Neurodesk Overview
A flexible, scalable and easy to use data analysis environment for reproducible neuroimaging.
What is Neurodesk?
Neurodesk provides a containerised data analysis environment to facilitate reproducible analysis of neuroimaging data. At Neurodesk, we believe that reproducibility should be a fundamental principle underlying neuroscientific data analysis (1). Analysis pipelines for neuroimaging data typically rely on specific versions of packages and software, and are dependent on their native operating system. These dependencies mean that a working analysis pipeline may fail or produce different results on a new computer, or even on the same computer after a software update. Neurodesk provides a platform in which anyone, anywhere, using any computer can reproduce your original research findings given the original data and analysis code.
The Neurodesk environment relies on software containers and allows users to build and use containers to analyse neuroimaging data. Containers can be compared to virtual machines, in that they allow users to create a virtual, isolated computing environment with an operating system separate to that of the host machine. However, containers differ from virtual machines in that they virtualise software rather than hardware. Practically, this means that container images require few system resources to install, start-up quickly, and are easily portable between computers.
We recommend watching this excellent short video from the Australian Research Data Commons (ARDC) on research applications of software containers.
To read more about Docker containers, visit the Docker webpage
What applications are included in Neurodesk?
You can check out the complete list of these applications
See here
Background
Neurodesk originates from various projects at the Centre for Advanced Imaging that enabled running neuroimaging tools on HPCs and Linux workstations using software containers. The ideas and code from projects like “DICOM2CLOUD”. “transparent singularity” and “CAID” were combined during a hackathon project to create a “Virtual Neuro Machine”. The project was later renamed to Neurodesk and further developments were funded through the ARDC platform project “AEDAPT” with the goal to create a national platform for reproducible electrophysiology data analysis and sharing, accessible to all Australian researchers across a wide range of disciplines that conduct electrophysiological research.
References
- National Academies of Sciences, Engineering, and Medicine. 2019. Reproducibility and Replicability in Science. Washington, DC: The National Academies Press. https://doi.org/10.17226/25303.
MIT License
Copyright (c) 2021 NeuroDesk
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the “Software”), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
2 - How to cite us
How should you cite Neurodesk and the tools you use through the platform?
Citing Neurodesk
Please cite the Neurodesk paper and if you used Neurodesktop, add the version that was used (e.g. v2025-04-08).
Renton, A.I., Dao, T.T., Johnstone, T., Civier, O., Sullivan, R. P., White, D. J., … Narayanan, A. & Bollmann, S. Neurodesk: an accessible, flexible and portable data analysis environment for reproducible neuroimaging. Nat Methods (2024). https://doi.org/10.1038/s41592-023-02145-x
@article{Renton2024,
author = {Renton, Angela I and Dao, Thuy T and Johnstone, Tom and Civier, Oren and Sullivan, Ryan P and White, David J and Lyons, Paris and Slade, Benjamin M and Abbott, David F and Amos, Toluwani J and Bollmann, Saskia and Botting, Andy and Campbell, Megan E J and Chang, Jeryn and Close, Thomas G and D{\"{o}}rig, Monika and Eckstein, Korbinian and Egan, Gary F and Evas, Stefanie and Flandin, Guillaume and Garner, Kelly G and Garrido, Marta I and Ghosh, Satrajit S and Grignard, Martin and Halchenko, Yaroslav O and Hannan, Anthony J and Heinsfeld, Anibal S and Huber, Laurentius and Hughes, Matthew E and Kaczmarzyk, Jakub R and Kasper, Lars and Kuhlmann, Levin and Lou, Kexin and Mantilla-Ramos, Yorguin-Jose and Mattingley, Jason B and Meier, Michael L and Morris, Jo and Narayanan, Akshaiy and Pestilli, Franco and Puce, Aina and Ribeiro, Fernanda L and Rogasch, Nigel C and Rorden, Chris and Schira, Mark M and Shaw, Thomas B and Sowman, Paul F and Spitz, Gershon and Stewart, Ashley W and Ye, Xincheng and Zhu, Judy D and Narayanan, Aswin and Bollmann, Steffen},
doi = {10.1038/s41592-023-02145-x},
issn = {1548-7105},
journal = {Nature Methods},
number = {5},
pages = {804--808},
title = {{Neurodesk: an accessible, flexible and portable data analysis environment for reproducible neuroimaging}},
url = {https://doi.org/10.1038/s41592-023-02145-x},
volume = {21},
year = {2024}
}
Check our preprint here Neurodesk Preprint
To guarantee reproducibility, we recommend specifying the name, version number and builddate for any containerised applications used through Neurodesk. You can find this information here.
In addition to citing Neurodesk, please remember to cite any papers associated with the tools you use. You find this information in the README.m for each tool, or when opening the application through the menu in Neurodesktop.
If you used any EEG/MEG/electrophysiology tools, please also cite the AEDAPT project.
Examples
Here are some example excerts of how to cite Neurodesk:
“TGV QSM (v1.0.0_20210629, Langkammer et al, 2015) was run in Neurodesk (v2025-04-08, Renton et al, 2022)”
“EEGlab (2020.0_20211026, Delorme & Makeig, 2004) was run in Neurodesk (v2025-04-08, Renton et al, 2022) part of the AEDAPT project.”
References
- Langkammer, C; Bredies, K; Poser, BA; Barth, M; Reishofer, G; Fan, AP; Bilgic, B; Fazekas, F; Mainero; C; Ropele, S. Fast Quantitative Susceptibility Mapping using 3D EPI and Total Generalized Variation. Neuroimage. 2015 May 1;111:622-30. doi: 10.1016/j.neuroimage.2015.02.041
- Renton, A.I., Dao, T.T., Johnstone, T., Civier, O., Sullivan, R. P., White, D. J., … Narayanan, A. & Bollmann, S. Neurodesk: an accessible, flexible and portable data analysis environment for reproducible neuroimaging. Nat Methods (2024). 10.1038/s41592-023-02145-x
- Delorme A & Makeig S (2004) EEGLAB: an open-source toolbox for analysis of single-trial EEG dynamics, Journal of Neuroscience Methods 134:9-21. doi: 10.1016/j.jneumeth.2003.10.009
4 - Neurodesk FAQ
Frequently Asked Questions.
General Overview
What is Neurodesk?
Neurodesk provides a containerised data analysis environment to facilitate reproducible analysis of neuroimaging data. Analysis pipelines for neuroimaging data typically rely on specific versions of packages and software, and are dependent on their native operating system. These dependencies mean that a working analysis pipeline may fail or produce different results on a new computer, or even on the same computer after a software update. Neurodesk provides a platform in which anyone, anywhere, using any computer can reproduce your original research findings given the original data and analysis code.
More information:
In-depth information:
What applications are included in Neurodesk?
You can check out the complete list of these applications
See here
Using Neurodesktop
Can I change the desktop resolution?
Yes.
In a Desktop RDP session, simply zoom or resize the window and the resolution will be adjusted.
In a Desktop VNC session, open a Terminal (LXTerminal) from the bottom toolbar and run lxrandr to change the desktop resolution

Can I run Neurodesk on an HPC without Docker?
Yes, our project aims to run on the hardware you have access to. However, without docker support you cannot use our desktop interface NeuroDesktop but you can still use the command line interface NeuroCommand on HPC. This works well for batch processing on HPCs once you developed your pipeline in our desktop interface. If your HPC provides a desktop interface you can use all our graphical applications without any issues and the GUIs even work via SSH x-forwarding - it’s not the most performant experience, but it works well enough.
How do I get my files into Neurodesk?
It depends where you are running Neurodesk and where your files are. We provide many different ways from drag-and-drop, to cloud storage to file mounts in Storage in Neurodesk.
Can I export a jupyter notebook as a PDF?
By default we don’t have all extensions installed to do this to keep the neurodesktop small. So you will see an error when trying to do File -> Save and Export Notebook As … -> Webpdf
To get webpdf export to work, open a terminal and run:
pip install nbconvert[webpdf]
playwright install chromium
If you are running containers on Linux there is no performance penalty - on an HPC with a Lustre filesystem it can even be faster to run our containers than running natively on the filesystem (because meta data operations are shifted to the compute node - more information can be found here:
Rioux, Pierre, Gregory Kiar, Alexandre Hutton, Alan C. Evans, and Shawn T. Brown. ‘Deploying Large Fixed File Datasets with SquashFS and Singularity’. ArXiv:2002.06129 [Cs], 14 February 2020. https://arxiv.org/abs/2002.06129 ).
However, running Neurodesktop on Windows and Mac will have a performance penalty, because Linux runs in a Hypervisor on these systems. Also, it is important to understand filesystem operation bottlenecks that can occur when accessing files across system boundaries (e.g. processing DICOM files stored on Windows inside Neurodesk).
How can I see how many compute resources Neurodesk containers need?
In Linux the containers run as normal processes and you can use htop and top to inspect the resource footprint. For Windows and Mac the information is not readily available, however, we have written a guide here: Troubleshooting
Can I just use the containers without neurodesktop or neurocommand?
Yes, there are multiple ways of using the containers directly and we provide an overview here: https://www.neurodesk.org/docs/getting-started/neurocontainers/
Can I keep my modifications in the container?
We designed neurodesk with reproducibility as a main goal, so the desktop containers should not be modified if one aims for full reproducibility. However, there is one good option to keep your settings across different container versions: You can write a shell script that installs additional packages and modifies the environment so it’s perfect for you. This script can then be re-executed in a new desktop version and will enable a reproducible customization.
Another option is to “save” your docker container including all changes you made. This could be useful when your changes are too difficult to write a shell script or when you do not care about reproducibility as much and you just want to get the job done. To do this you can commit (https://docs.docker.com/engine/reference/commandline/commit/) your container and by uploading the container to your own docker hub you could even share it.
Can I force a complete container download to my system?
To increase speed and reliability of Neurodesktop we mount the application containers from a CVMFS mount and download only the files required to run your current task. Although we aim to keep everything on there reproducible, there might be a reason that you want to fully download the containers to your system. You can force this behaviour by adding another parameter to the docker call: -e CVMFS_DISABLE=true
For windows an example would look like this:
docker run --shm-size=1gb -it --privileged --user=root --name neurodesktop -v C:/neurodesktop-storage:/neurodesktop-storage -e CVMFS_DISABLE=true -p 8888:8888 -e NEURODESKTOP_VERSION=2025-04-08 vnmd/neurodesktop:2025-04-08
How to restart a Neurodesk Lab session?
In the jupyter menu, click on File -> Hub Control Panel:
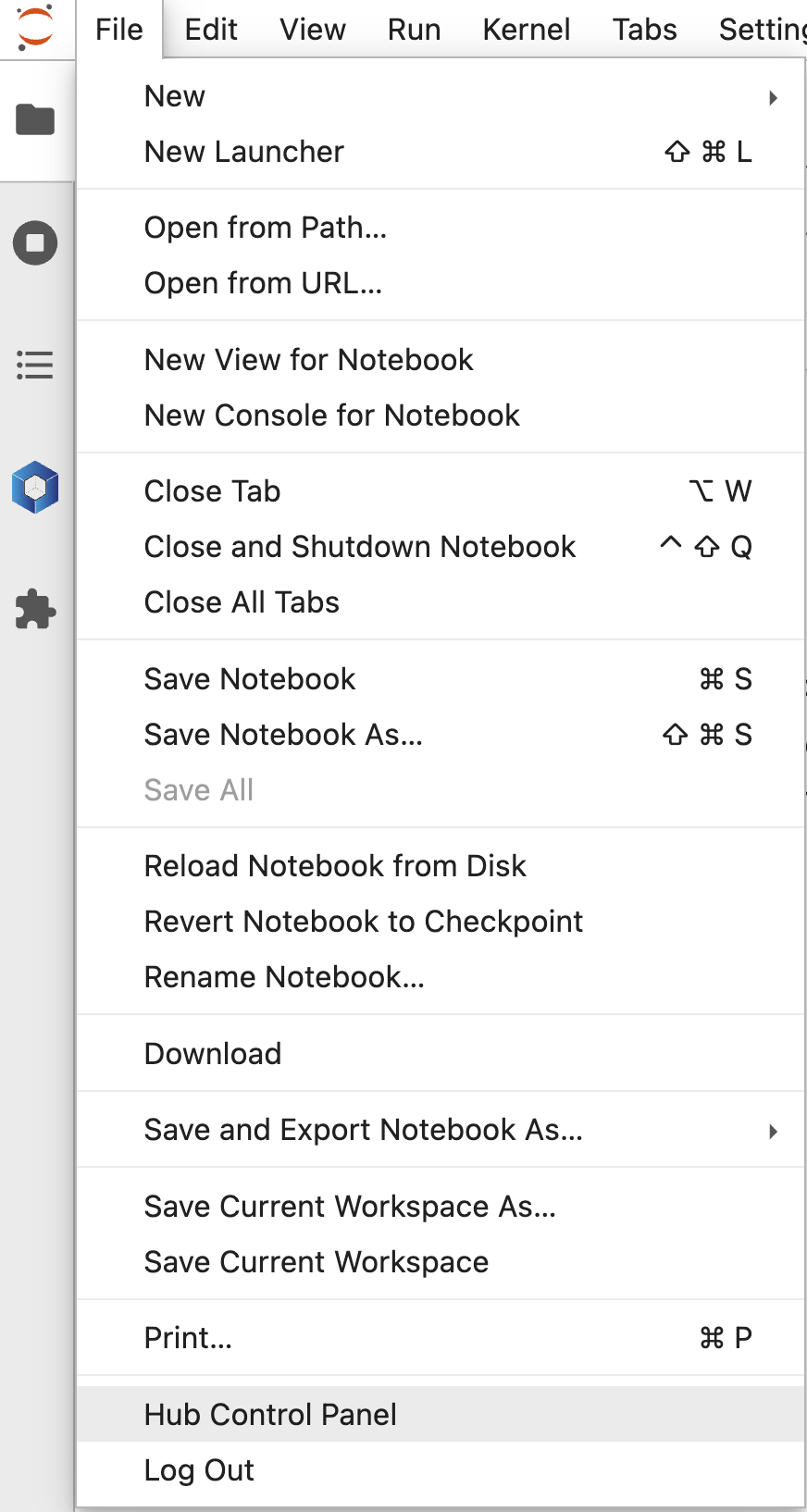
Then “Stop My Server”

Then “Start My Server”

How to restart a stuck Neurodesk when running locally in Docker or using the NeurodeskApp?
Open Docker Desktop Dashboard. Find the container under “Containers” and delete the Container there.
Troubleshooting Common Issues
Freeview 7.2.0 crashes when I open files
Freeview (and Freesurfer!) need a valid license to work and we are not allowed to distribute a license with Neurodesk!
You can follow these steps to run freeview 7.2.0 and open your files:
- apply for a license (https://surfer.nmr.mgh.harvard.edu/registration.html) and paste this license in ~/.license
then run
echo "export FS_LICENSE=~/.license" >> ~/.bashrc
then start freeview 7.2.0 and it should all work perfectly.
Matlab asks me to reactivate after restarting the desktop
There is one workaround for this problem. Fix the mac address for your session by including this in your docker command:
--mac-address 02:42:ac:11:00:02
Trouble installing neurodesk images
This may be a memory issue. First, ensure that there is enough free space on the disk. If there is, try resetting docker settings and data. To do this:
- Open the docker engine
- Navigate to “Troubleshooting” (the bug icon in the top right).
- Click “Reset to factory defaults”
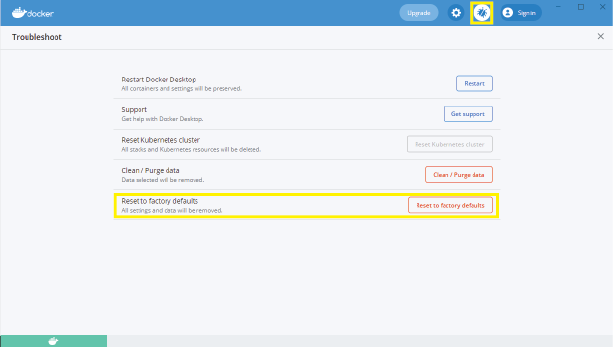
If you are still experiencing issues after this, you may need to update docker to the latest version. This can be achieved through “settings” in the docker engine, or (on windows) by right clicking on the docker tray icon:
I got an error message ‘X killed’ or not enough memory
This may be due to Docker not having access to enough RAM from your host computer.
How to empty trash bin in Neurodesktop
If you have deleted files using the Neurodesktop GUI but your storage is not yet emptied, execute the following command in the terminal.
rm -rf ~/.local/share/Trash/
Keyboard, Clipboard, and Language Support
Keyboard and Multi-Language support
I am using a European keyboard layout and I cannot type symbols that require the ALT-GR key (like @ or \)
This seems to be a bug in Guacamole and RDP in combination with certain browsers. There are a few workarounds you can try:
- Use the connection option “Desktop Fixed-Resolution (VNC)” instead of Desktop Auto-Resolution (RDP)
- connect to the desktop using an RDP client instead of the browser (https://www.neurodesk.org/docs/getting-started/neurodesktop/windows/#using-an-rdp-client)
Clipboard issues
How do I copy and paste text?
You can copy and paste text within Neurodesktop and between Neurodektop and your host computer using the regular keyboard shortcuts (CTRL+C, CTRL+X, and CTRL+V). Note however that some applications (e.g., command-line terminal) are using other keyboard shortcuts. You can usually find them in the “Edit” menu of the relevant application.
Note for Mac Users: The Safari Browser is currently not compatible for copy and paste with Guacamole. Please either use the Neurodeskapp or a Edge/Firefox on MacOS. You will need to use a combination of CTRL and Command shortcuts in order to copy and paste text between Neurodesktop and the host computer. For example, copy text from your Mac with Command+C and then paste it into Neurodesktop using CTRL+V. For the other way around, you’d use CTRL+C in Neurodesktop and then Command+V on the Mac.
I cannot copy and paste text within Neurodesktop using keyboard shortcuts
If you’re using Mac, you might be trying to use Mac keyboard shortcuts, but Neurodesktop is using Linux keyboard shortcuts (CTRL+C and CTRL+V)
If you use the terminal, please see “I fixed my internet browser clipboard, but copy or paste still do not work in the terminal” below.
Copying text from my host computer and pasting it inside Neurodesktop doesn’t work in Firefox
This is a “feature” of firefox and you can disable this “feature”:
- navigate to
about:config and “Accept the Risk and Continue” (“about:config” needs to be entered in the address bar of firefox and hit enter) - now search for clipboard and then set the following to “true”:
- dom.events.asyncClipboard.clipboardItem
- dom.events.asyncClipboard.read
- dom.events.testing.asyncClipboard
Then close firefox and restart. Then the clipboard should work as one would expect.
If the clipboard still does not work, check “I fixed my internet browser clipboard, but …” sections below.
Copy and paste between my host computer and Neurodesktop (or vice versa) doesn’t work in Chrome or Edge
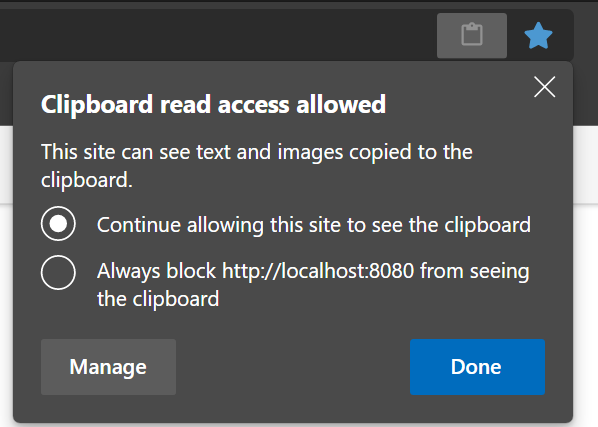
The browsers have a security feature to protect you from something stealing your clipboard content. Depending on your security settings you have to enable it explicitly - it’s a little icon in the browser address bar that looks like a clipboard.
After pressing the icon, you should choose the option shown below in the dialog that opens. After pressing “Done”, close the current browser tab and open a new one for the changes to take effect.
If the clipboard still does not work, check “I fixed my internet browser clipboard, but …” sections below.
I fixed my internet browser clipboard, but copy or paste still do not work in the terminal
The terminal is using special keyboard shortcuts, Shift+CTRL+C for copy, and Shift+CTRL+V for paste. Alternatively, you can copy and paste text by using the terminal’s “Edit” menu.
I fixed my internet browser clipboard, but copy or paste still do not work in the file browser
The copy and paste options in the “Edit” menu of the file browser are used to copy and paste files, not text. To copy and paste text from/into the file browser application (e.g., copy a path into the path field in the top), use the CTRL+C and CTRL+V keyboard shortcuts.
I fixed my internet browser clipboard, but copy or paste still do not work in a specific/all applications
If you’re using Mac, you might be trying to use Mac keyboard shortcuts, but Neurodesktop is using Linux keyboard shortcuts.
For more details, read the “Note for Mac users” here.
It is also possible that the text you are trying to copy includes special characters (e.g., non-English characters), which may cause Neurodesktop to not execute your paste command (including the non specials characters). Give special attention to the following characters:
- Dash: the en (short) dash is the normal one and copies ok, but the em (long) dash is considered a special character. When in doubt, replace all the dashes in the text you want to copy with en dashes.
- Quotation mark: a strictly vertical quotation mark is fine, but a slightly leaning quotation mark / an apostrophe are special characters. When in doubt, replace all quotation marks in the text you want to copy with vertical quotation marks.
If it still does not work, please report the problem and we will do our best to help you:
- Copy some text from your host computer (CTRL+C, or Command+C)
- Open the Mousepad text editor in Neurodesktop (Start button → “Accessories” → “Mousepad”)
- Try to paste the text using the menu option “Edit” → “Paste”
- Try to paste the text again using CTRL+V
- If you don’t have one already, please create a Github account here
- Go to our discussion forum here
- If you are not logged into Github, please log in (upper right corner)
- Press “New Discussion” button
- In the message that you write, please specify your operating system, your internet browser, the application in question, and if you can copy/paste to Mousepad and how?
Docker, WSL, Memory
Docker
Docker: Shows an error that unprivileged user namespaces are not enabled.
This can be activated by running
echo "kernel.apparmor_restrict_unprivileged_userns = 0" >/etc/sysctl.d/99-userns.conf
sysctl --system
Docker: Cannot connect to the Docker daemon at unix:///var/run/docker.sock. Is the docker daemon running?.
This is usually a docker-related error, not related to neurodesktop itself. To troubleshoot docker, we can try a simpler container first:
Try the following solutions (in this order, until the above command works)
- Win/Mac: Open docker GUI and accept T&Cs. Check that the docker engine is running
- Linux: Make sure you started the docker daemon (sudo systemctl start docker)
- Add your user to the OS docker group (sudo usermod -aG docker $USER)
- docker.sock permissions need to be changed (sudo chown root:docker /var/run/docker.sock)
- Close and open the terminal and retry.
- Log out and login again, or restart the machine (current user environment does not fully know docker is running)
Windows
Windows users: WSL not installed properly
The Docker engine relies on the Windows subsystem for Linux (WSL) to run on a windows machine.
Note
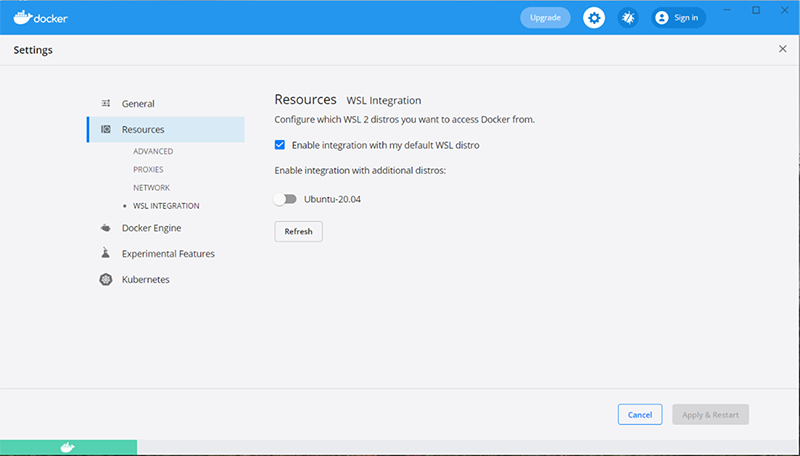
We recommend the manual install instructions, as the simplified install requires an upgrade to a preview build of Windows that may be unstable.If WSL is properly installed, the Resources tab of the Docker settings should look something like this:
However, if WSL is missing or incorrectly configured, the Resources tab of the Docker settings may look something like this:
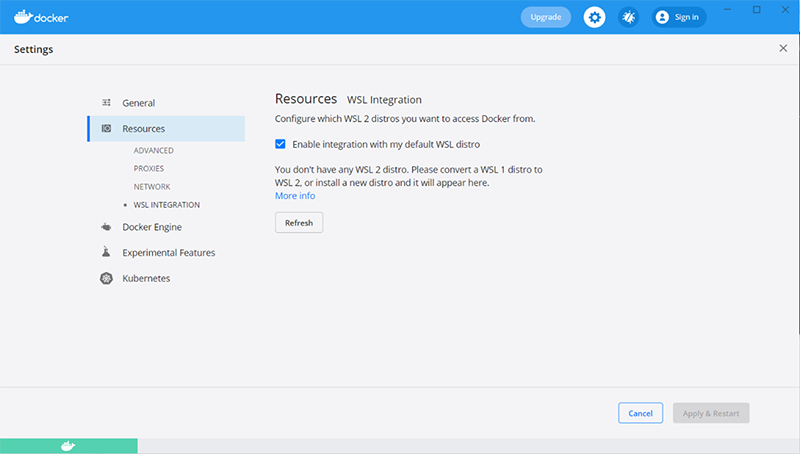
If this is the case, follow the manual install instructions to install WSL 2 (including installation of Ubuntu through Microsoft Store).
Windows users: Not enough free space on the partition in Windows and WSL2
This could help: https://yjmantilla.github.io/tutorials/wsl2-move-vhdx.html
Windows users: Failure to connect to Neurodesktop in Firefox
We recommend using Microsoft Edge or Google Chrome to access Neurodesktop.
If you are using Docker on Windows 10 with the WSL2 backend
then this is managed by Windows settings. Try the following steps to check how much RAM Docker has access to and increase the amount if necessary.
- Run Docker
- Open a terminal (ie. Powershell) in the PC you want to use to run Neurodesktop (not in Neurodesktop itself) and type the following command:
This will generate information about your Docker installation (make sure Docker is running during this step)
- Look for the line that says
- If this value is ~2GB, try increasing it*:
- Quit Docker (make sure it’s not running in the background, ie. system tray, check task manager)
- In the terminal, run the following command
This will list any running distributions. For the update to be successful, WSL needs to have completely stopped running (ie. no distributions running)- Restart Docker and rerun steps 1-3 to confirm it was successful
If you are not using WSL2, you can check and manage your RAM allocation in the Docker desktop application.
- Open the Docker application and navigate to settings > resources > advances
- Scroll down to the Memory option and use the sliding bar to adjust the setting
- Click apply and restart
*RAM requirements will vary based on the tools/data you are using. If the system you’re using has limited RAM, test out a few different amounts by running the above steps and then your analyses in Neurodesktop. Version 20220208 onwards has a memory monitor in the taskbar - you can use this to check how much memory Neurodesktop has access to and how much is being used by the analyses being run.
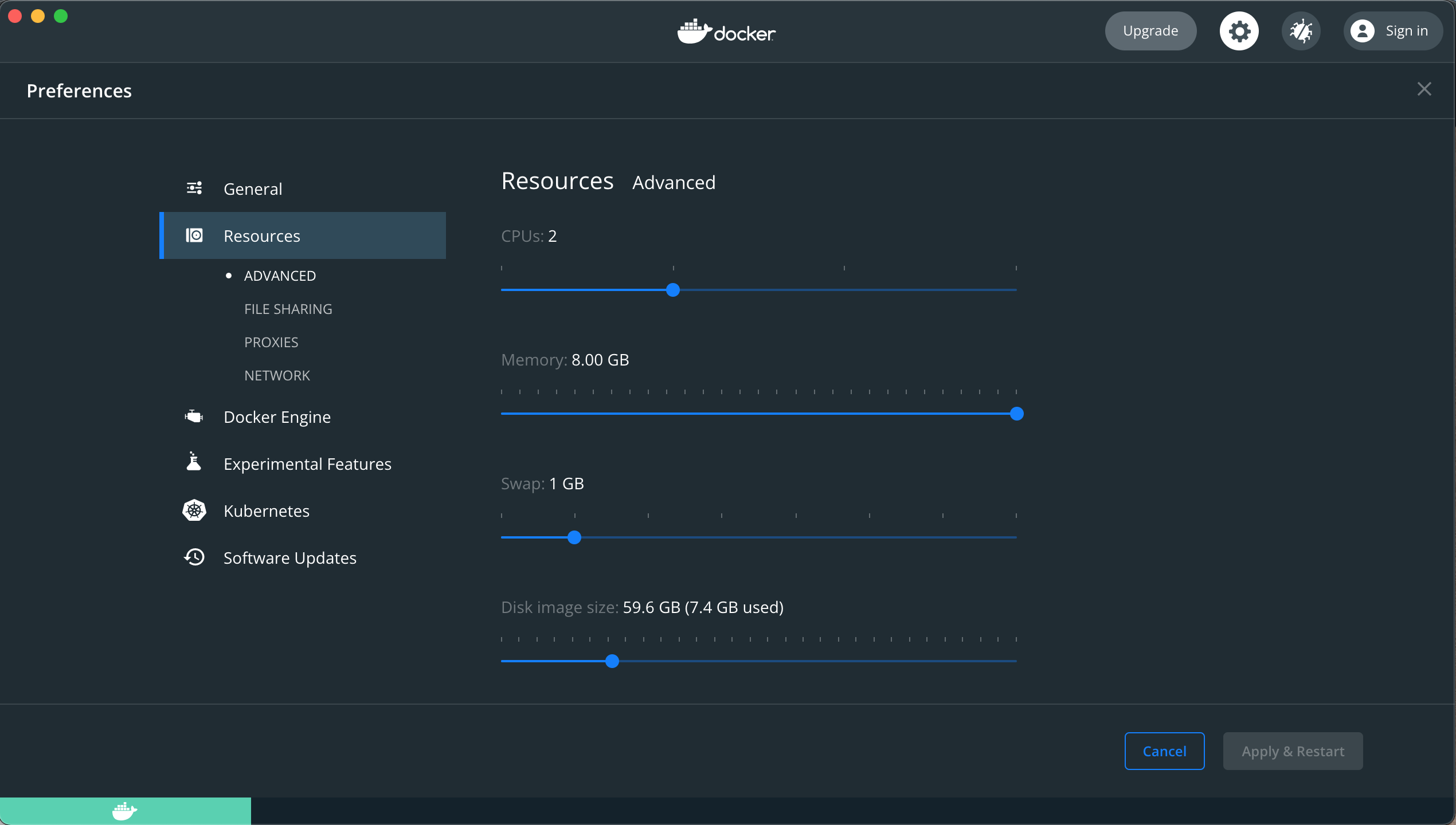
MacOS
If you are using Docker on MacOS
- The memory amount is managed via the Docker settings:
VScode
Vscode is not starting up (or starts many many copies of itself …)
This problem seems to be caused by a corrupted config directory. To fix this, run:
Contributing & Support
How can I contribute new containers?
We are still working on making this easier, but here is the current workflow to add new applications.
Neurodesk is an open-source project that is always evolving. If you are experiencing an issue not listed here, you can reach out to us on Github.
Post your question at https://github.com/orgs/NeuroDesk/discussions or open a new issue, so that we can aim to solve it and update our help documentation.
7 - Acknowledgments
Recognition of contributors, supporters, and the lands on which we work.
We are deeply grateful to all the individuals, institutions, and communities that support the Neurodesk project through their time, expertise, infrastructure, and resources.
Cultural and Land Acknowledgement
We acknowledge and honour the Traditional Custodians and Indigenous Peoples of all lands on which we live, work, and connect around the world. These lands have been, and continue to be, places of deep cultural, spiritual, and ancestral significance for their First Peoples. We honour their resilience, knowledge, and contributions, and commit to respectful collaboration wherever our work reaches.
This project is not tied to a single place, but we honour Indigenous sovereignty wherever our work reaches — including digital spaces. We invite all who engage with this work to reflect on their own relationship with the lands they are on and the histories they are a part of.
Grant and Program Support
This work is supported by the Wellcome Trust with a Discretionary Award as part of the Chan Zuckerberg Initiative (CZI), The Kavli Foundation, and Wellcome’s Essential Open Source Software for Science (Cycle 6) Program (Grant Ref: [313306/Z/24/Z])
This project is also supported by the Australian Research Data Commons (ARDC) via the AEDAPT platform project:
Australian Electrophysiology Data Analytics Platform.

Infrastructure and Cloud Support
This work benefits from services and infrastructure provided by:
- The EGI Federation with support from CESNET-MCC
- Indiana University’s Jetstream2 Cloud
- ARDC Nectar Cloud
We are also grateful for:
- AWS Cloud Credits (providing cloud credits since 2024)
- Google Cloud Platform (support from 2024–2025)
- Oracle for Research (support from 2022–2023)
Citations and Credit
If you use Neurodesk in your work, please cite us and the tools you use appropriately. See the full citation guide here: How to Cite Neurodesk