DIAGNOSTIC, 34, -4.719590829411e-01, 9.357680072590e-07, 5.6349e+01, 1.1465e+00,
2DIAGNOSTIC, 35, -4.719590829411e-01, 9.090267250288e-07, 5.7855e+01, 1.5061e+00,
2DIAGNOSTIC, 36, -4.719590829411e-01, 8.837713425837e-07, 5.8882e+01, 1.0272e+00,
2DIAGNOSTIC, 37, -4.719590829411e-01, 8.598813600325e-07, 5.9880e+01, 9.9783e-01,
2DIAGNOSTIC, 38, -4.719590829411e-01, 8.372489638565e-07, 6.0997e+01, 1.1169e+00,
2DIAGNOSTIC, 39, -4.719590829411e-01, 8.157774002600e-07, 6.2093e+01, 1.0957e+00,
2DIAGNOSTIC, 40, -4.719590829411e-01, 7.953795923867e-07, 6.3092e+01, 9.9935e-01,
2DIAGNOSTIC, 41, -4.719590829411e-01, 7.759769600768e-07, 6.4397e+01, 1.3049e+00,
2DIAGNOSTIC, 42, -4.719590829411e-01, 7.574984081521e-07, 6.5624e+01, 1.2271e+00,
2DIAGNOSTIC, 43, -4.719590829411e-01, 7.398794559903e-07, 6.6780e+01, 1.1560e+00,
2DIAGNOSTIC, 44, -4.719590829411e-01, 7.230614856961e-07, 6.7851e+01, 1.0712e+00,
2DIAGNOSTIC, 45, -4.719590829411e-01, 7.069910906328e-07, 6.9085e+01, 1.2338e+00,
2DIAGNOSTIC, 46, -4.719590829411e-01, 6.916195088788e-07, 7.0908e+01, 1.8225e+00,
2DIAGNOSTIC, 47, -4.719590829411e-01, 6.769021290431e-07, 7.2656e+01, 1.7483e+00,
2DIAGNOSTIC, 48, -4.719590829411e-01, 6.627980578856e-07, 7.3748e+01, 1.0923e+00,
2DIAGNOSTIC, 49, -4.719590829411e-01, 6.492697408218e-07, 7.4763e+01, 1.0145e+00,
2DIAGNOSTIC, 50, -4.719590829411e-01, 6.362826280304e-07, 7.5797e+01, 1.0339e+00,
2DIAGNOSTIC, 51, -4.719590829411e-01, 6.238048798381e-07, 7.6787e+01, 9.9039e-01,
2DIAGNOSTIC, 52, -4.719590829411e-01, 6.118071060809e-07, 7.7786e+01, 9.9876e-01,
2DIAGNOSTIC, 53, -4.719590829411e-01, 6.002621350185e-07, 7.8797e+01, 1.0116e+00,
2DIAGNOSTIC, 54, -4.719590829411e-01, 5.891448078624e-07, 7.9787e+01, 9.8992e-01,
2DIAGNOSTIC, 55, -4.719590829411e-01, 5.784317957530e-07, 8.0744e+01, 9.5626e-01,
2DIAGNOSTIC, 56, -4.719590829411e-01, 5.681014364212e-07, 8.1755e+01, 1.0118e+00,
2DIAGNOSTIC, 57, -4.719590829411e-01, 5.581335879128e-07, 8.2762e+01, 1.0071e+00,
2DIAGNOSTIC, 58, -4.719590829411e-01, 5.485094975131e-07, 8.3763e+01, 1.0005e+00,
2DIAGNOSTIC, 59, -4.719590829411e-01, 5.392116840391e-07, 8.4785e+01, 1.0217e+00,
2DIAGNOSTIC, 60, -4.719590829411e-01, 5.302238318509e-07, 8.5775e+01, 9.9054e-01,
2DIAGNOSTIC, 61, -4.719590829411e-01, 5.215306953001e-07, 8.6729e+01, 9.5418e-01,
2DIAGNOSTIC, 62, -4.719590829411e-01, 5.131180124268e-07, 8.7718e+01, 9.8874e-01,
2DIAGNOSTIC, 63, -4.719590829411e-01, 5.049724268909e-07, 8.8713e+01, 9.9447e-01,
2DIAGNOSTIC, 64, -4.719590829411e-01, 4.970814172084e-07, 8.9677e+01, 9.6487e-01,
2DIAGNOSTIC, 65, -4.719590829411e-01, 4.894332325429e-07, 9.0641e+01, 9.6385e-01,
2DIAGNOSTIC, 66, -4.719590829411e-01, 4.820168342697e-07, 9.1658e+01, 1.0165e+00,
2DIAGNOSTIC, 67, -4.719590829411e-01, 4.748218428823e-07, 9.2682e+01, 1.0244e+00,
2DIAGNOSTIC, 68, -4.719590829411e-01, 4.678384894746e-07, 9.3803e+01, 1.1207e+00,
2DIAGNOSTIC, 69, -4.719590829411e-01, 4.610575715016e-07, 9.4827e+01, 1.0243e+00,
2DIAGNOSTIC, 70, -4.719590829411e-01, 4.544704123605e-07, 9.5855e+01, 1.0280e+00,
2DIAGNOSTIC, 71, -4.719590829411e-01, 4.480688242815e-07, 9.6905e+01, 1.0500e+00,
2DIAGNOSTIC, 72, -4.719590829411e-01, 4.418450744361e-07, 9.7906e+01, 1.0013e+00,
2DIAGNOSTIC, 73, -4.719590829411e-01, 4.357918537267e-07, 9.8900e+01, 9.9379e-01,
2DIAGNOSTIC, 74, -4.719590829411e-01, 4.299022482099e-07, 9.9905e+01, 1.0046e+00,
2DIAGNOSTIC, 75, -4.719590829411e-01, 4.241697126837e-07, 1.0095e+02, 1.0444e+00,
2DIAGNOSTIC, 76, -4.719590829411e-01, 4.185880464937e-07, 1.0203e+02, 1.0797e+00,
2DIAGNOSTIC, 77, -4.719590829411e-01, 4.131513710939e-07, 1.0304e+02, 1.0068e+00,
2DIAGNOSTIC, 78, -4.719590829411e-01, 4.078541094542e-07, 1.0401e+02, 9.7360e-01,
2DIAGNOSTIC, 79, -4.719590829411e-01, 4.026909669313e-07, 1.0496e+02, 9.5444e-01,
2DIAGNOSTIC, 80, -4.719590829411e-01, 3.976569136425e-07, 1.0592e+02, 9.5437e-01,
2DIAGNOSTIC, 81, -4.719590829411e-01, 3.927471681157e-07, 1.0696e+02, 1.0389e+00,
2DIAGNOSTIC, 82, -4.719590829411e-01, 3.879571821436e-07, 1.0791e+02, 9.5736e-01,
2DIAGNOSTIC, 83, -4.719590829411e-01, 3.832826267133e-07, 1.0886e+02, 9.4829e-01,
2DIAGNOSTIC, 84, -4.719590829411e-01, 3.787193789909e-07, 1.0982e+02, 9.5969e-01,
2DIAGNOSTIC, 85, -4.719590829411e-01, 3.742635101573e-07, 1.1078e+02, 9.6001e-01,
2DIAGNOSTIC, 86, -4.719590829411e-01, 3.699112741552e-07, 1.1174e+02, 9.5766e-01,
2DIAGNOSTIC, 87, -4.719590829411e-01, 3.656590971540e-07, 1.1274e+02, 9.9589e-01,
2DIAGNOSTIC, 88, -4.719590829411e-01, 3.615035678098e-07, 1.1375e+02, 1.0172e+00,
2DIAGNOSTIC, 89, -4.719590829411e-01, 3.574414280845e-07, 1.1477e+02, 1.0126e+00,
2DIAGNOSTIC, 90, -4.719590829411e-01, 3.534695647654e-07, 1.1575e+02, 9.7952e-01,
2DIAGNOSTIC, 91, -4.719590829411e-01, 3.495850014936e-07, 1.1672e+02, 9.7264e-01,
2DIAGNOSTIC, 92, -4.719590829411e-01, 3.457848913140e-07, 1.1770e+02, 9.8520e-01,
2DIAGNOSTIC, 93, -4.719590829411e-01, 3.420665097564e-07, 1.1865e+02, 9.4925e-01,
2DIAGNOSTIC, 94, -4.719590829411e-01, 3.384272482584e-07, 1.1961e+02, 9.5593e-01,
2DIAGNOSTIC, 95, -4.719590829411e-01, 3.348646081328e-07, 1.2060e+02, 9.8836e-01,
2DIAGNOSTIC, 96, -4.719590829411e-01, 3.313761947864e-07, 1.2164e+02, 1.0433e+00,
2DIAGNOSTIC, 97, -4.719590829411e-01, 3.279597123977e-07, 1.2265e+02, 1.0097e+00,
2DIAGNOSTIC, 98, -4.719590829411e-01, 3.246129588411e-07, 1.2362e+02, 9.7477e-01,
2DIAGNOSTIC, 99, -4.719590829411e-01, 3.213338209893e-07, 1.2460e+02, 9.7495e-01,
2DIAGNOSTIC, 100, -4.719590829411e-01, 3.181202702270e-07, 1.2555e+02, 9.5462e-01,
Elapsed time (stage 1): 1.2694e+02
Stage 2
iterations = 50x10x0
convergence threshold = 1.0000e-09
convergence window size = 15
number of levels = 3
using the CC metric (radius = 4, weight = 5.0000e-01)
preprocessing: winsorizing the image intensities
preprocessing: histogram matching the images
using the CC metric (radius = 4, weight = 5.0000e-01)
preprocessing: winsorizing the image intensities
preprocessing: histogram matching the images
Shrink factors (level 1 out of 3): [4, 4, 4]
Shrink factors (level 2 out of 3): [2, 2, 2]
Shrink factors (level 3 out of 3): [1, 1, 1]
smoothing sigmas per level: [2, 1, 0]
Using default NONE metricSamplingStrategy
*** Running SyN registration (varianceForUpdateField = 3.0000e+00, varianceForTotalField = 0.0000e+00) ***
XXDIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
1DIAGNOSTIC, 1, -4.237949235954e-01, 1.797693134862e+308, 5.0319e+00, 5.0319e+00,
1DIAGNOSTIC, 2, -4.282599218742e-01, 1.797693134862e+308, 6.9136e+00, 1.8817e+00,
1DIAGNOSTIC, 3, -4.328208074055e-01, 1.797693134862e+308, 8.4367e+00, 1.5231e+00,
1DIAGNOSTIC, 4, -4.374425534367e-01, 1.797693134862e+308, 1.4326e+01, 5.8890e+00,
1DIAGNOSTIC, 5, -4.415208329598e-01, 1.797693134862e+308, 1.5812e+01, 1.4858e+00,
1DIAGNOSTIC, 6, -4.459613784006e-01, 1.797693134862e+308, 1.7004e+01, 1.1921e+00,
1DIAGNOSTIC, 7, -4.492045680540e-01, 1.797693134862e+308, 2.0669e+01, 3.6656e+00,
1DIAGNOSTIC, 8, -4.520287728292e-01, 1.797693134862e+308, 2.1949e+01, 1.2794e+00,
1DIAGNOSTIC, 9, -4.539549239509e-01, 1.797693134862e+308, 2.3224e+01, 1.2749e+00,
1DIAGNOSTIC, 10, -4.562956181430e-01, 1.797693134862e+308, 2.4418e+01, 1.1941e+00,
1DIAGNOSTIC, 11, -4.582374762772e-01, 1.797693134862e+308, 2.6020e+01, 1.6023e+00,
1DIAGNOSTIC, 12, -4.601852880586e-01, 1.797693134862e+308, 2.7220e+01, 1.2001e+00,
1DIAGNOSTIC, 13, -4.619250729265e-01, 1.797693134862e+308, 2.8588e+01, 1.3679e+00,
1DIAGNOSTIC, 14, -4.638908765775e-01, 1.797693134862e+308, 2.9865e+01, 1.2773e+00,
1DIAGNOSTIC, 15, -4.651489886577e-01, 3.717704021784e-03, 3.1079e+01, 1.2133e+00,
1DIAGNOSTIC, 16, -4.664988832599e-01, 3.151787152635e-03, 3.2308e+01, 1.2292e+00,
1DIAGNOSTIC, 17, -4.678077552041e-01, 2.667069996373e-03, 3.3596e+01, 1.2884e+00,
1DIAGNOSTIC, 18, -4.691149562469e-01, 2.263717307992e-03, 3.5574e+01, 1.9774e+00,
1DIAGNOSTIC, 19, -4.702502132739e-01, 1.936303913130e-03, 3.7154e+01, 1.5801e+00,
1DIAGNOSTIC, 20, -4.712752545299e-01, 1.667509466475e-03, 3.9136e+01, 1.9819e+00,
1DIAGNOSTIC, 21, -4.722927197849e-01, 1.463825440369e-03, 4.0337e+01, 1.2019e+00,
1DIAGNOSTIC, 22, -4.730405026743e-01, 1.293373445145e-03, 4.1996e+01, 1.6587e+00,
1DIAGNOSTIC, 23, -4.740938601155e-01, 1.154419073875e-03, 4.4065e+01, 2.0685e+00,
1DIAGNOSTIC, 24, -4.750684402206e-01, 1.028721666762e-03, 4.7046e+01, 2.9811e+00,
1DIAGNOSTIC, 25, -4.759524596217e-01, 9.234250540542e-04, 4.9118e+01, 2.0724e+00,
1DIAGNOSTIC, 26, -4.772390944700e-01, 8.387602907328e-04, 5.1295e+01, 2.1772e+00,
1DIAGNOSTIC, 27, -4.779657133318e-01, 7.656519489656e-04, 5.3241e+01, 1.9454e+00,
1DIAGNOSTIC, 28, -4.788041920138e-01, 7.033339347179e-04, 5.4781e+01, 1.5407e+00,
1DIAGNOSTIC, 29, -4.795846456086e-01, 6.548831112698e-04, 5.6265e+01, 1.4841e+00,
1DIAGNOSTIC, 30, -4.803184738065e-01, 6.092592739870e-04, 5.7443e+01, 1.1779e+00,
1DIAGNOSTIC, 31, -4.810060076628e-01, 5.677259087847e-04, 5.8627e+01, 1.1838e+00,
1DIAGNOSTIC, 32, -4.818296277018e-01, 5.319893466413e-04, 6.0085e+01, 1.4579e+00,
1DIAGNOSTIC, 33, -4.823695607924e-01, 4.986396379492e-04, 6.1269e+01, 1.1844e+00,
1DIAGNOSTIC, 34, -4.829966229942e-01, 4.672823340962e-04, 6.2669e+01, 1.4000e+00,
1DIAGNOSTIC, 35, -4.836421198188e-01, 4.374011323817e-04, 6.3884e+01, 1.2144e+00,
1DIAGNOSTIC, 36, -4.843352157449e-01, 4.100798353019e-04, 6.5676e+01, 1.7925e+00,
1DIAGNOSTIC, 37, -4.845752135092e-01, 3.774947210786e-04, 6.6926e+01, 1.2494e+00,
1DIAGNOSTIC, 38, -4.848168494351e-01, 3.444872335594e-04, 6.8919e+01, 1.9937e+00,
1DIAGNOSTIC, 39, -4.851343028193e-01, 3.120385334514e-04, 7.0101e+01, 1.1814e+00,
1DIAGNOSTIC, 40, -4.854870377122e-01, 2.803922156895e-04, 7.2040e+01, 1.9390e+00,
1DIAGNOSTIC, 41, -4.857705860638e-01, 2.539661531251e-04, 7.3308e+01, 1.2681e+00,
1DIAGNOSTIC, 42, -4.860014098278e-01, 2.271320238424e-04, 7.4531e+01, 1.2229e+00,
1DIAGNOSTIC, 43, -4.862790655738e-01, 2.023645813410e-04, 7.5844e+01, 1.3127e+00,
1DIAGNOSTIC, 44, -4.865606284917e-01, 1.798600059114e-04, 7.7077e+01, 1.2329e+00,
1DIAGNOSTIC, 45, -4.867534031454e-01, 1.589955914072e-04, 7.8385e+01, 1.3080e+00,
1DIAGNOSTIC, 46, -4.869892290898e-01, 1.403543450454e-04, 7.9641e+01, 1.2570e+00,
1DIAGNOSTIC, 47, -4.872732755681e-01, 1.260865839886e-04, 8.1278e+01, 1.6364e+00,
1DIAGNOSTIC, 48, -4.875373315888e-01, 1.137042974742e-04, 8.2975e+01, 1.6975e+00,
1DIAGNOSTIC, 49, -4.878213885254e-01, 1.043729293915e-04, 8.4262e+01, 1.2869e+00,
1DIAGNOSTIC, 50, -4.878354371080e-01, 9.616367916878e-05, 8.5740e+01, 1.4775e+00,
XXDIAGNOSTIC,Iteration,metricValue,convergenceValue,ITERATION_TIME_INDEX,SINCE_LAST
1DIAGNOSTIC, 1, -4.954169991543e-01, 1.797693134862e+308, 1.0255e+02, 1.6809e+01,
1DIAGNOSTIC, 2, -4.974579296616e-01, 1.797693134862e+308, 1.2930e+02, 2.6748e+01,
1DIAGNOSTIC, 3, -4.995222683030e-01, 1.797693134862e+308, 1.4096e+02, 1.1663e+01,
1DIAGNOSTIC, 4, -5.016385544297e-01, 1.797693134862e+308, 1.7351e+02, 3.2549e+01,
1DIAGNOSTIC, 5, -5.034849535398e-01, 1.797693134862e+308, 1.8530e+02, 1.1791e+01,
1DIAGNOSTIC, 6, -5.051476916116e-01, 1.797693134862e+308, 1.9597e+02, 1.0670e+01,
1DIAGNOSTIC, 7, -5.066164232876e-01, 1.797693134862e+308, 2.0710e+02, 1.1136e+01,
1DIAGNOSTIC, 8, -5.079946217073e-01, 1.797693134862e+308, 2.1815e+02, 1.1049e+01,
1DIAGNOSTIC, 9, -5.091540690922e-01, 1.797693134862e+308, 2.2898e+02, 1.0825e+01,
1DIAGNOSTIC, 10, -5.104437255052e-01, 1.797693134862e+308, 2.4586e+02, 1.6881e+01,
Elapsed time (stage 2): 264
Total elapsed time: 489.1
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//antsApplyTransforms -d 3 -i OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz -o ANTS_Stripped_BrainExtractionPriorWarped.nii.gz -r ds000102/sub-08/anat/sub-08_T1w.nii.gz -n Gaussian -t [ANTS_Stripped_BrainExtractionPrior0GenericAffine.mat,1] -t ANTS_Stripped_BrainExtractionPrior1InverseWarp.nii.gz --float 0 --verbose 1
Using double precision for computations.
Input scalar image: OASIS/MICCAI2012-Multi-Atlas-Challenge-Data/T_template0_BrainCerebellumProbabilityMask.nii.gz
Reference image: ds000102/sub-08/anat/sub-08_T1w.nii.gz
=============================================================================
The composite transform comprises the following transforms (in order):
1. ANTS_Stripped_BrainExtractionPrior1InverseWarp.nii.gz (type = DisplacementFieldTransform)
2. inverse of ANTS_Stripped_BrainExtractionPrior0GenericAffine.mat (type = AffineTransform)
=============================================================================
Default pixel value: 0
Interpolation type: GaussianInterpolateImageFunction
Output warped image: ANTS_Stripped_BrainExtractionPriorWarped.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionPriorWarped.nii.gz ANTS_Stripped_BrainExtractionPriorWarped.nii.gz 0.5 1 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz MD ANTS_Stripped_BrainExtractionPriorWarped.nii.gz 2
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz GetLargestComponent ANTS_Stripped_BrainExtractionMask.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//Atropos -d 3 -o ANTS_Stripped_BrainExtractionSegmentation.nii.gz -a ANTS_Stripped_N4Corrected0.nii.gz -x ANTS_Stripped_BrainExtractionMask.nii.gz -i kmeans[3] -c [3,0.0] -m [0.1,1x1x1] -k Gaussian -r 1 --verbose 1
Running Atropos for 3-dimensional images.
Progress:
Iteration 0 (of 3): posterior probability = 0 (annealing temperature = 1)
Iteration 1 (of 3): posterior probability = 0.953862 (annealing temperature = 1)
Writing output:
AtroposSegmentationImageFilter (0x7ee83f0)
RTTI typeinfo: itk::ants::AtroposSegmentationImageFilter<itk::Image<float, 3u>, itk::Image<unsigned int, 3u>, itk::Image<unsigned int, 3u> >
Reference Count: 1
Modified Time: 9132831
Debug: Off
Object Name:
Observers:
IterationEvent(Command)
Inputs:
Primary: (0x7ef5aa0) *
_1: (0x7ef0b90)
Indexed Inputs:
0: Primary (0x7ef5aa0)
1: _1 (0x7ef0b90)
Required Input Names: Primary
NumberOfRequiredInputs: 1
Outputs:
Primary: (0x7eed070)
Indexed Outputs:
0: Primary (0x7eed070)
NumberOfRequiredOutputs: 1
Number Of Threads: 32
ReleaseDataFlag: Off
ReleaseDataBeforeUpdateFlag: Off
AbortGenerateData: Off
Progress: 0
Multithreader:
RTTI typeinfo: itk::PoolMultiThreader
Reference Count: 1
Modified Time: 207
Debug: Off
Object Name:
Observers:
none
DynamicMultiThreading: On
CoordinateTolerance: 1e-06
DirectionTolerance: 1e-06
Maximum number of iterations: 3
Convergence threshold: 0
Number of tissue classes: 3
Number of partial volume classes: 0
Minimize memory usage: false
Initialization strategy: K means clustering
Posterior probability formulation: Socrates
initial annealing temperature = 1
annealing rate = 1
minimum annealing temperature = 0.1
MRF parameters
MRF smoothing factor = 0.1
MRF radius = [1, 1, 1]
Use asynchronous updating of the labels.
ICM parameters
maximum ICM code = 13
maximum number of ICM iterations = 1
No outlier handling.
Tissue class 1: proportion = 0.21748
GaussianListSampleFunction (0x7ef33c0)
mean = [62.82458487650495], covariance = [898.368]
Tissue class 2: proportion = 0.418772
GaussianListSampleFunction (0x7ef0ed0)
mean = [138.78809473825126], covariance = [535.745]
Tissue class 3: proportion = 0.363905
GaussianListSampleFunction (0x7ef4af0)
mean = [220.97805075402698], covariance = [369.979]
Elapsed time: 44.9657
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz PadImage ANTS_Stripped_BrainExtractionSegmentation.nii.gz 10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionPriorWarped.nii.gz PadImage ANTS_Stripped_BrainExtractionPriorWarped.nii.gz 10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz ANTS_Stripped_BrainExtractionWM.nii.gz 3 3 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz ANTS_Stripped_BrainExtractionGM.nii.gz 2 2 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz ANTS_Stripped_BrainExtractionCSF.nii.gz 1 1 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionWM.nii.gz GetLargestComponent ANTS_Stripped_BrainExtractionWM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionGM.nii.gz GetLargestComponent ANTS_Stripped_BrainExtractionGM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionTmp.nii.gz FillHoles ANTS_Stripped_BrainExtractionGM.nii.gz 2
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//MultiplyImages 3 ANTS_Stripped_BrainExtractionGM.nii.gz ANTS_Stripped_BrainExtractionTmp.nii.gz ANTS_Stripped_BrainExtractionGM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//MultiplyImages 3 ANTS_Stripped_BrainExtractionWM.nii.gz 3 ANTS_Stripped_BrainExtractionWM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionTmp.nii.gz ME ANTS_Stripped_BrainExtractionCSF.nii.gz 10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionGM.nii.gz addtozero ANTS_Stripped_BrainExtractionGM.nii.gz ANTS_Stripped_BrainExtractionTmp.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//MultiplyImages 3 ANTS_Stripped_BrainExtractionGM.nii.gz 2 ANTS_Stripped_BrainExtractionGM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz addtozero ANTS_Stripped_BrainExtractionWM.nii.gz ANTS_Stripped_BrainExtractionGM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz ANTS_Stripped_BrainExtractionMask.nii.gz 3 3 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ThresholdImage 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz ANTS_Stripped_BrainExtractionTmp.nii.gz 2 2 1 0
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz addtozero ANTS_Stripped_BrainExtractionMask.nii.gz ANTS_Stripped_BrainExtractionTmp.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz ME ANTS_Stripped_BrainExtractionMask.nii.gz 2
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz GetLargestComponent ANTS_Stripped_BrainExtractionMask.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz MD ANTS_Stripped_BrainExtractionMask.nii.gz 4
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz FillHoles ANTS_Stripped_BrainExtractionMask.nii.gz 2
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz addtozero ANTS_Stripped_BrainExtractionMask.nii.gz ANTS_Stripped_BrainExtractionPriorWarped.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz MD ANTS_Stripped_BrainExtractionMask.nii.gz 5
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz ME ANTS_Stripped_BrainExtractionMask.nii.gz 5
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionSegmentation.nii.gz PadImage ANTS_Stripped_BrainExtractionSegmentation.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionMask.nii.gz PadImage ANTS_Stripped_BrainExtractionMask.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionWM.nii.gz PadImage ANTS_Stripped_BrainExtractionWM.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionGM.nii.gz PadImage ANTS_Stripped_BrainExtractionGM.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionCSF.nii.gz PadImage ANTS_Stripped_BrainExtractionCSF.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//ImageMath 3 ANTS_Stripped_BrainExtractionPriorWarped.nii.gz PadImage ANTS_Stripped_BrainExtractionPriorWarped.nii.gz -10
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
/opt/ants-2.3.1//MultiplyImages 3 ANTS_Stripped_N4Corrected0.nii.gz ANTS_Stripped_BrainExtractionMask.nii.gz ANTS_Stripped_BrainExtractionBrain.nii.gz
END <<<<<<<<<<<<<<<<<<<<
--------------------------------------------------------------------------------------
Done with brain extraction: 0h 13m 11s
--------------------------------------------------------------------------------------
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionPriorWarped.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionPrior1Warp.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionPrior1InverseWarp.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionTmp.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionGM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionCSF.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionSegmentation.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionInitialAffine.mat
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionInitialAffineMoving.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionInitialAffineFixed.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionLaplacian.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionTemplateLaplacian.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_BrainExtractionWM.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_N4Truncated0.nii.gz
END <<<<<<<<<<<<<<<<<<<<
BEGIN >>>>>>>>>>>>>>>>>>>>
rm ANTS_Stripped_N4Corrected0.nii.gz
END <<<<<<<<<<<<<<<<<<<<
The option -d 3 means that it is a three-dimensional image; -a indicates the anatomical image to be stripped; and -e is used to supply a an anatomical template (with skull) and -m to provide a brain probability mask for skull-stripping , and -o is the label (prefix) for the output.
2. AFNI#
Next, we will use AFNI’s 3dSkullStrip for brain extraction
! 3dSkullStrip -help
Usage: A program to extract the brain from surrounding.
tissue from MRI T1-weighted images.
The simplest command would be:
3dSkullStrip <-input DSET>
Also consider the script @SSwarper, which combines the use of
3dSkullStrip and nonlinear warping to an MNI template to produce
a skull-stripped dataset in MNI space, plus the nonlinear warp
that can used to transform other datasets from the same subject
(e.g., EPI) to MNI space. (This script only applies to human brain
images.)
The fully automated process consists of three steps:
1- Preprocessing of volume to remove gross spatial image
non-uniformity artifacts and reposition the brain in
a reasonable manner for convenience.
** Note that in many cases, using 3dUnifize before **
** using 3dSkullStrip will give better results. **
2- Expand a spherical surface iteratively until it envelopes
the brain. This is a modified version of the BET algorithm:
Fast robust automated brain extraction,
by Stephen M. Smith, HBM 2002 v 17:3 pp 143-155
Modifications include the use of:
. outer brain surface
. expansion driven by data inside and outside the surface
. avoidance of eyes and ventricles
. a set of operations to avoid the clipping of certain brain
areas and reduce leakage into the skull in heavily shaded
data
. two additional processing stages to ensure convergence and
reduction of clipped areas.
. use of 3d edge detection, see Deriche and Monga references
in 3dedge3 -help.
3- The creation of various masks and surfaces modeling brain
and portions of the skull
Common examples of usage:
-------------------------
o 3dSkullStrip -input VOL -prefix VOL_PREFIX
Vanilla mode, should work for most datasets.
o 3dSkullStrip -input VOL -prefix VOL_PREFIX -push_to_edge
Adds an aggressive push to brain edges. Use this option
when the chunks of gray matter are not included. This option
might cause the mask to leak into non-brain areas.
o 3dSkullStrip -input VOL -surface_coil -prefix VOL_PREFIX -monkey
Vanilla mode, for use with monkey data.
o 3dSkullStrip -input VOL -prefix VOL_PREFIX -ld 30
Use a denser mesh, in the cases where you have lots of
csf between gyri. Also helps when some of the brain is clipped
close to regions of high curvature.
Tips:
-----
I ran the program with the default parameters on 200+ datasets.
The results were quite good in all but a couple of instances, here
are some tips on fixing trouble spots:
Clipping in frontal areas, close to the eye balls:
+ Try -push_to_edge option first.
Can also try -no_avoid_eyes option.
Clipping in general:
+ Try -push_to_edge option first.
Can also use lower -shrink_fac, start with 0.5 then 0.4
Problems down below:
+ Piece of cerebellum missing, reduce -shrink_fac_bot_lim
from default value.
+ Leakage in lower areas, increase -shrink_fac_bot_lim
from default value.
Some lobules are not included:
+ Use a denser mesh. Start with -ld 30. If that still fails,
try even higher density (like -ld 50) and increase iterations
(say to -niter 750).
Expect the program to take much longer in that case.
+ Instead of using denser meshes, you could try blurring the data
before skull stripping. Something like -blur_fwhm 2 did
wonders for some of my data with the default options of 3dSkullStrip
Blurring is a lot faster than increasing mesh density.
+ Use also a smaller -shrink_fac is you have lots of CSF between
gyri.
Massive chunks missing:
+ If brain has very large ventricles and lots of CSF between gyri,
the ventricles will keep attracting the surface inwards.
This often happens with older brains. In such
cases, use the -visual option to see what is happening.
For example, the options below did the trick in various
instances.
-blur_fwhm 2 -use_skull
or for more stubborn cases increase csf avoidance with this cocktail
-blur_fwhm 2 -use_skull -avoid_vent -avoid_vent -init_radius 75
+ Too much neck in the volume might throw off the initialization
step. You can fix this by clipping tissue below the brain with
@clip_volume -below ZZZ -input INPUT
where ZZZ is a Z coordinate somewhere below the brain.
Large regions outside brain included:
+ Usually because noise level is high. Try @NoisySkullStrip.
Make sure that brain orientation is correct. This means the image in
AFNI's axial slice viewer should be close to the brain's axial plane.
The same goes for the other planes. Otherwise, the program might do a lousy
job removing the skull.
Eye Candy Mode:
---------------
You can run 3dSkullStrip and have it send successive iterations
to SUMA and AFNI. This is very helpful in following the
progression of the algorithm and determining the source
of trouble, if any.
Example:
afni -niml -yesplugouts &
suma -niml &
3dSkullStrip -input Anat+orig -o_ply anat_brain -visual
Help section for the intrepid:
------------------------------
3dSkullStrip < -input VOL >
[< -o_TYPE PREFIX >] [< -prefix VOL_PREFIX >]
[< -spatnorm >] [< -no_spatnorm >] [< -write_spatnorm >]
[< -niter N_ITER >] [< -ld LD >]
[< -shrink_fac SF >] [< -var_shrink_fac >]
[< -no_var_shrink_fac >] [< -shrink_fac_bot_lim SFBL >]
[< -pushout >] [< -no_pushout >] [< -exp_frac FRAC]
[< -touchup >] [< -no_touchup >]
[< -fill_hole R >] [< -NN_smooth NN_SM >]
[< -smooth_final SM >] [< -avoid_vent >] [< -no_avoid_vent >]
[< -use_skull >] [< -no_use_skull >]
[< -avoid_eyes >] [< -no_avoid_eyes >]
[< -use_edge >] [< -no_use_edge >]
[< -push_to_edge >] [<-no_push_to_edge>]
[< -perc_int PERC_INT >]
[< -max_inter_iter MII >] [-mask_vol | -orig_vol | -norm_vol]
[< -debug DBG >] [< -node_debug NODE_DBG >]
[< -demo_pause >]
[< -monkey >] [< -marmoset >] [<-rat>]
NOTE: Please report bugs and strange failures
to saadz@mail.nih.gov
Mandatory parameters:
-input VOL: Input AFNI (or AFNI readable) volume.
Optional Parameters:
-monkey: the brain of a monkey.
-marmoset: the brain of a marmoset.
this one was tested on one dataset
and may not work with non default
options. Check your results!
-rat: the brain of a rat.
By default, no_touchup is used with the rat.
-surface_coil: Data acquired with a surface coil.
-o_TYPE PREFIX: prefix of output surface.
where TYPE specifies the format of the surface
and PREFIX is, well, the prefix.
TYPE is one of: fs, 1d (or vec), sf, ply.
More on that below.
-skulls: Output surface models of the skull.
-4Tom: The output surfaces are named based
on PREFIX following -o_TYPE option below.
-prefix VOL_PREFIX: prefix of output volume.
If not specified, the prefix is the same
as the one used with -o_TYPE.
The output volume is skull stripped version
of the input volume. In the earlier version
of the program, a mask volume was written out.
You can still get that mask volume instead of the
skull-stripped volume with the option -mask_vol .
NOTE: In the default setting, the output volume does not
have values identical to those in the input.
In particular, the range might be larger
and some low-intensity values are set to 0.
If you insist on having the same range of values as in
the input, then either use option -orig_vol, or run:
3dcalc -nscale -a VOL+VIEW -b VOL_PREFIX+VIEW \
-expr 'a*step(b)' -prefix VOL_SAME_RANGE
With the command above, you can preserve the range
of values of the input but some low-intensity voxels would
still be masked. If you want to preserve them, then use
-mask_vol in the 3dSkullStrip command that would produce
VOL_MASK_PREFIX+VIEW. Then run 3dcalc masking with voxels
inside the brain surface envelope:
3dcalc -nscale -a VOL+VIEW -b VOL_MASK_PREFIX+VIEW \
-expr 'a*step(b-3.01)' -prefix VOL_SAME_RANGE_KEEP_LOW
-norm_vol: Output a masked and somewhat intensity normalized and
thresholded version of the input. This is the default,
and you can use -orig_vol to override it.
-orig_vol: Output a masked version of the input AND do not modify
the values inside the brain as -norm_vol would.
-mask_vol: Output a mask volume instead of a skull-stripped
volume.
The mask volume contains:
0: Voxel outside surface
1: Voxel just outside the surface. This means the voxel
center is outside the surface but inside the
bounding box of a triangle in the mesh.
2: Voxel intersects the surface (a triangle), but center
lies outside.
3: Voxel contains a surface node.
4: Voxel intersects the surface (a triangle), center lies
inside surface.
5: Voxel just inside the surface. This means the voxel
center is inside the surface and inside the
bounding box of a triangle in the mesh.
6: Voxel inside the surface.
-spat_norm: (Default) Perform spatial normalization first.
This is a necessary step unless the volume has
been 'spatnormed' already.
-no_spatnorm: Do not perform spatial normalization.
Use this option only when the volume
has been run through the 'spatnorm' process
-spatnorm_dxyz DXYZ: Use DXY for the spatial resolution of the
spatially normalized volume. The default
is the lowest of all three dimensions.
For human brains, use DXYZ of 1.0, for
primate brain, use the default setting.
-write_spatnorm: Write the 'spatnormed' volume to disk.
-niter N_ITER: Number of iterations. Default is 250
For denser meshes, you need more iterations
N_ITER of 750 works for LD of 50.
-ld LD: Parameter to control the density of the surface.
Default is 20 if -no_use_edge is used,
30 with -use_edge. See CreateIcosahedron -help
for details on this option.
-shrink_fac SF: Parameter controlling the brain vs non-brain
intensity threshold (tb). Default is 0.6.
tb = (Imax - t2) SF + t2
where t2 is the 2 percentile value and Imax is the local
maximum, limited to the median intensity value.
For more information on tb, t2, etc. read the BET paper
mentioned above. Note that in 3dSkullStrip, SF can vary across
iterations and might be automatically clipped in certain areas.
SF can vary between 0 and 1.
0: Intensities < median inensity are considered non-brain
1: Intensities < t2 are considered non-brain
-var_shrink_fac: Vary the shrink factor with the number of
iterations. This reduces the likelihood of a surface
getting stuck on large pools of CSF before reaching
the outer surface of the brain. (Default)
-no_var_shrink_fac: Do not use var_shrink_fac.
-shrink_fac_bot_lim SFBL: Do not allow the varying SF to go
below SFBL . Default 0.65, 0.4 when edge detection is used.
This option helps reduce potential for leakage below
the cerebellum.
In certain cases where you have severe non-uniformity resulting
in low signal towards the bottom of the brain, you will need to
reduce this parameter.
-pushout: Consider values above each node in addition to values
below the node when deciding on expansion. (Default)
-no_pushout: Do not use -pushout.
-exp_frac FRAC: Speed of expansion (see BET paper). Default is 0.1.
-touchup: Perform touchup operations at end to include
areas not covered by surface expansion.
Use -touchup -touchup for aggressive makeup.
(Default is -touchup)
-no_touchup: Do not use -touchup
-fill_hole R: Fill small holes that can result from small surface
intersections caused by the touchup operation.
R is the maximum number of pixels on the side of a hole
that can be filled. Big holes are not filled.
If you use -touchup, the default R is 10. Otherwise
the default is 0.
This is a less than elegant solution to the small
intersections which are usually eliminated
automatically.
-NN_smooth NN_SM: Perform Nearest Neighbor coordinate interpolation
every few iterations. Default is 72
-smooth_final SM: Perform final surface smoothing after all iterations.
Default is 20 smoothing iterations.
Smoothing is done using Taubin's method,
see SurfSmooth -help for detail.
-avoid_vent: avoid ventricles. Default.
Use this option twice to make the avoidance more
aggressive. That is at times needed with old brains.
-no_avoid_vent: Do not use -avoid_vent.
-init_radius RAD: Use RAD for the initial sphere radius.
For the automatic setting, there is an
upper limit of 100mm for humans.
For older brains with lots of CSF, you
might benefit from forcing the radius
to something like 75mm
-avoid_eyes: avoid eyes. Default
-no_avoid_eyes: Do not use -avoid_eyes.
-use_edge: Use edge detection to reduce leakage into meninges and eyes.
Default.
-no_use_edge: Do no use edges.
-push_to_edge: Perform aggressive push to edge at the end.
This option might cause leakage.
-no_push_to_edge: (Default).
-use_skull: Use outer skull to limit expansion of surface into
the skull due to very strong shading artifacts.
This option is buggy at the moment, use it only
if you have leakage into skull.
-no_use_skull: Do not use -use_skull (Default).
-send_no_skull: Do not send the skull surface to SUMA if you are
using -talk_suma
-perc_int PERC_INT: Percentage of segments allowed to intersect
surface. Ideally this should be 0 (Default).
However, few surfaces might have small stubborn
intersections that produce a few holes.
PERC_INT should be a small number, typically
between 0 and 0.1. A -1 means do not do
any testing for intersection.
-max_inter_iter N_II: Number of iteration to remove intersection
problems. With each iteration, the program
automatically increases the amount of smoothing
to get rid of intersections. Default is 4
-blur_fwhm FWHM: Blur dset after spatial normalization.
Recommended when you have lots of CSF in brain
and when you have protruding gyri (finger like)
Recommended value is 2..4.
-interactive: Make the program stop at various stages in the
segmentation process for a prompt from the user
to continue or skip that stage of processing.
This option is best used in conjunction with options
-talk_suma and -feed_afni
-demo_pause: Pause at various step in the process to facilitate
interactive demo while 3dSkullStrip is communicating
with AFNI and SUMA. See 'Eye Candy' mode below and
-talk_suma option.
-fac FAC: Multiply input dataset by FAC if range of values is too
small.
Specifying output surfaces using -o or -o_TYPE options:
-o_TYPE outSurf specifies the output surface,
TYPE is one of the following:
fs: FreeSurfer ascii surface.
fsp: FeeSurfer ascii patch surface.
In addition to outSurf, you need to specify
the name of the parent surface for the patch.
using the -ipar_TYPE option.
This option is only for ConvertSurface
sf: SureFit surface.
For most programs, you are expected to specify prefix:
i.e. -o_sf brain. In some programs, you are allowed to
specify both .coord and .topo file names:
i.e. -o_sf XYZ.coord TRI.topo
The program will determine your choice by examining
the first character of the second parameter following
-o_sf. If that character is a '-' then you have supplied
a prefix and the program will generate the coord and topo names.
vec (or 1D): Simple ascii matrix format.
For most programs, you are expected to specify prefix:
i.e. -o_1D brain. In some programs, you are allowed to
specify both coord and topo file names:
i.e. -o_1D brain.1D.coord brain.1D.topo
coord contains 3 floats per line, representing
X Y Z vertex coordinates.
topo contains 3 ints per line, representing
v1 v2 v3 triangle vertices.
ply: PLY format, ascii or binary.
stl: STL format, ascii or binary (see also STL under option -i_TYPE).
byu: BYU format, ascii or binary.
mni: MNI obj format, ascii only.
gii: GIFTI format, ascii.
You can also enforce the encoding of data arrays
by using gii_asc, gii_b64, or gii_b64gz for
ASCII, Base64, or Base64 Gzipped.
If AFNI_NIML_TEXT_DATA environment variable is set to YES, the
the default encoding is ASCII, otherwise it is Base64.
obj: No support for writing OBJ format exists yet.
Note that if the surface filename has the proper extension,
it is enough to use the -o option and let the programs guess
the type from the extension.
SUMA communication options:
-talk_suma: Send progress with each iteration to SUMA.
-refresh_rate rps: Maximum number of updates to SUMA per second.
The default is the maximum speed.
-send_kth kth: Send the kth element to SUMA (default is 1).
This allows you to cut down on the number of elements
being sent to SUMA.
-sh <SumaHost>: Name (or IP address) of the computer running SUMA.
This parameter is optional, the default is 127.0.0.1
-ni_text: Use NI_TEXT_MODE for data transmission.
-ni_binary: Use NI_BINARY_MODE for data transmission.
(default is ni_binary).
-feed_afni: Send updates to AFNI via SUMA's talk.
-np PORT_OFFSET: Provide a port offset to allow multiple instances of
AFNI <--> SUMA, AFNI <--> 3dGroupIncorr, or any other
programs that communicate together to operate on the same
machine.
All ports are assigned numbers relative to PORT_OFFSET.
The same PORT_OFFSET value must be used on all programs
that are to talk together. PORT_OFFSET is an integer in
the inclusive range [1025 to 65500].
When you want to use multiple instances of communicating programs,
be sure the PORT_OFFSETS you use differ by about 50 or you may
still have port conflicts. A BETTER approach is to use -npb below.
-npq PORT_OFFSET: Like -np, but more quiet in the face of adversity.
-npb PORT_OFFSET_BLOC: Similar to -np, except it is easier to use.
PORT_OFFSET_BLOC is an integer between 0 and
MAX_BLOC. MAX_BLOC is around 4000 for now, but
it might decrease as we use up more ports in AFNI.
You should be safe for the next 10 years if you
stay under 2000.
Using this function reduces your chances of causing
port conflicts.
See also afni and suma options: -list_ports and -port_number for
information about port number assignments.
You can also provide a port offset with the environment variable
AFNI_PORT_OFFSET. Using -np overrides AFNI_PORT_OFFSET.
-max_port_bloc: Print the current value of MAX_BLOC and exit.
Remember this value can get smaller with future releases.
Stay under 2000.
-max_port_bloc_quiet: Spit MAX_BLOC value only and exit.
-num_assigned_ports: Print the number of assigned ports used by AFNI
then quit.
-num_assigned_ports_quiet: Do it quietly.
Port Handling Examples:
-----------------------
Say you want to run three instances of AFNI <--> SUMA.
For the first you just do:
suma -niml -spec ... -sv ... &
afni -niml &
Then for the second instance pick an offset bloc, say 1 and run
suma -niml -npb 1 -spec ... -sv ... &
afni -niml -npb 1 &
And for yet another instance:
suma -niml -npb 2 -spec ... -sv ... &
afni -niml -npb 2 &
etc.
Since you can launch many instances of communicating programs now,
you need to know wich SUMA window, say, is talking to which AFNI.
To sort this out, the titlebars now show the number of the bloc
of ports they are using. When the bloc is set either via
environment variables AFNI_PORT_OFFSET or AFNI_PORT_BLOC, or
with one of the -np* options, window title bars change from
[A] to [A#] with # being the resultant bloc number.
In the examples above, both AFNI and SUMA windows will show [A2]
when -npb is 2.
-visual: Equivalent to using -talk_suma -feed_afni -send_kth 5
-debug DBG: debug levels of 0 (default), 1, 2, 3.
This is no Rick Reynolds debug, which is oft nicer
than the results, but it will do.
-node_debug NODE_DBG: Output lots of parameters for node
NODE_DBG for each iteration.
The next 3 options are for specifying surface coordinates
to keep the program from having to recompute them.
The options are only useful for saving time during debugging.
-brain_contour_xyz_file BRAIN_CONTOUR_XYZ.1D
-brain_hull_xyz_file BRAIN_HULL_XYZ.1D
-skull_outer_xyz_file SKULL_OUTER_XYZ.1D
-help: The help you need
[-novolreg]: Ignore any Rotate, Volreg, Tagalign,
or WarpDrive transformations present in
the Surface Volume.
[-noxform]: Same as -novolreg
[-setenv "'ENVname=ENVvalue'"]: Set environment variable ENVname
to be ENVvalue. Quotes are necessary.
Example: suma -setenv "'SUMA_BackgroundColor = 1 0 1'"
See also options -update_env, -environment, etc
in the output of 'suma -help'
Common Debugging Options:
[-trace]: Turns on In/Out debug and Memory tracing.
For speeding up the tracing log, I recommend
you redirect stdout to a file when using this option.
For example, if you were running suma you would use:
suma -spec lh.spec -sv ... > TraceFile
This option replaces the old -iodbg and -memdbg.
[-TRACE]: Turns on extreme tracing.
[-nomall]: Turn off memory tracing.
[-yesmall]: Turn on memory tracing (default).
NOTE: For programs that output results to stdout
(that is to your shell/screen), the debugging info
might get mixed up with your results.
Global Options (available to all AFNI/SUMA programs)
-h: Mini help, at time, same as -help in many cases.
-help: The entire help output
-HELP: Extreme help, same as -help in majority of cases.
-h_view: Open help in text editor. AFNI will try to find a GUI editor
-hview : on your machine. You can control which it should use by
setting environment variable AFNI_GUI_EDITOR.
-h_web: Open help in web browser. AFNI will try to find a browser.
-hweb : on your machine. You can control which it should use by
setting environment variable AFNI_GUI_EDITOR.
-h_find WORD: Look for lines in this programs's -help output that match
(approximately) WORD.
-h_raw: Help string unedited
-h_spx: Help string in sphinx loveliness, but do not try to autoformat
-h_aspx: Help string in sphinx with autoformatting of options, etc.
-all_opts: Try to identify all options for the program from the
output of its -help option. Some options might be missed
and others misidentified. Use this output for hints only.
Compile Date:
Apr 9 2024
Ziad S. Saad SSCC/NIMH/NIH saadz@mail.nih.gov
# Get skull-stripped output and a mask-volume
! 3dSkullStrip -input $input_image -prefix AFNI_ss.nii.gz -push_to_edge
! 3dSkullStrip -input $input_image -prefix AFNI_mask.nii.gz -push_to_edge -mask_vol
3dSkullStrip: Pushing to Edge ...
The intensity in the output dataset is a modified version
of the intensity in the input volume.
To obtain a masked version of the input with identical values inside
the brain, you can either use 3dSkullStrip's -orig_vol option
or run the following command:
3dcalc -a ds000102/sub-08/anat/sub-08_T1w.nii.gz -b ./AFNI_ss.nii.gz+orig -expr 'a*step(b)' \
-prefix ./AFNI_ss.nii.gz_orig_vol
to generate a new masked version of the input.
3dSkullStrip: Pushing to Edge ...
The output dataset is a mask reflecting where voxels in the
input dataset lie in the brain.
To obtain a masked version of the input with identical values inside
the brain, you can either use 3dSkullStrip's -orig_vol option
or run the following command:
3dcalc -a ds000102/sub-08/anat/sub-08_T1w.nii.gz -b ./AFNI_mask.nii.gz+orig -expr 'a*step(b)' \
-prefix ./AFNI_mask.nii.gz_orig_vol
to generate a new masked version of the input.
From AFNI’s documentation:
-push_to_edge: Adds an aggressive push to brain edges. Use this option when the chunks of gray matter are not included. This option might cause the mask to leak into non-brain areas.
3. FreeSurfer#
FreeSurfer’s SynthStrip is a skull-stripping tool that extracts brain voxels from a landscape of image types, ranging across imaging modalities, resolutions, and subject populations. It leverages a deep learning strategy to synthesize arbitrary training images from segmentation maps, yielding a robust model agnostic to acquisition specifics.
! mri_synthstrip --help
usage: mri_synthstrip [-h] -i file [-o file] [-m file] [-g] [-b BORDER]
[--no-csf] [--model file]
Robust, universal skull-stripping for brain images of any type. If you use
SynthStrip in your analysis, please cite: SynthStrip: Skull-Stripping for Any
Brain Image. A Hoopes, JS Mora, AV Dalca, B Fischl, M Hoffmann. NeuroImage 206
(2022), 119474. https://doi.org/10.1016/j.neuroimage.2022.119474
optional arguments:
-h, --help show this help message and exit
-i file, --image file
Input image to skullstrip.
-o file, --out file Save stripped image to path.
-m file, --mask file Save binary brain mask to path.
-g, --gpu Use the GPU.
-b BORDER, --border BORDER
Mask border threshold in mm. Default is 1.
--no-csf Exclude CSF from brain border.
--model file Alternative model weights.
Next, you can run SynthStrip using the following syntax. In this command, “synth_stripped.nii.gz” will be the skull-stripped version of the input image “sub-08_T1w.nii.gz.” Use the -m flag to save a binary brain mask:
! mri_synthstrip -i $input_image -o synth_stripped.nii.gz -m synth_mask.nii.gz
Configuring model on the CPU
Running SynthStrip model version 1
Input image read from: ds000102/sub-08/anat/sub-08_T1w.nii.gz
Masked image saved to: synth_stripped.nii.gz
Binary brain mask saved to: synth_mask.nii.gz
If you use SynthStrip in your analysis, please cite:
----------------------------------------------------
SynthStrip: Skull-Stripping for Any Brain Image.
A Hoopes, JS Mora, AV Dalca, B Fischl, M Hoffmann.
NeuroImage 206 (2022), 119474.
4. FSL#
Another option for brain extraction is FSL’s BET (Brain Extraction Tool):
! bet
Usage: bet <input> <output> [options]
Main bet2 options:
-o generate brain surface outline overlaid onto original image
-m generate binary brain mask
-s generate approximate skull image
-n don't generate segmented brain image output
-f <f> fractional intensity threshold (0->1); default=0.5; smaller values give larger brain outline estimates
-g <g> vertical gradient in fractional intensity threshold (-1->1); default=0; positive values give larger brain outline at bottom, smaller at top
-r <r> head radius (mm not voxels); initial surface sphere is set to half of this
-c <x y z> centre-of-gravity (voxels not mm) of initial mesh surface.
-t apply thresholding to segmented brain image and mask
-e generates brain surface as mesh in .vtk format
Variations on default bet2 functionality (mutually exclusive options):
(default) just run bet2
-R robust brain centre estimation (iterates BET several times)
-S eye & optic nerve cleanup (can be useful in SIENA - disables -o option)
-B bias field & neck cleanup (can be useful in SIENA)
-Z improve BET if FOV is very small in Z (by temporarily padding end slices)
-F apply to 4D FMRI data (uses -f 0.3 and dilates brain mask slightly)
-A run bet2 and then betsurf to get additional skull and scalp surfaces (includes registrations)
-A2 <T2> as with -A, when also feeding in non-brain-extracted T2 (includes registrations)
Miscellaneous options:
-v verbose (switch on diagnostic messages)
-h display this help, then exits
-d debug (don't delete temporary intermediate images)
! bet $input_image anat_bet.nii.gz -m
Results#
We will begin by visualizing the brain extraction results from each tool individually to assess the quality of the extracted brain images. Then, to better highlight differences between the methods, we will generate an overlay of the brain edges, allowing a direct comparison of how the algorithms define brain boundaries.
display(Markdown("### ANTs BrainExtraction"))
nv_ANTS = NiiVue()
nv_ANTS.load_volumes([{"path": "ANTS_Stripped_BrainExtractionBrain.nii.gz"}])
nv_ANTS
ANTs BrainExtraction
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/ANTS_brain_extraction.png')
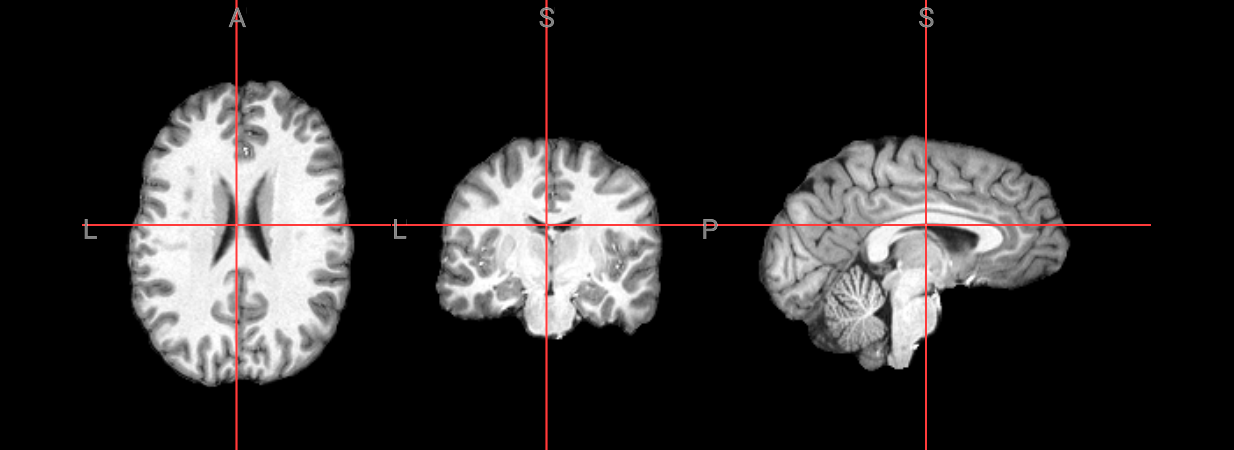
display(Markdown("### AFNI 3dSkullStrip"))
nv_AFNI = NiiVue()
nv_AFNI.load_volumes([{"path": "AFNI_ss.nii.gz"}])
nv_AFNI
AFNI 3dSkullStrip
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/AFNI_brain_extraction.png')
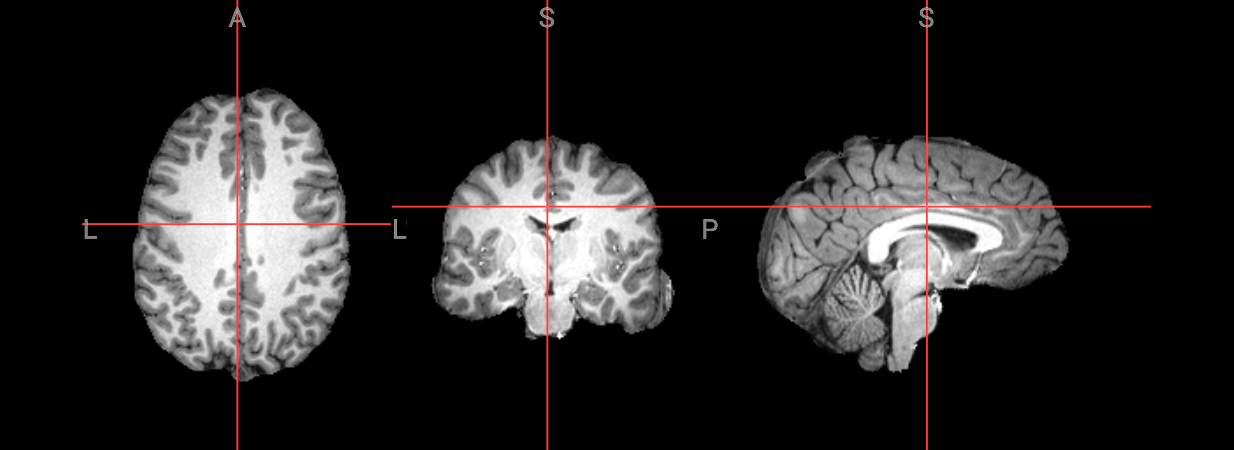
display(Markdown("### FreeSurfer SynthStrip"))
nv_FreeSurfer = NiiVue()
nv_FreeSurfer.load_volumes([{"path": "synth_stripped.nii.gz"}])
nv_FreeSurfer
FreeSurfer SynthStrip
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/FreeSurfer_brain_extraction.png')
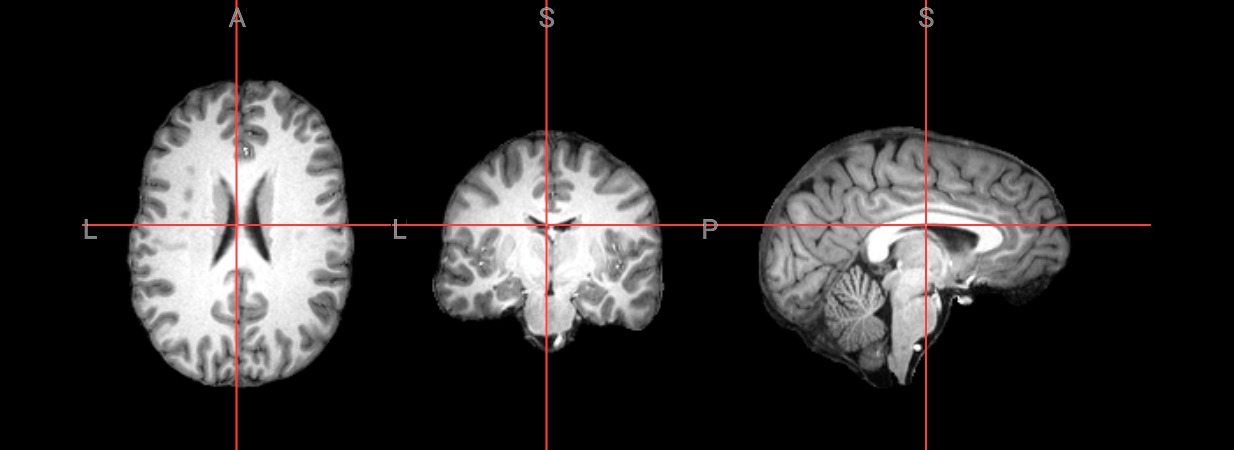
display(Markdown("### FSL BET"))
nv_FSL = NiiVue()
nv_FSL.load_volumes([{"path": "anat_bet.nii.gz"}])
nv_FSL
FSL BET
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/FSL_brain_extraction.png')
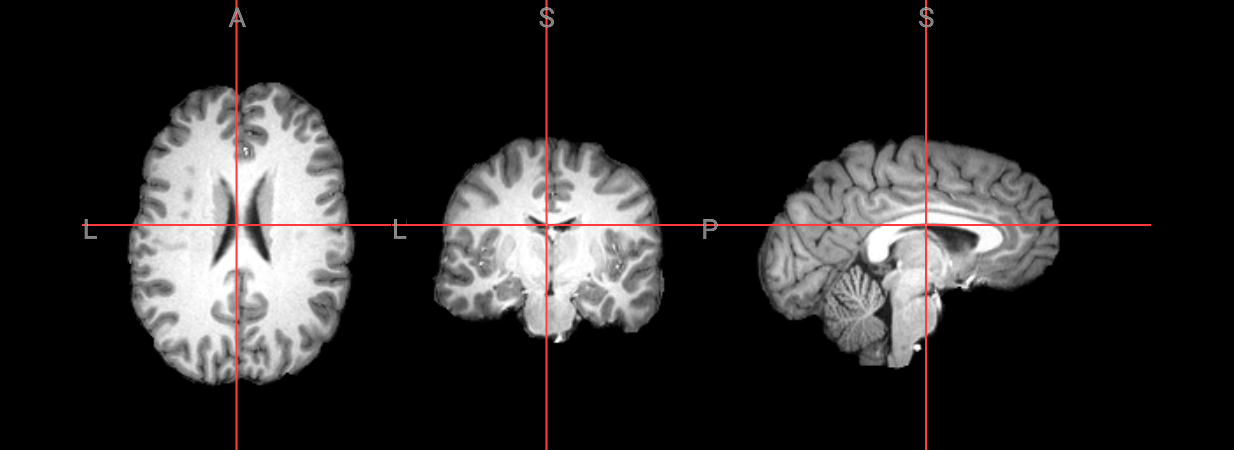
Comparison of the different brain extraction methods#
def extract_edges_from_mask(mask_path):
"""
Detect edges from a binary brain mask using 3D Sobel operator.
Parameters:
mask_path (str): path to the binary brain mask (.nii.gz)
Returns:
nib.Nifti1Image: NIfTI image of the edge mask
"""
img = nib.load(mask_path)
data = img.get_fdata()
# Ensure it's binary
binary = (data > 0).astype(float)
# Compute 3D gradient magnitude using Sobel
grad_x = ndimage.sobel(binary, axis=0)
grad_y = ndimage.sobel(binary, axis=1)
grad_z = ndimage.sobel(binary, axis=2)
grad_mag = np.sqrt(grad_x**2 + grad_y**2 + grad_z**2)
# Threshold to detect edge voxels
edges = (grad_mag > 0).astype(float)
return nib.Nifti1Image(edges, img.affine)
def create_combined_plot(edge_imgs, labels, colors, bg_img, title):
"""Combined plot """
# Initialize combined data
combined_data = np.zeros_like(edge_imgs[0].get_fdata())
# Combine the edge data
for i, edge_img in enumerate(edge_imgs):
edge_data = edge_img.get_fdata()
# Assign different values (1, 2, 3, etc.) for each edge mask
combined_data[edge_data > 0] = i + 1
# Create the combined image
combined_img = nib.Nifti1Image(combined_data, edge_imgs[0].affine)
custom_cmap = ListedColormap(colors)
fig = plt.figure(figsize=(14, 8))
plotting.plot_stat_map(combined_img, bg_img=bg_img,
cmap=custom_cmap,
transparency=0.8,
dim=-1,
threshold=0.5, # Only show actual edge values
title=title,
colorbar=False,
vmin=1,
vmax=len(edge_imgs)) # 4 edge images = values 1-4
# Add legend
legend_elements = [mpatches.Patch(color=colors[i], label=labels[i])
for i in range(len(labels))]
plt.legend(handles=legend_elements, loc='upper left', bbox_to_anchor=(1.02, 1))
plt.subplots_adjust(right=0.82)
# Generate edges from the binary brain masks
ants_edges = extract_edges_from_mask("ANTS_Stripped_BrainExtractionMask.nii.gz")
bet_edges = extract_edges_from_mask("anat_bet_mask.nii.gz")
afni_edges = extract_edges_from_mask("AFNI_mask.nii.gz")
synth_edges = extract_edges_from_mask("synth_mask.nii.gz")
#Check edges
plotting.plot_anat(synth_edges)
<nilearn.plotting.displays._slicers.OrthoSlicer at 0x7f5993867d10>
edge_imgs = [ants_edges, bet_edges, afni_edges, synth_edges]
labels = ['ANTS', 'FSL', 'AFNI', 'FreeSurfer']
colors = ['#FF0000', '#0000FF', '#00FF00', '#FFA500']
create_combined_plot(edge_imgs, labels, colors, input_image, "Brain Extraction Method Comparison")
<Figure size 1400x800 with 0 Axes>
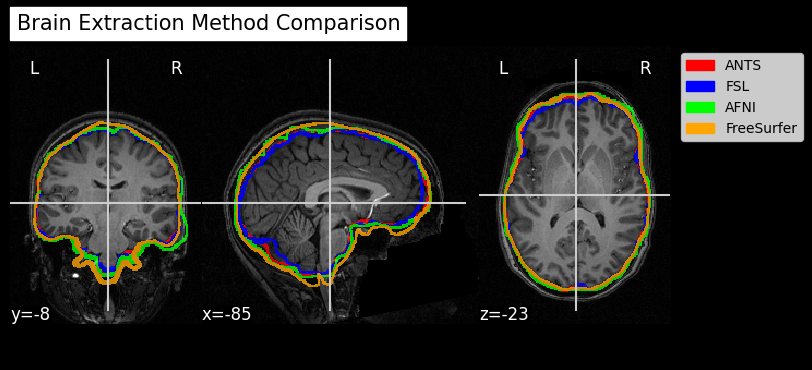
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-10-30T00:56:49.507799+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
nilearn : 0.12.1
IPython : 8.16.1
ipyniivue : 2.3.2
nibabel : 5.2.1
matplotlib: 3.8.4
numpy : 2.2.6
scipy : 1.13.0
