This is the multi-page printable view of this section. Click here to print.
MRI phase Processing
1 - Quantitative Susceptibility Mapping
This tutorial was created by Steffen Bollmann and Ashley Stewart.
Github: @stebo85; @astewartau Web: mri.sbollmann.net Twitter: @sbollmann_MRI
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereAn example notebook can be found here: https://neurodesk.org/example-notebooks/structural_imaging/qsmxt_example.html
Please see the above example notebook which provides a detailed QSM tutorial using QSMxT.
2 - SWI
This tutorial was created by Steffen Bollmann.
Github: @stebo85 Web: mri.sbollmann.net Twitter: @sbollmann_MRI
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereDownload demo data
Open a terminal and run:
pip install osfclient
cd /neurodesktop-storage/
osf -p ru43c fetch 01_bids.zip /neurodesktop-storage/swi-demo/01_bids.zip
unzip /neurodesktop-storage/swi-demo/01_bids.zip -d /neurodesktop-storage/swi-demo/Open the CLEARSWI tool from the application menu:
paste this julia script in a julia file and execute:
cd /neurodesktop-storage/
vi clearswi.jlhit a or i and then paste this:
using CLEARSWI
TEs = [20]
nifti_folder = "/neurodesktop-storage/swi-demo/01_bids/sub-170705134431std1312211075243167001/ses-1/anat"
magfile = joinpath(nifti_folder, "sub-170705134431std1312211075243167001_ses-1_acq-qsm_run-1_magnitude.nii.gz")
phasefile = joinpath(nifti_folder, "sub-170705134431std1312211075243167001_ses-1_acq-qsmPH00_run-1_phase.nii.gz")
mag = readmag(magfile);
phase = readphase(phasefile);
data = Data(mag, phase, mag.header, TEs);
swi = calculateSWI(data);
# mip = createIntensityProjection(swi, minimum); # minimum intensity projection, other Julia functions can be used instead of minimum
mip = createMIP(swi); # shorthand for createIntensityProjection(swi, minimum)
savenii(swi, "/neurodesktop-storage/swi-demo/swi.nii"; header=mag.header)
savenii(mip, "/neurodesktop-storage/swi-demo/mip.nii"; header=mag.header)hit SHIFT-Z-Z and run:
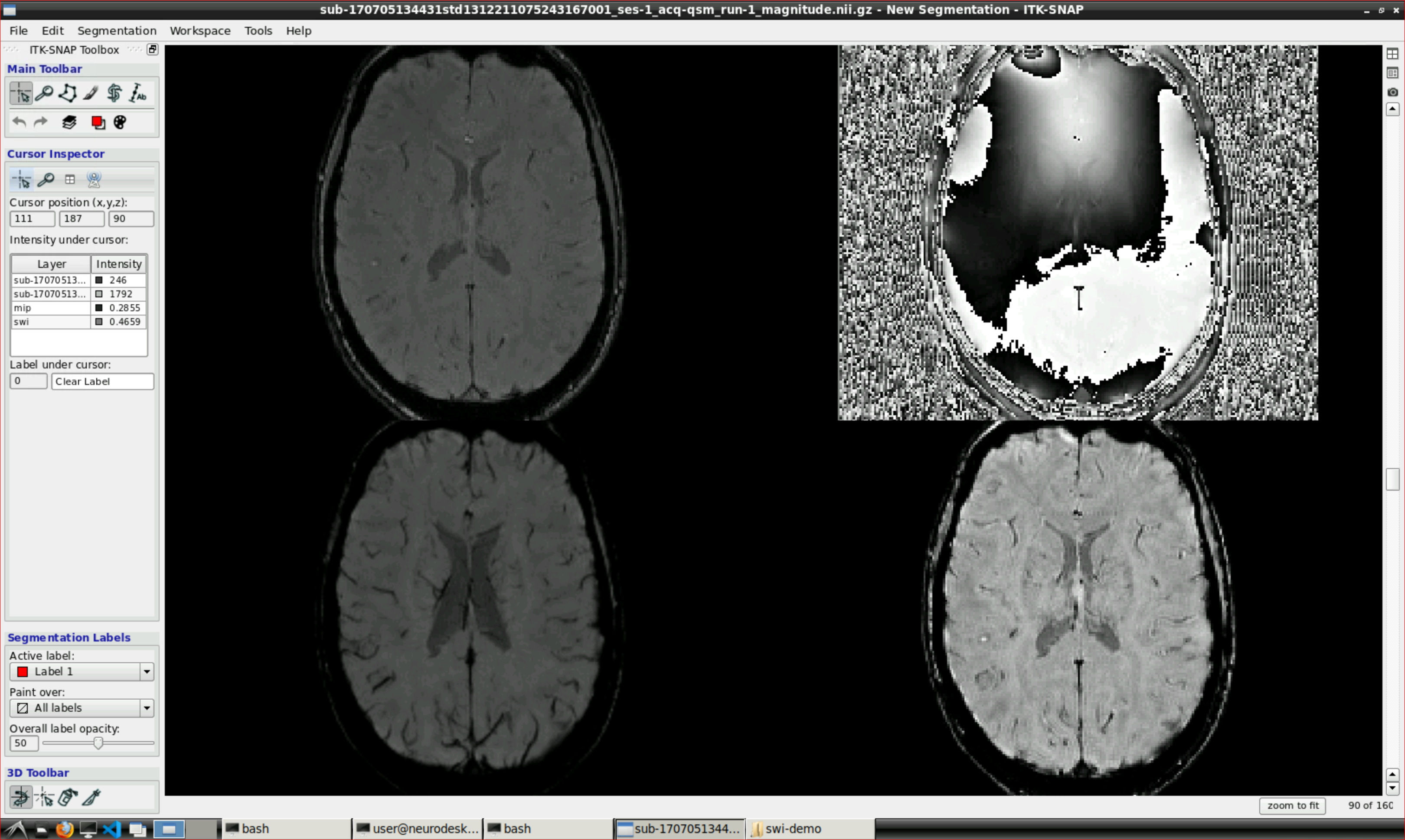
julia clearswi.jlOpen ITK snap from the Visualization Application’s menu and inspect the results (the outputs are in swi-demo/swi.nii and mip.nii)
3 - Unwrapping
This tutorial was created by Steffen Bollmann.
Github: @stebo85 Web: mri.sbollmann.net Twitter: @sbollmann_MRI
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereDownload demo data
Open a terminal and run:
pip install osfclient
cd /neurodesktop-storage/
osf -p ru43c fetch 01_bids.zip /neurodesktop-storage/swi-demo/01_bids.zip
unzip /neurodesktop-storage/swi-demo/01_bids.zip -d /neurodesktop-storage/swi-demo/
mkdir /neurodesktop-storage/romeo-demo/
cp /neurodesktop-storage/swi-demo/01_bids/sub-170705134431std1312211075243167001/ses-1/anat/sub-170705134431std1312211075243167001_ses-1_acq-qsmPH00_run-1_phase.nii.gz /neurodesktop-storage/romeo-demo/phase.nii.gz
cp /neurodesktop-storage/swi-demo/01_bids/sub-170705134431std1312211075243167001/ses-1/anat/sub-170705134431std1312211075243167001_ses-1_acq-qsm_run-1_magnitude.nii.gz /neurodesktop-storage/romeo-demo/mag.nii.gz
gunzip /neurodesktop-storage/romeo-demo/mag.nii.gz
gunzip /neurodesktop-storage/romeo-demo/phase.nii.gzUsing ROMEO for phase unwrapping
Open the ROMEO tool from the application menu and run:
romeo -p /neurodesktop-storage/romeo-demo/phase.nii -m /neurodesktop-storage/romeo-demo/mag.nii -k nomask -o /neurodesktop-storage/romeo-demo/