A batch scripting example for PhysIO toolbox
This tutorial was created by Kelly G. Garner.
Github: @kel-github
Twitter: @garner_theory
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereThis tutorial walks through 1 way to batch script the use of the PhysIO toolbox with Neurodesk. The goal is to use the toolbox to generate physiological regressors to use when modelling fMRI data. The output format of the regressor files are directly compatible for use with SPM, and can be adapted to fit the specifications of other toolboxes.
Getting started
This tutorial assumes the following:
- Your data are (largely) in BIDS format
- That you have converted your .zip files containing physiological data to .log files. For example, if you’re using a CMRR multi-band sequence, then you can use this function
- That your .log files are in the subject derivatives/…/sub-…/ses-…/‘func’ folders of aforementioned BIDs structured data
- That you have a file that contains the motion regressors you plan to use in your GLM. I’ll talk below a bit about what I did with the output given by fmriprep (e.g. …_desc-confounds_timeseries.tsv’)
- That you can use SPM12 and the PhysIO GUI to initialise your batch code
NB. You can see the code generated from this tutorial here
1. Generate an example script for batching
First you will create an example batch script that is specific to one of your participants. To achieve this I downloaded locally the relevant ‘.log’ files for one participant, as well as the ‘…desc-confounds_timeseries.tsv’ output for fmriprep for each run. PhysIO is nice in that it will append the regressors from your physiological data to your movement parameters, so that you have a single file of regressors to add to your design matrix in SPM etc (other toolboxes are available).
To work with PhysIO toolbox, your motion parameters need to be in the .txt format as required by SPM.
I made some simple functions in python that would extract my desired movement regressors and save them to the space separated .txt file as is required by SPM. They can be found here.
Once I had my .log files and .txt motion regressors file, I followed the instructions here to get going with the Batch editor, and used this paper to aid my understanding of how to complete the fields requested by the Batch editor.
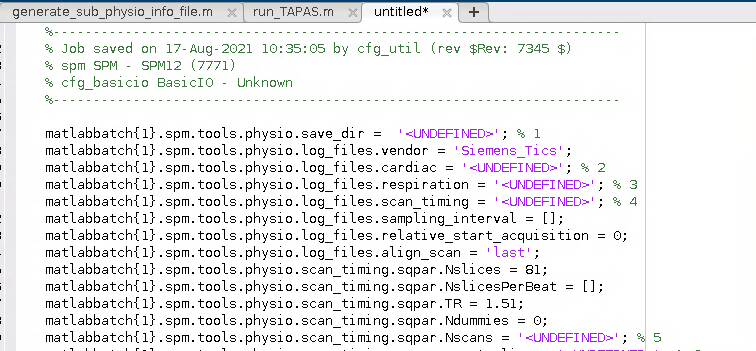
I wound up with a Batch script for the PhysIO toolbox that looked a little bit like this:
2. Generalise the script for use with any participant
Now that you have an example script that contains the specific details for a single participant, you are ready to generalise this code so that you can run it for any participant you choose. I decided to do this by doing the following:
- First I generate an ‘info’ structure for each participant. This is a structure saved as a matfile for each participant under ‘derivatives’, in the relevant sub-z/ses-y/func/ folder. This structure contains the subject specific details that PhysIO needs to know to run. Thus I wrote a matlab function that saves a structure called info with the following fields:
% -- outputs: a matfile containing a structure called info with the
% following fields:
% -- sub_num = subject number: [string] of form '01' '11' or '111'
% -- sess = session number: [integer] e.g. 2
% -- nrun = [integer] number of runs for that participant
% -- nscans = number of scans (volumes) in the design matrix for each
% run [1, nrun]
% -- cardiac_files = a cell of the cardiac files for that participant
% (1,n = nrun) - attained by using extractCMRRPhysio()
% -- respiration_files = same as above but for the resp files - attained by using extractCMRRPhysio()
% -- scan_timing = info file from Siemens - attained by using extractCMRRPhysio()
% -- movement = a cell of the movement regressor files for that
% participant (.txt, formatted for SPM)To see the functions that produce this information, you can go to this repo here
- Next I amended the batch script to load a given participant’s info file and to retrieve this information for the required fields in the batch. The batch script winds up looking like this:
%% written by K. Garner, 2022
% uses batch info:
%-----------------------------------------------------------------------
% Job saved on 17-Aug-2021 10:35:05 by cfg_util (rev $Rev: 7345 $)
% spm SPM - SPM12 (7771)
% cfg_basicio BasicIO - Unknown
%-----------------------------------------------------------------------
% load participant info, and print into the appropriate batch fields below
% before running spm jobman
% assumes data is in BIDS format
%% load participant info
sub = '01';
dat_path = '/file/path/to/top-level/of-your-derivatives-fmriprep/folder';
task = 'attlearn';
load(fullfile(dat_path, sprintf('sub-%s', sub), 'ses-02', 'func', ...
sprintf('sub-%s_ses-02_task-%s_desc-physioinfo', sub, task)))
% set variables
nrun = info.nrun;
nscans = info.nscans;
cardiac_files = info.cardiac_files;
respiration_files = info.respiration_files;
scan_timing = info.scan_timing;
movement = info.movement;
%% initialise spm
spm_jobman('initcfg'); % check this for later
spm('defaults', 'FMRI');
%% run through runs, print info and run
for irun = 1:nrun
clear matlabbatch
matlabbatch{1}.spm.tools.physio.save_dir = cellstr(fullfile(dat_path, sprintf('sub-%s', sub), 'ses-02', 'func')); % 1
matlabbatch{1}.spm.tools.physio.log_files.vendor = 'Siemens_Tics';
matlabbatch{1}.spm.tools.physio.log_files.cardiac = cardiac_files(irun); % 2
matlabbatch{1}.spm.tools.physio.log_files.respiration = respiration_files(irun); % 3
matlabbatch{1}.spm.tools.physio.log_files.scan_timing = scan_timing(irun); % 4
matlabbatch{1}.spm.tools.physio.log_files.sampling_interval = [];
matlabbatch{1}.spm.tools.physio.log_files.relative_start_acquisition = 0;
matlabbatch{1}.spm.tools.physio.log_files.align_scan = 'last';
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.Nslices = 81;
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.NslicesPerBeat = [];
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.TR = 1.51;
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.Ndummies = 0;
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.Nscans = nscans(irun); % 5
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.onset_slice = 1;
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.time_slice_to_slice = [];
matlabbatch{1}.spm.tools.physio.scan_timing.sqpar.Nprep = [];
matlabbatch{1}.spm.tools.physio.scan_timing.sync.nominal = struct([]);
matlabbatch{1}.spm.tools.physio.preproc.cardiac.modality = 'PPU';
matlabbatch{1}.spm.tools.physio.preproc.cardiac.filter.no = struct([]);
matlabbatch{1}.spm.tools.physio.preproc.cardiac.initial_cpulse_select.auto_template.min = 0.4;
matlabbatch{1}.spm.tools.physio.preproc.cardiac.initial_cpulse_select.auto_template.file = 'initial_cpulse_kRpeakfile.mat';
matlabbatch{1}.spm.tools.physio.preproc.cardiac.initial_cpulse_select.auto_template.max_heart_rate_bpm = 90;
matlabbatch{1}.spm.tools.physio.preproc.cardiac.posthoc_cpulse_select.off = struct([]);
matlabbatch{1}.spm.tools.physio.preproc.respiratory.filter.passband = [0.01 2];
matlabbatch{1}.spm.tools.physio.preproc.respiratory.despike = true;
matlabbatch{1}.spm.tools.physio.model.output_multiple_regressors = 'mregress.txt';
matlabbatch{1}.spm.tools.physio.model.output_physio = 'physio';
matlabbatch{1}.spm.tools.physio.model.orthogonalise = 'none';
matlabbatch{1}.spm.tools.physio.model.censor_unreliable_recording_intervals = true; %false;
matlabbatch{1}.spm.tools.physio.model.retroicor.yes.order.c = 3;
matlabbatch{1}.spm.tools.physio.model.retroicor.yes.order.r = 4;
matlabbatch{1}.spm.tools.physio.model.retroicor.yes.order.cr = 1;
matlabbatch{1}.spm.tools.physio.model.rvt.no = struct([]);
matlabbatch{1}.spm.tools.physio.model.hrv.no = struct([]);
matlabbatch{1}.spm.tools.physio.model.noise_rois.no = struct([]);
matlabbatch{1}.spm.tools.physio.model.movement.yes.file_realignment_parameters = {fullfile(dat_path, sprintf('sub-%s', sub), 'ses-02', 'func', sprintf('sub-%s_ses-02_task-%s_run-%d_desc-motion_timeseries.txt', sub, task, irun))}; %8
matlabbatch{1}.spm.tools.physio.model.movement.yes.order = 6;
matlabbatch{1}.spm.tools.physio.model.movement.yes.censoring_method = 'FD';
matlabbatch{1}.spm.tools.physio.model.movement.yes.censoring_threshold = 0.5;
matlabbatch{1}.spm.tools.physio.model.other.no = struct([]);
matlabbatch{1}.spm.tools.physio.verbose.level = 2;
matlabbatch{1}.spm.tools.physio.verbose.fig_output_file = '';
matlabbatch{1}.spm.tools.physio.verbose.use_tabs = false;
spm_jobman('run', matlabbatch);
end3. Ready to run on Neurodesk!
Now we have a batch script, we’re ready to run this on Neurodesk - yay!
First make sure the details at the top of the script are correct. You can see that this script could easily be amended to run multiple subjects.
On Neurodesk, go to the PhysIO toolbox, but select the command line tool rather than the GUI interface (‘physio r7771 instead of physioGUI r7771). This will take you to the container for the PhysIO toolbox
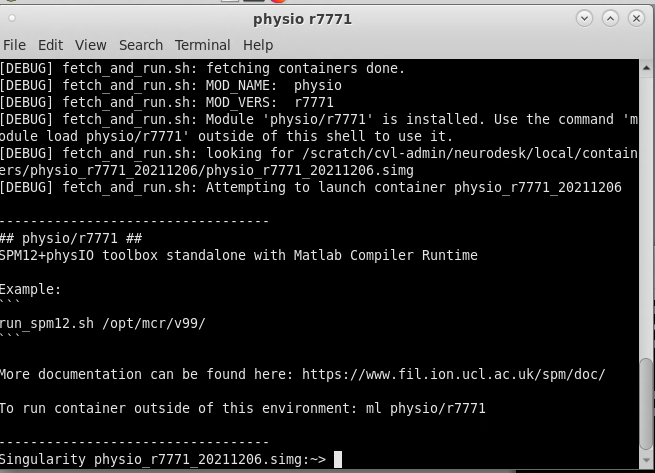
Now to run your PhysIO batch script, type the command:
run_spm12.sh /opt/mcr/v99/ batch /your/batch/script/named_something.mEt Voila! Physiological regressors are now yours - mua ha ha!