Neurodesk offers several options to suit different needs and computing environments. Here we show examples of different Neurodesk installations that could be good starting point for you.
This is the multi-page printable view of this section. Click here to print.
Installation Examples
1 - Ubuntu 24.04
On this page we show specific examples of the different ways of how Neurodesk can be installed on a local computer. We start from the highest level using the Neurodesk app, then go lower level via docker, neurocommand and down to the lowest level using neurocontainers. We also show on each level how containers can be streamed via CVMFS or downloaded locally.
Running Neurodesk on a Ubuntu 24.04 computer
On a linux machine you have mutliple options to use Neurodesk:
- Highest abstraction level, and easiest option: Neurodeskapp - this provides you a full Linux desktop with everything configured you need. You do not need to think about Docker or Singularity containers and you can just get your work done. This is the recommended option.
- High abstraction level: Running Neurodesktop via Docker manually - you still get the desktop with everything configured, but you now have to manage the docker container yourself. This is useful when the app doesn’t work well - for example in a remote SSH setup.
- Middle abstraction level: Use the containers through wrapper scripts on the terminal through Neurocommand - this is great if you don’t need a full desktop environment and you want to use the neurodesk tools in your scripts. Neurocommand handles multiple containers for you and you just run your tools as you are used to without having to think about the fact that they are running in singularity/apptainer containers
- Low abstraction level: Use the containers on the terminal directly - if you just want ot use the containers directly and you want to do everything yourself, that’s the best option for you :)
Highest abstraction level, and easiest option: Neurodeskapp
Download Neurodeskapp: https://github.com/neurodesk/neurodesk-app/releases/latest/download/NeurodeskApp-Setup-Debian-x64.deb
and Install:
sudo apt install ./NeurodeskApp-Setup-Debian-x64.debInstall Docker:
# Add Docker's official GPG key:
sudo apt-get update
sudo apt-get install ca-certificates curl
sudo install -m 0755 -d /etc/apt/keyrings
sudo curl -fsSL https://download.docker.com/linux/ubuntu/gpg -o /etc/apt/keyrings/docker.asc
sudo chmod a+r /etc/apt/keyrings/docker.asc
# Add the repository to Apt sources:
echo \
"deb [arch=$(dpkg --print-architecture) signed-by=/etc/apt/keyrings/docker.asc] https://download.docker.com/linux/ubuntu \
$(. /etc/os-release && echo "${UBUNTU_CODENAME:-$VERSION_CODENAME}") stable" | \
sudo tee /etc/apt/sources.list.d/docker.list > /dev/null
sudo apt-get update
sudo apt-get install docker-ce docker-ce-cli containerd.io docker-buildx-plugin docker-compose-plugin
sudo groupadd docker
sudo usermod -aG docker $USER
newgrp docker
sudo chown root:docker /var/run/docker.sock
sudo chmod 666 /var/run/docker.sockAfter installation run the following command to verify that Docker is working correctly:
docker run hello-worldThe Neurodeskapp can be launched directly from the application menu, or by running the neurodeskapp command in the command line.
In the Neurodeskapp settings you can choose if you want to stream or download containers to your system.
more information can be found here: https://neurodesk.org/docs/getting-started/local/neurodeskapp/
High abstraction level: Running Neurodesktop via Docker manually
If you run Ubuntu > 23.10 and you haven’t installed the Neurodeskapp before you need to create this apparmor profile under /etc/apparmor.d/neurodeskapp
echo -e "# This profile allows everything and only exists to give the\n# application a name instead of having the label \"unconfined\"\n\nabi <abi/4.0>,\ninclude <tunables/global>\n\nprofile neurodeskapp \"/opt/NeurodeskApp/neurodeskapp\" flags=(unconfined) {\n userns,\n\n # Site-specific additions and overrides. See local/README for details.\n include if exists <local/neurodeskapp>\n}" | sudo tee /etc/apparmor.d/neurodeskappyou also need to create the ~/neurodesktop-storage folder if you haven’t used the app before:
mkdir -p ~/neurodesktop-storageMake sure you have Docker installed and configured correctly (see Neurodeskapp for instructions), then run in a terminal:
docker volume create neurodesk-home &&
sudo docker run \
--shm-size=1gb -it --security-opt apparmor=neurodeskapp --privileged --user=root --name neurodesktop \
-v ~/neurodesktop-storage:/neurodesktop-storage \
--mount source=neurodesk-home,target=/home/jovyan \
-e NB_UID="$(id -u)" -e NB_GID="$(id -g)" \
-p 8888:8888 \
-e NEURODESKTOP_VERSION=2025-12-20 vnmd/neurodesktop:2025-12-20Then open the jupyter link with the token displayed in your browser. Make sure it starts with 127.0.0.1:8888/lab&token=…
You can also add a flag to the docker command to activate the offline mode: -e CVMFS_DISABLE=true
when finished, make sure to delete the container - otherwise, you will get an error the next time you run the docker command:
docker rm neurodesktopIf you want to pass your GPU into the desktop, first install this on the host:
# Manually set the distribution to ubuntu22.04 (works with Ubuntu 24.04) - because it doens't exist yet for 24.04
distribution="ubuntu22.04"
# Add the NVIDIA container toolkit repo using the 22.04 version
curl -s -L https://nvidia.github.io/libnvidia-container/$distribution/libnvidia-container.list | \
sed 's|^deb |deb [signed-by=/usr/share/keyrings/nvidia-container-toolkit.gpg] |' | \
sudo tee /etc/apt/sources.list.d/nvidia-container-toolkit.list > /dev/null
# Update package lists
sudo apt update
sudo apt install -y nvidia-container-toolkit
sudo nvidia-ctk runtime configure --runtime=docker
sudo systemctl restart dockerThen start the neurodesktop container with the GPU flag:
sudo docker run \
--shm-size=1gb -it --privileged --user=root --name neurodesktop \
-v ~/neurodesktop-storage:/neurodesktop-storage \
-e NB_UID="$(id -u)" -e NB_GID="$(id -g)" \
--gpus all \
-p 8888:8888 \
-e NEURODESKTOP_VERSION=2025-12-20 vnmd/neurodesktop:2025-12-20then export the –nv flag
export neurodesk_singularity_opts="--nv" more information can be found here: https://neurodesk.org/docs/getting-started/neurodesktop/linux/
Middle abstraction level: Use the containers through wrapper scripts on the terminal through Neurocommand
For this you do not need Docker, but rather Apptainer or Singularity:
sudo apt-get install -y software-properties-common
sudo add-apt-repository -y ppa:apptainer/ppa
sudo apt-get update
sudo apt-get install -y apptainer
sudo apt-get install -y apptainer-suidMake sure you have Python configured on your system with pip3:
sudo apt install python3-pipThen install neurocommand:
cd ~
git clone https://github.com/neurodesk/neurocommand.git
cd neurocommand
python3 -m venv ./venv
./venv/bin/pip3 install -r neurodesk/requirements.txt
bash build.sh --cli
export APPTAINER_BINDPATH=`pwd -P`now you can search and install containers:
# this searches for containers and you can install individual containers by running the install commands displayed
bash containers.sh itksnap
# this installs all containers matching the pattern itksnap
bash containers.sh --itksnapthen link the containers directory to the neurodesktop-storage:
ln -s $PWD/local/containers/ ~/neurodesktop-storage/ then you can install lmod:
sudo apt install lmodand configure lmod:
# Create the module.sh file
sudo bash -c 'cat > /usr/share/module.sh << "EOL"
# system-wide profile.modules #
# Initialize modules for all sh-derivative shells #
#----------------------------------------------------------------------#
trap "" 1 2 3
case "$0" in
-bash|bash|*/bash) . /usr/share/lmod/8.6.19/init/bash ;;
-ksh|ksh|*/ksh) . /usr/share/lmod/8.6.19/init/ksh ;;
-zsh|zsh|*/zsh) . /usr/share/lmod/8.6.19/init/zsh ;;
-sh|sh|*/sh) . /usr/share/lmod/8.6.19/init/sh ;;
*) . /usr/share/lmod/8.6.19/init/sh ;; # default for scripts
esac
trap - 1 2 3
EOL'then add the module setup to your ~/.bashrc:
cat >> ~/.bashrc << 'EOL'
if [ -f '/usr/share/module.sh' ]; then source /usr/share/module.sh; fi
if [ -d /cvmfs/neurodesk.ardc.edu.au/neurodesk-modules ]; then
module use /cvmfs/neurodesk.ardc.edu.au/neurodesk-modules/*
else
export MODULEPATH="~/neurodesktop-storage/containers/modules"
module use $MODULEPATH
fi
EOLmake sure you have set the APPTAINER_BINDPATH to all directories that you want the containers to access
export APPTAINER_BINDPATH='/data,/scratch'you can also add this to your ~/.bashrc
then restart the terminal you can load and run the software using:
ml itksnap
itksnapIf you need nvidia gpu support activate via exporting this environment variable:
export neurodesk_singularity_opts='--nv'If you get errors like this:
/opt/itksnap-4.0.2/lib/snap-4.0.2/ITK-SNAP: /lib/x86_64-linux-gnu/libc.so.6: version `GLIBC_2.38' not found (required by /.singularity.d/libs/libGLX.so.0)
/opt/itksnap-4.0.2/lib/snap-4.0.2/ITK-SNAP: /lib/x86_64-linux-gnu/libc.so.6: version `GLIBC_2.38' not found (required by /.singularity.d/libs/libEGL.so.1)
/opt/itksnap-4.0.2/lib/snap-4.0.2/ITK-SNAP: /lib/x86_64-linux-gnu/libc.so.6: version `GLIBC_2.38' not found (required by /.singularity.d/libs/libGLdispatch.so.0)This means that the glibc versions inside the container and outside the container are not compatible. You can either disable the GPU flag –nv or use a newer version of the container.
If you do not want to download the containers you can also stream the containers using CVMFS: https://neurodesk.org/docs/getting-started/neurocontainers/cvmfs/
more information: https://neurodesk.org/docs/getting-started/neurocommand/linux-and-hpc/
Low abstraction level: Use the containers on the terminal directly
for this you only need apptainer or singularity installed. See above for installation instructions.
Then you can download a container and run it directly:
#find out which containers are available:
curl -s https://raw.githubusercontent.com/neurodesk/neurocommand/main/cvmfs/log.txt
#select a container and download it:
export container=itksnap_3.8.0_20201208
curl -X GET https://neurocontainers.s3.us-east-2.amazonaws.com/$container.simg -O
singularity shell itksnap_3.8.0_20201208.simg
itksnapif you need nvidia GPU support, add –nv:
singularity shell --nv itksnap_3.8.0_20201208.simg
itksnapIf you want to stream the containers, follow these instructions for setting up CVMFS: https://neurodesk.org/docs/getting-started/neurocontainers/cvmfs/
then you can run:
singularity shell /cvmfs/neurodesk.ardc.edu.au/containers/itksnap_3.8.0_20201208/itksnap_3.8.0_20201208.simgand the container will be streamed to you :)
More information about Download (offline) mode: https://neurodesk.org/docs/getting-started/neurocontainers/singularity/
2 - Bunya
Neurodesk is installed at the University of Queensland’s supercomputer “Bunya”. To access neurodesk tools you need to be in an interactive job (so either start a virtual desktop via Open On-Demand: https://bunya-ondemand.rcc.uq.edu.au/pun/sys/dashboard) or run:
salloc --nodes=1 --ntasks-per-node=1 --cpus-per-task=1 --mem=5G --job-name=TinyInteractive --time=01:00:00 --partition=debug --account=REPLACE_THIS_WITH_YOUR_AccountString srun --export=PATH,TERM,HOME,LANG --pty /bin/bash -lThen load the neurodesk modules:
module use /sw/local/rocky8/noarch/neuro/software/neurocommand/local/containers/modules/
export APPTAINER_BINDPATH=/scratch,/QRISdataNow you can list all modules (Neurodesk modules are the first ones in the list):
ml avOr you can module load any tool you need:
ml qsmxt/6.4.1If you want to use GUI applications (fsleyes, afni, suma, matlab, …) you need to overwrite the temporary directory to be /tmp (otherwise you get an error that it cannot connect to the DISPLAY):
export TMPDIR=/tmp For matlab you also need to create a network license file in your ~/Downloads/network.lic:
cat <<EOF > ~/Downloads/network.lic
SERVER uq-matlab.research.dc.uq.edu.au ANY 27000
USE_SERVER
EOFNOTE: If you are using AFNI on Bunya then the default detach behavior will cause SIGBUS errors and a crash. To fix this run AFNI with:
afni -no_detachNOTE: MRIQC has its $HOME variable hardcoded to be /home/mriqc. This leads to problems on Bunya. A workaround is to run this before mriqc:
export neurodesk_singularity_opts="--home $HOME:/home"If you are missing an application, please contact mail.neurodesk@gmail.com and ask for the neurodesk installation to be updated on Bunya :)
Using this inside a jupyter notebook
You need to install this in addtion:
pip install jupyterlab_niivue ipyniivue jupyterlmod jupyterlab_slurmThen start a notebook and run these commands:
import module
await module.load('niimath')3 - Greatlakes
Setup on a desktop
module load singularity
# now change to a directory with enough storage, e.g. /nfs/turbo/username
git clone https://github.com/neurodesk/neurocommand.git
cd neurocommand
pip3 install -r neurodesk/requirements.txt --user
bash build.sh --cli
bash containers.sh
export SINGULARITY_BINDPATH=`pwd -P`
bash containers.sh itksnap
# now select a version of itksnap to install. For this copy and paste the installation
echo "module load singularity" >> ~/.bashrc
echo "module use $PWD/local/containers/modules/" >> ~/.bashrc
echo "export SINGULARITY_BINDPATH=/nfs/,/scratch/" >> ~/.bashrcSetup on with a jupyter notebook
Start new Jupyter notebook by entering “load singularity” in the Module Commands field:
Then run these commands:
!pip install jupyterlmod
# Restart the kernel by clicking Kernel -> Restart Kernel
import module
await module.load('niimath')4 - Sherlock
Neurodesk runs on Stanfords supercomputer “Sherlock” and below are different ways of accessing it.
Using Neurodesk on Sherlock via ssh
Using Neurodesk containers
Setup your ~/.ssh/config
Host sherlock
ControlMaster auto
ForwardX11 yes
ControlPath ~/.ssh/%l%r@%h:%p
HostName login.sherlock.stanford.edu
User <sunetid>
ControlPersist yesand then connect to sherlock
ssh sherlockYou can module use the neurodesk modules (if they have been installed before - see instructions for installing and updating at the end of this page below):
module use $GROUP_HOME/modules
export APPTAINER_BINDPATH=/scratch,/tmpYou can also add these to your ~/.bashrc:
echo "module use $GROUP_HOME/modules/" >> ~/.bashrc
echo "export APPTAINER_BINDPATH=/scratch,/tmp" >> ~/.bashrcNow you can list all modules (Neurodesk modules are the first ones in the list):
ml avOr you can module load any tool you need:
ml fsl/6.0.7.18Submitting a job
put this in a file, e.g. submit.sbatch:
#!/bin/bash
#
#SBATCH --job-name=test
#SBATCH --time=01:00:00
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --mem-per-cpu=2G
#SBATCH --output=logs/%x_%j.out
#SBATCH --error=logs/%x_%j.err
#SBATCH -p normal
module purge
module use $GROUP_HOME/modules/
module load ants/2.6.0
ants.... $1use sh_part to see which partitions and limits are available:
sh_partthen submit:
sbatch submit.sbatchto size jobs you can use ruse https://www.sherlock.stanford.edu/docs/user-guide/running-jobs/#sizing-a-job
module load system ruse
ruse ./myappor parallize across subjects:
for file in `ls sub*.nii`;
do echo "submitting job for $file";
sbatch submit.sbatch $file;
doneif you need lots of jobs, consider using array jobs: https://www.sherlock.stanford.edu/docs/advanced-topics/job-management/?h=array+jobs
starting a matlab job:
#!/bin/bash
#SBATCH --job-name=invert
#SBATCH --time=00:03:00
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=3
#SBATCH --mem-per-cpu=4G
#SBATCH --output=logs/%x_%j.out
#SBATCH --error=logs/%x_%j.err
#SBATCH --partition=normal
#SBATCH --mail-type=ALL
module purge
module load matlab
matlab -batch matlab_file_without_the_dot_m_endingcheck:
squeue -u $USER
# or
squeue --me
# or to watch it continuesly:
watch -n 5 "squeue -u $USER"
# or get more details:
squeue --me -o "%.18i %.9P %.30j %.8u %.8T %.10M %.9l %.6D %.4C %.10m"
# or create an alias:
echo 'alias sq="squeue --me -o \"%.18i %.9P %.30j %.8u %.8T %.10M %.9l %.6D %.4C %.10m\""' >> ~/.bashrccancel jobs:
scancel <jobid>
scancel --name=my_job_namemore details https://www.sherlock.stanford.edu/docs/user-guide/running-jobs/#example-sbatch-script
Using GUI applications
First you need to connect to Sherlock with SSH forwarding (e.g. from a Linux machine or from your local neurodesk or from a mac with https://www.xquartz.org/ installed, or from windows using Mobaxterm)
and then request an interactive job and start the software:
sh_dev
ml mrtrix3
mrviewThis runs via x-forwarding and doesn’t work well, for a better experience see below how to start a full neurodesktop on Sherlock.
GPU support
request a GPU and then add –nv option:
sh_dev -g 1
module load fsl
export neurodesk_singularity_opts='--nv'
git clone https://github.com/neurolabusc/gpu_test.git
cd gpu_test/etest/
bash runme_gpu.shUsing Neurodesk on Sherlock via Ondemand
Open a jupyterlab session via Open On-Demand: https://ondemand.sherlock.stanford.edu/
Make sure to select python 3.9 - otherwise the HPC slurm plugin for jupyterlab will not work
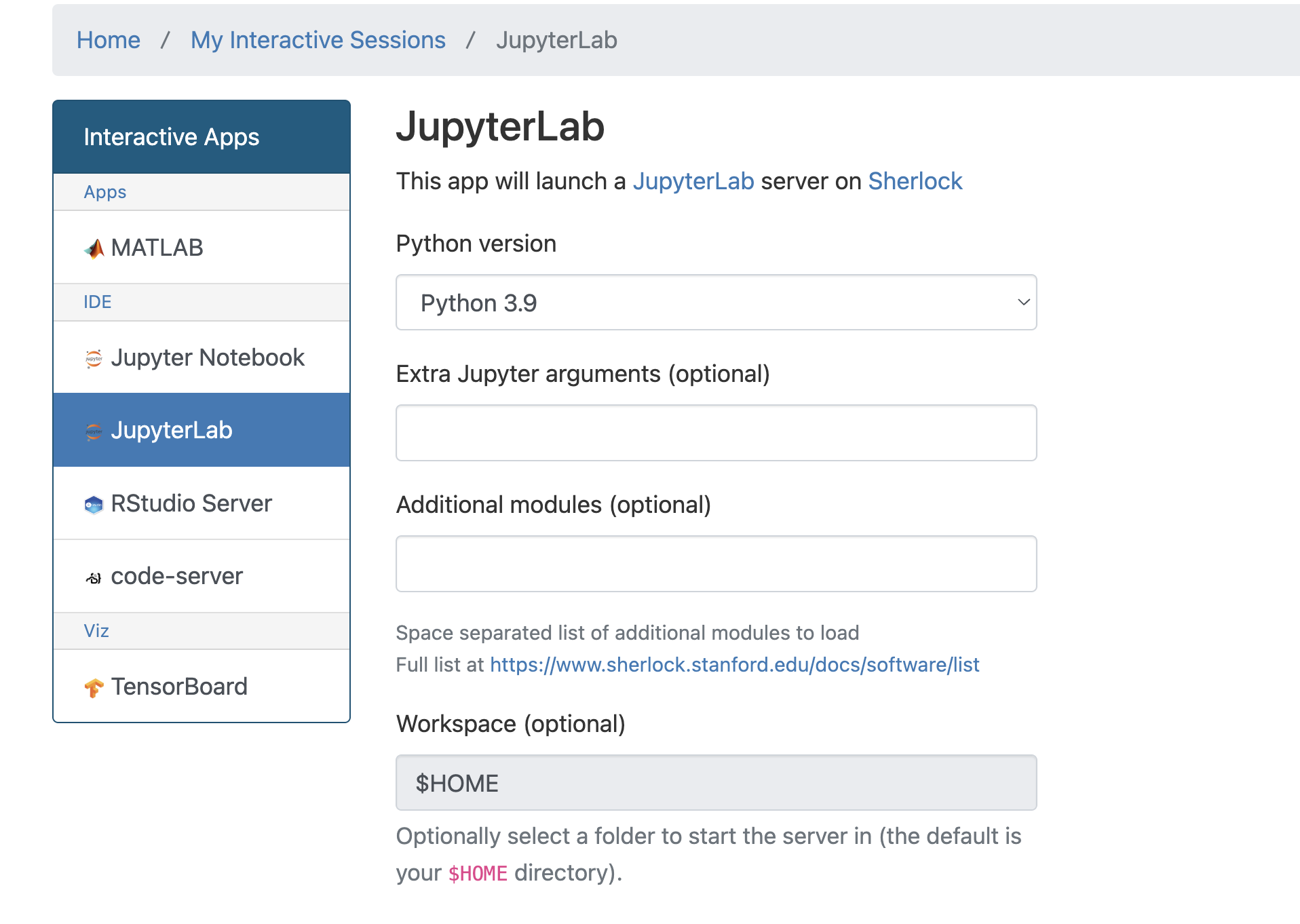
Installing jupyterlab plugins
open a terminal in jupyterlab and install:
pip install jupyterlab_niivue ipyniivue jupyterlmod jupyterlab_slurmAfter the installation finished restart the jupyterlab session in Ondemand.
Neuroimaging Visualization in the File Browser and notebooks of Jupyter Lab
The pip install jupyterlab_niivue added an extension to jupyterlab that visualizes neuroimaging data directly via a double-click in the filebrowser in jupyterlab:
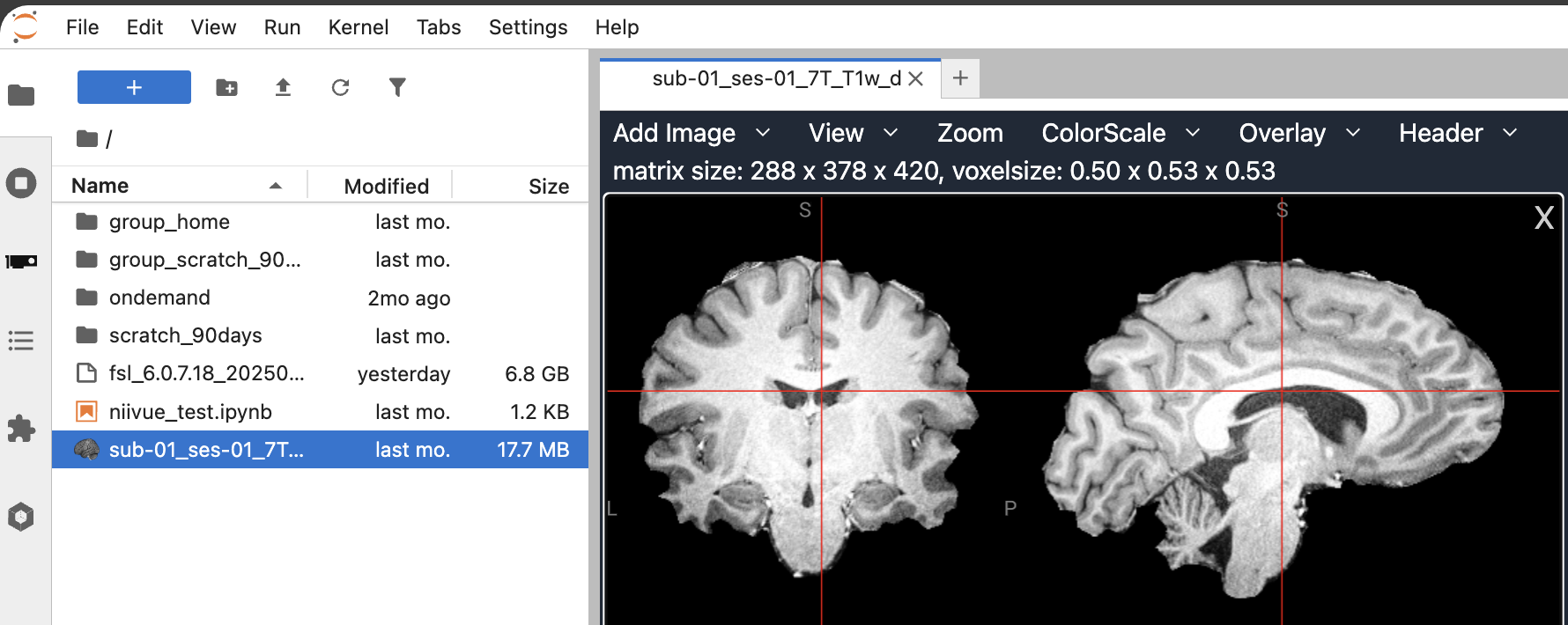
Using containers inside a jupyter notebook
The install of pip install jupyterlmod made the following possible inside a jupyter notebook:
import os
import lmod
group_home = os.environ.get("GROUP_HOME", "")
os.environ["MODULEPATH"] = os.path.abspath(f"{group_home}/neurodesk/local/containers/modules/")
await lmod.load('fsl')now you can run command line tools in a notebook
!betUsing niivue inside a jupyter notebook
The install of pip install ipyniivue allows interactive visualizations inside jupyter notebooks: See examples here https://niivue.github.io/ipyniivue/gallery/index.html
e.g.:
from ipyniivue import NiiVue
nv = NiiVue()
nv.load_volumes([{'path': 'sub-01_ses-01_7T_T1w_defaced_brain.nii.gz'}])
nvChecking on SLURM inside jupyter lab
The install of pip install jupyterlab_slurm added a plugin that allows monitoring slurm jobs.
Using Neurodesk via a full neurodesktop session
This is an ideal setup for visualizing results on Sherlock and for running GUI applications.
downloading startup script
curl -J -O https://raw.githubusercontent.com/neurodesk/neurodesk.github.io/refs/heads/main/content/en/Getting-Started/Installations/connectSherlock.shstarting session
bash connectSherlock.shstart desktop manually when already inside a job
apptainer run \
--fakeroot \
--nv \
--overlay $SCRATCH/neurodesktop-overlay.img \
--bind $GROUP_HOME/neurodesk/local/containers/:/neurodesktop-storage/containers \
--no-home \
--env CVMFS_DISABLE=true \
--env NB_UID=$(id -u) \
--env NB_GID=$(id -g) \
--env NEURODESKTOP_VERSION=latest \
$GROUP_HOME/neurodesk/neurodesktop-neurodesktop_latest.sif \
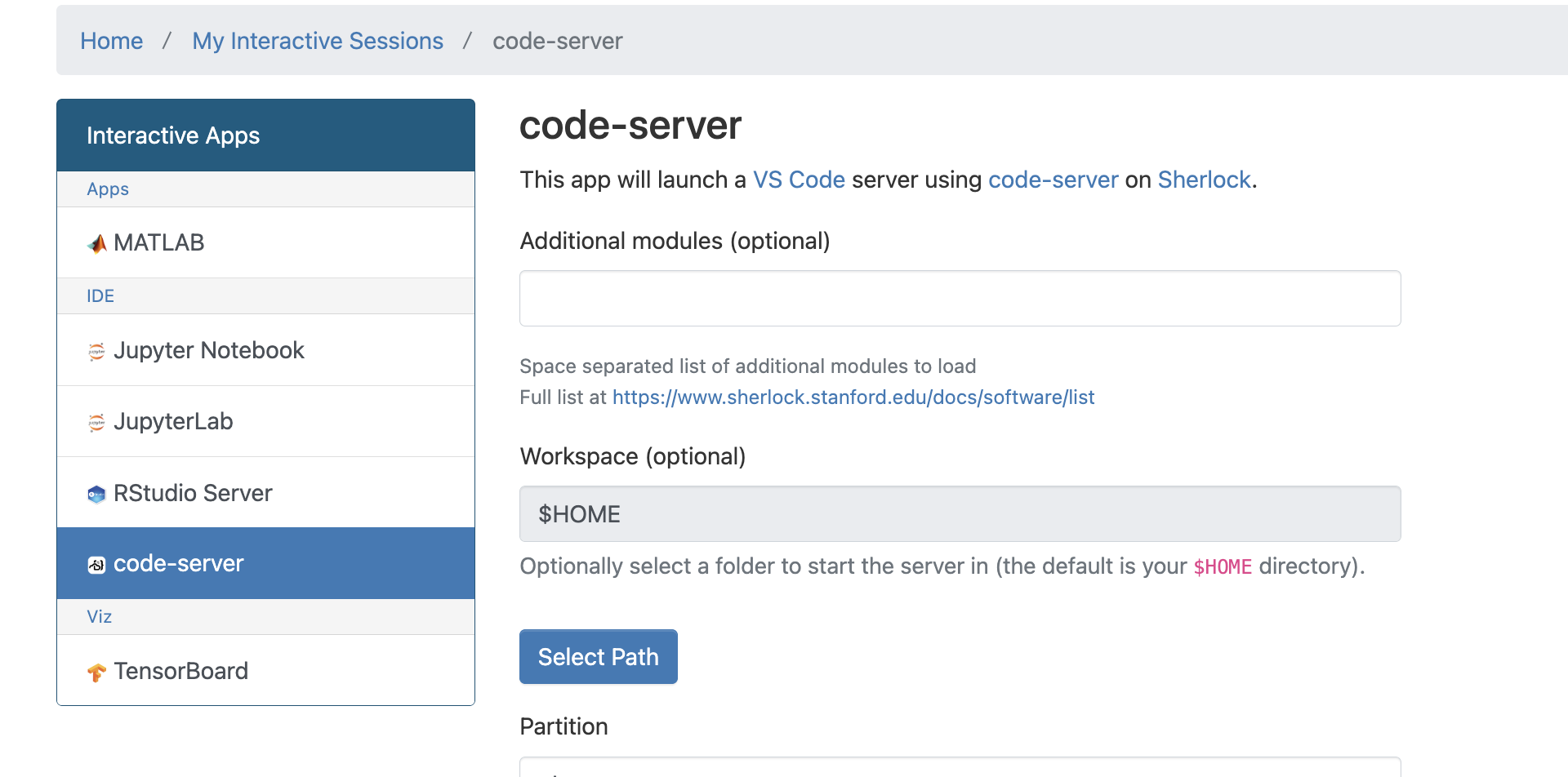
start-notebook.py --allow-rootconnecting with VScode
VScode server does not work on he login nodes due to resource restrictions. It might be possible to run it inside a compute job and inside a container. However, it is possible to run vscode server through ondemand:
A great extension to install is niivue for vscode which allows visualizing neuroimaging data in vscode:
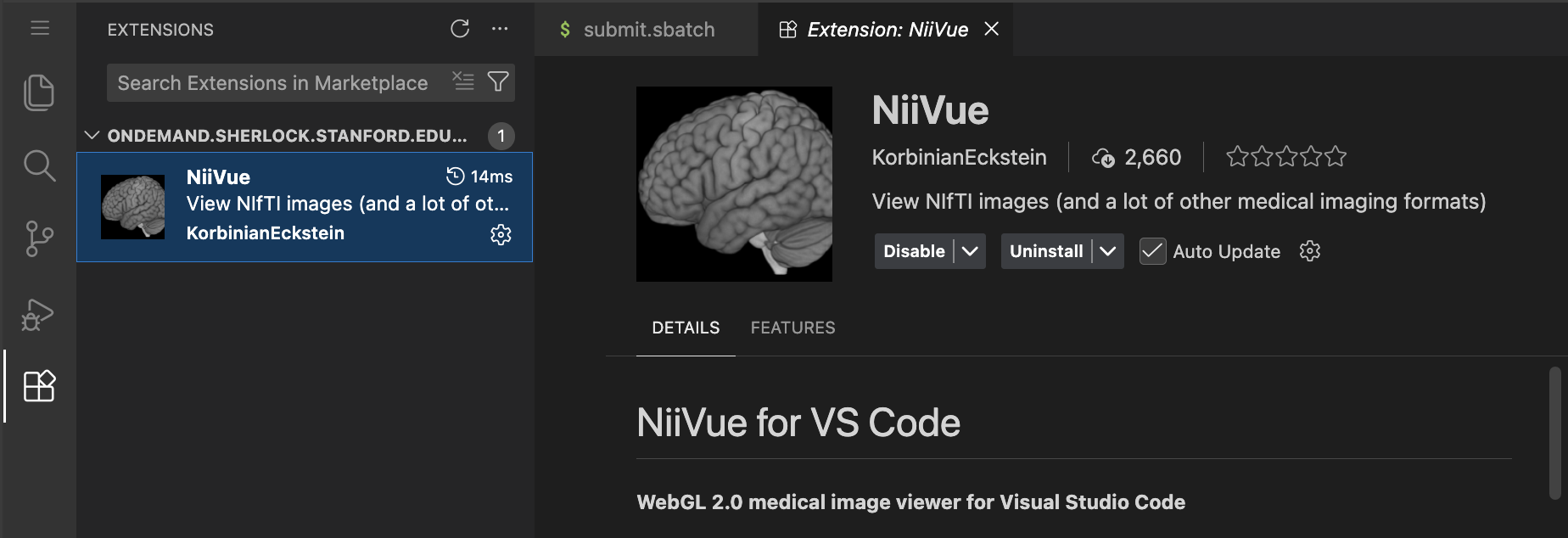
and for AI coding:
- claude code
- gemini CLI companion
- gemini code assist
and for checking on slurm jobs in vscode:
- slurm–
and for matlab scripts:
- MATLAB Extension for Windsurf – path is: /share/software/user/restricted/matlab/R2022b/
useful shortcuts:
- you can execute a line from your scripts on the terminal via setting a keyboard shortcut to “Terminal: Run Selected Text in Active Terminal” - that makes testing scripts and debugging them quite quick
connecting with Cursor
Cursor does not work on the login nodes due to resource restrictions. It might be possible to run it inside a compute job and inside a container.
using coding agents on sherlock
Copilot CLI — an extension of GitHub Copilot that answers natural-language prompts and generates shell commands and code snippets interactively in the CLI. Integrates with developer workflow and git metadata, good at scaffolding repo-level changes. Use this for drafting Slurm scripts, shell-based data-movement commands, Makefiles, container entrypoints, and succinct code edits from the terminal. Caution: always validate generated shell commands before running on Oak.
ml copilot-cli
copilotGemini CLI — a CLI assistant that can generate code from Google’s Gemini family of models (via Google Cloud/Vertex AI or client tooling). Provides strong multilingual reasoning and contextual code completion. Use this for translating research intent into cloud and hybrid workflows, generating code for TPU/GPU workloads, and producing infrastructure-as-code snippets that tie to GCP resources. Caution: always confirm data residency and compliance requirements for sensitive data.
ml gemini-cli
geminiClaude Code (Claude family) — a coding-specialized variant in the Anthropic Claude model family aimed at code generation, refactoring, and reasoning tasks. Provides conversational reasoning about code, multi-step planning for algorithmic tasks, and safer-response tuning relative to generic models. Caution: check private endpoints/dedicated instances before sending sensitive datasets.
ml claude-code
claudeCodex — an OpenAI model family good at producing short code snippets, language translations, and API glue, historically the basis for many coding assistants. Use this for scaffold code, translating pseudocode to working scripts, and generating wrappers for system calls and schedulers. Caution: watch out for API hallucinations and insecure shell usage suggestions; verification in GPT-4 (which often supersedes Codex in capability and safety) advised.
ml codex
codexCrush CLI — an all-around CLI assistant from the Charmbracelet Go-based “ecosystem” intended to improve interactive developer workflows and scripting. Use it for interactive shells or task runners, pipeline composition for local data preprocessing, productivity (nicer prompts, piping primitives, nicer output formatting), or small automation tasks such as repo tooling and glue scripts.
ml crush
crushMisc
note on miniconda
we need an older version of Miniconda on Sherlock due to the outdated glibc:
wget https://repo.anaconda.com/miniconda/Miniconda3-py310_23.3.1-0-Linux-x86_64.sh
bash Miniconda3-py310_23.3.1-0-Linux-x86_64.shnote on MRIQC
NOTE: MRIQC has its $HOME variable hardcoded to be /home/mriqc. This leads to problems. A workaround is to run this before mriqc:
export neurodesk_singularity_opts="--home $HOME:/home"note on AFNI
NOTE: If you are using AFNI then the default detach behavior will cause SIGBUS errors and a crash. To fix this run AFNI with:
afni -no_detachData transfer
Transfer files to and from Onedrive
First install rclone on your computer and set it up for onedrive. Then copy the config file ~/.config/rclone/rclone.conf to sherlock. Then run rclone on sherlock:
ml system
ml rclone
rclone ls
rclone copysetting up rclone for onedrive (needs to be done on a computer with a browser, so not sherlock):
rclone config
# select n for new remote
# enter a name, e.g. onedrive
# select one drive from the list, depending on the rclone version this could be 38
# hit enter for default client_id
# hit enter for default client_secret
# select region 1 Microsoft Global
# hit enter for default tenant
# enter n to skip advanced config
# enter y to open a webbrowser and authenticate with onedrive
# enter 1 for config type OneDrive Personal or Business
# hit enter for default config_driveid
# enter y to accept
# enter y again to confirm
# then quit config q
# now test:
rclone ls onedrive:
# if it's not showing the files from your onedrive, change the config_driveid in ~/.config/rclone/rclone.conf
vi ~/.config/rclone/rclone.confmounting sherlock files on your computer through sshfs
install sshfs for your operating system, e.g. on MacOS:
brew tap macos-fuse-t/homebrew-cask
brew install fuse-t-sshfsthen mount for macos:
mkdir ~/sherlock_scratch
sshfs sciget@dtn.sherlock.stanford.edu:./ ~/sherlock_scratch -o subtype=fuse-ton linux:
mkdir ~/sherlock_scratch
sshfs <sunetid>@dtn.sherlock.stanford.edu:./ ~/sherlock_scratchTransfer files using datalad
ml contribs
ml poldrack
ml datalad-uv
dataladTransfer files via scp
# this will transfer a file from your computer to your scratch space
scp foo <sunetid>@dtn.sherlock.stanford.edu:
# this will transfer a directory from sherlock to your computer:
scp -r <sunetid>@dtn.sherlock.stanford.edu:/scratch/groups/<your_group_here>/<your_directory_here> .Managing Neurodesk on Sherlock
Installing Neurodesk for a lab
This is already done and doesn’t need to be run again!
cd $GROUP_HOME/
git clone https://github.com/neurodesk/neurocommand.git neurodesk
cd neurodesk
pip3 install -r neurodesk/requirements.txt --user
bash build.sh --cli
bash containers.sh
export APPTAINER_BINDPATH=`pwd -P`Installing additional containers
Everyone has write permissions and can download and install new containers.
cd $GROUP_HOME/neurodesk
git pull
bash build.sh
bash containers.sh
# to search for a container:
bash containers.sh freesurfer
# then install the choosen version by copy and pasting the specific command install command displayedIf a new container was installed from Neurodesktop, the paths will need to be adjusted to work outside of Neurodesktop for the rest of sherlock:
sh_dev
#First, test if that happened:
cd $GROUP_HOME/neurodesk/local/containers/
find . -maxdepth 2 -type f -exec grep -l '/home/jovyan/' {} \; 2>/dev/null
cd $GROUP_HOME/neurodesk/local/containers/modules
find . -maxdepth 2 -type f -exec grep -l '/home/jovyan/' {} \; 2>/dev/null
#Then fix for modules:
cd $GROUP_HOME/neurodesk/local/containers/modules
find . -maxdepth 2 -type f -exec sh -c 'if grep -q "/home/jovyan/neurodesktop-storage/containers/" "$1"; then sed -i "s|/home/jovyan/neurodesktop-storage/containers/|${GROUP_HOME}/neurodesk/local/containers/|g" "$1" && echo "Updated: $1"; fi' sh {} \;
#Then fix for containers:
cd $GROUP_HOME/neurodesk/local/containers
find . -maxdepth 2 -type f -exec sh -c 'if grep -q "/home/jovyan/neurodesktop-storage/containers/" "$1"; then sed -i "s|/home/jovyan/neurodesktop-storage/containers/|${GROUP_HOME}/neurodesk/local/containers/|g" "$1" && echo "Updated: $1"; fi' sh {} \;Updating Neurodesktop image
ssh sherlock
sh_dev -m 32 -p normal -c 4
export VERSION="2026-01-30"
cd ${GROUP_HOME}/neurodesk
export APPTAINER_TMPDIR=$SCRATCH/apptainer_temp
mkdir -p $APPTAINER_TMPDIR
apptainer pull docker://ghcr.io/neurodesk/neurodesktop/neurodesktop:${VERSION}
rm ${GROUP_HOME}/neurodesk/neurodesktop_latest.sif
ln -s ${GROUP_HOME}/neurodesk/neurodesktop_${VERSION}.sif ${GROUP_HOME}/neurodesk/neurodesktop_latest.sif Or submit the update as a single Slurm job:
sbatch -p normal -c 4 --mem=32G --job-name=neurodesktop-update --wrap 'export VERSION="2026-01-30"; cd ${GROUP_HOME}/neurodesk; export APPTAINER_TMPDIR=$SCRATCH/apptainer_temp; mkdir -p $APPTAINER_TMPDIR; apptainer pull docker://ghcr.io/neurodesk/neurodesktop/neurodesktop:${VERSION}; rm ${GROUP_HOME}/neurodesk/neurodesktop_latest.sif; ln -s ${GROUP_HOME}/neurodesk/neurodesktop_${VERSION}.sif ${GROUP_HOME}/neurodesk/neurodesktop_latest.sif'5 - Nectar Virtual Desktop Service
Note
Link to Nectar Virtual Desktop Service: https://desktop.rc.nectar.org.auThere are a few differences between the open-source version of Neurodesk and what’s hosted on Nectar VDI:
- There is no
/neurodesktop-storagefolder (the folder on the Desktop does not lead anywhere). - Files uploaded via drag and drop do not get stored on the desktop but in
/home/vdiuser/thinclient_drives/GUACFS
Instructions for use
Click on “Sign in”.
Choose the AAF option.
Choose your institution from the list.
Provide your email address and password.
Click on create a Workspace (you only need to do this when you sign in for the first time).

Fill form ‘Apply for new Workspace’ and submit.

Click on “EXPLORE”.

Click “VIEW DETAILS” under Neurodesktop:
Click “CREATE DESKTOP +” button on the top right corner.
Choose the desired availability zone.
Wait until everything is completed:
Click “OPEN DESKTOP ->”:

For a general guide on using the ARDC virtual desktops, click here: https://tutorials.rc.nectar.org.au/virtual-desktop-service/01-overview
For a specific explanation on how to launch the various applications available in the Neurodesktop desktop, follow the instructions here: https://neurodesk.org/docs/getting-started/neurodesktop/